Molecular marker closely linked with melon pseudoperonospora cubensis resisting gene and application thereof
A downy mildew resistance and molecular marker technology, applied in the field of agricultural biology, can solve the problems of long genetic distance and difficult application of molecular marker-assisted breeding, so as to shorten the breeding cycle and improve the selection accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Embodiment one: the molecular marker closely linked with melon downy mildew resistance gene
[0063] A kind of molecular marker closely linked with melon downy mildew resistance gene specifically comprises the following steps:
[0064] (1) Crossbreed the susceptible farm variety "Karaxai" as the female parent and the disease-resistant resource PI438685 as the male parent to obtain the hybrid F 1 .
[0065] (2) by Hybrid F 1 Self-pollination to obtain F 2 generation group, F 1 Crossed with "Karaxai" to obtain backcross population BC S , F 1 Crossed with PI438685 to obtain backcross population BC r .
[0066] (3) To PI438685, "Karaxai", F 1 、BC S 、BC r and F 2 Identification of downy mildew resistance.
[0067] Preparation of downy mildew inoculum: collect natural early diseased leaves from the field, use a soft brush to pick up the sporangia on the back of the leaves, put them in a beaker filled with sterile water, and inoculate them with the common spot meth...
Embodiment 2
[0081] Embodiment two: the molecular marker SSR-666 closely linked with the melon downy mildew resistance gene 230 Applications
[0082] The invention provides a molecular marker SSR-666 closely linked to the melon downy mildew resistance gene 230 application of the molecular marker SSR-666 230 Be used for the genotype detection of muskmelon variety or line, be used for judging whether this kind or line is resistant to melon downy mildew, comprises the following steps:
[0083] (1) Using the disease-resistant resource PI438685 to cross with other melons and propagate to F 2 generation above.
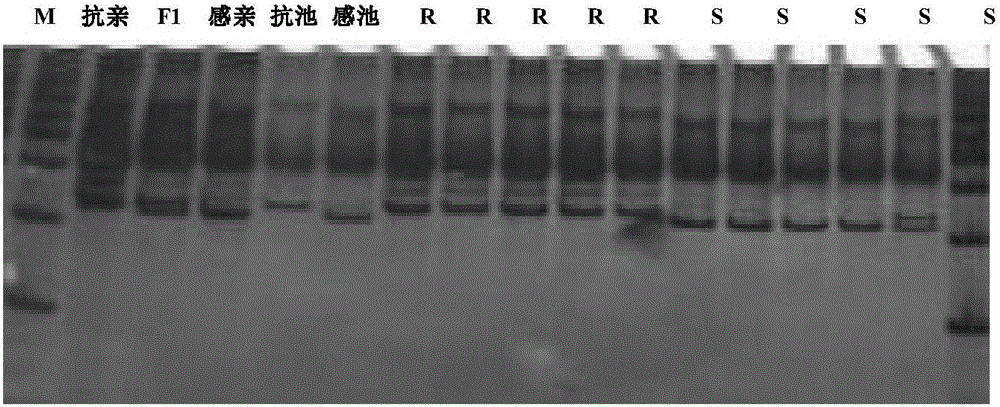
[0084] (2) Genomic DNA is extracted from the muskmelon individual plant obtained by step (1), and SSR-666 is used to 230 The marker is amplified by PCR to detect whether only a unique 230bp specific band occurs, and if only this specific band occurs, it is predicted that the melon plant is resistant to downy mildew.
Embodiment 3
[0085] Embodiment three: the SSR molecular marker SSR-666 linked with the melon downy mildew resistance gene 230 obtained.
[0086] (1) F 1 , F 1 F 2 , F 1 Hybridized with PI438685 to get BCr, F 1 Cross with "Karaxai" to get BCs. By pair of parents, F 1、 f 2 , BCr, and BCs were artificially inoculated and the incidence of each individual plant was counted: BCs, F 1 and "Karaxai" were all susceptible, PI438685 was resistant, and the ratio of resistant to susceptible in BCr was approximately 1:1, and F 2 The population statistics of generation segregation showed that among the 197 strain populations, 53 strains were disease-resistant and 144 strains were susceptible, and the resistance-sensitivity ratio was 1:3, so it was determined that the downy mildew resistance gene of PI438685 was single-gene recessive.
[0087] (2) Inoculation of downy mildew and disease classification: adopt the method of spraying inoculation with conidial spore suspension and 6-level disease cla...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com