Grain weight molecular marker for wheat and application of grain weight molecular marker in breeding
A heavy molecule, wheat grain technology, applied in the fields of genetic engineering and wheat breeding, can solve the problems of QTLs loci that have not been verified by the breeding process or variety validity, poor practicability, and poor accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] The leaf DNA of embodiment 1 wheat is extracted
[0023] (1) Take about 0.3-0.5 g leaves into a 5mL centrifuge tube, freeze them in liquid nitrogen and grind them into powder;
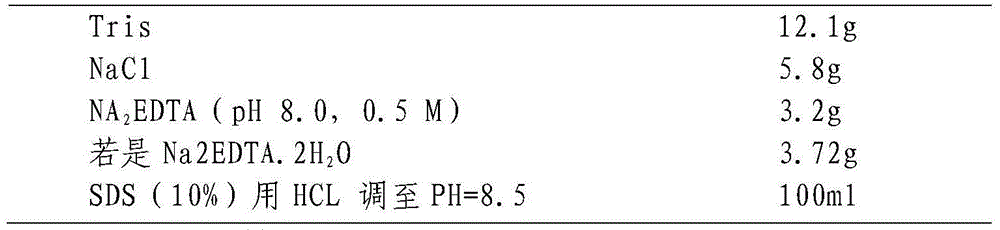
[0024] (2) Add about 1600 μL of buffer S that has been preheated to 65°C, mix by inversion several times, bathe in water for 0.5-1 hour, and shake gently several times during this period to fully mix;
[0025] (3) Cool down to room temperature, wait for 10 minutes, add 10-15μL RNase (10mg / mL) in 37℃ water bath for 30 minutes, shake gently several times to fully mix, about once / 10 min;
[0026] (4) Take out the centrifuge tube, add an equal volume of 1600 μL, extract at 4°C with phenol (Tris-balanced phenol):chloroform:isoamyl alcohol (25:24:1 (volume ratio), mix gently for 10 min, and place in a refrigerator at 4°C Let stand for 5 minutes, then centrifuge at 8000 rpm for 10 minutes;
[0027] (5) Take the supernatant in another tube, about 1300 μL, add an equal volume of cold chloroform (placed...
Embodiment 2
[0043] Embodiment 2 target product amplification
[0044] Forward primer sequence: 5'-CCTTCCATATGTTTTTTTAATGAGCCGCC-3' (as shown in SEQ ID NO:3)
[0045] Reverse primer sequence: 5'-GCTTCTTTTCCAACTCAATAAATGAGCC-3' (as shown in SEQ ID NO:4)
[0046] PCR amplification: the PCR amplification system is 20μL
[0047]
[0048]Note: Mix available: or (Taq enzyme 0.25μL, DNK 2.0μL, Buffer1.5μL, MgCl 0.4μL configuration.)
[0049] Amplification conditions:
[0050]
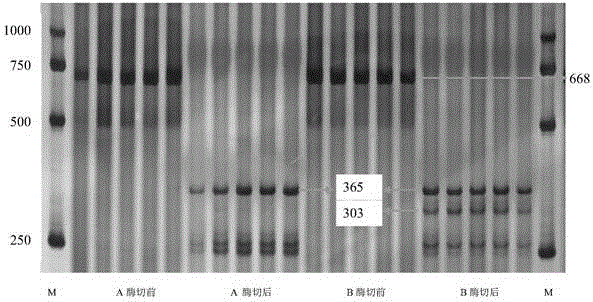
[0051] A 668bp fragment can be obtained through the above amplification, and its nucleotide sequence is shown in SEQ ID NO:2.
Embodiment 4
[0052] Example 4 Specific enzyme digestion of PCR products:
[0053] Enzyme digestion system 10μL:
[0054] Specific enzyme HaeIII: 0.3 μL
[0055] PCR product: 3 μL
[0056] 10×NE buffer: 0.7μL
[0057] wxya 2 O: 6μL
[0058] Enzyme digestion reaction conditions: add 0.3 μL of HaeIII specific enzyme (obtained from the market) to the PCR amplification product, bathe in water at 37°C for 2 hours, then inactivate the enzyme digestion system at 65°C for 5 min.
[0059] After the above-mentioned amplified products were separated by electrophoresis on 8% polyacrylamide gel, the molecular weight of the amplified product was 668bp. A 303bp electrophoresis band appeared. However, in varieties or lines with an increased thousand-grain weight gene, the segment is chopped and the segment is deleted.
[0060]
[0061]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com