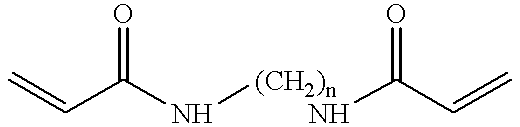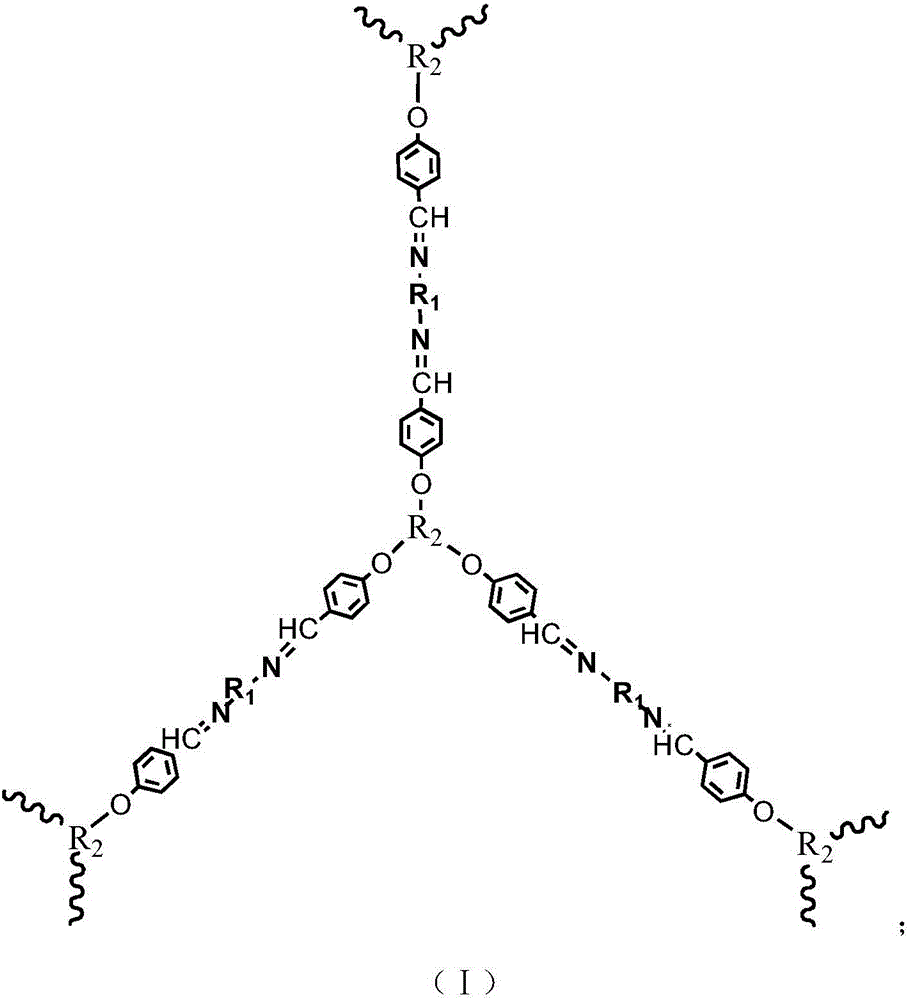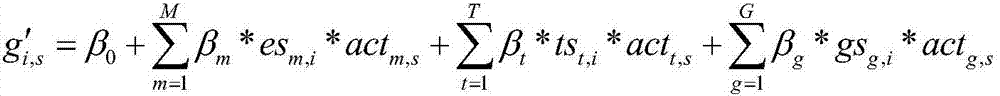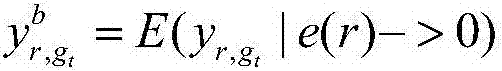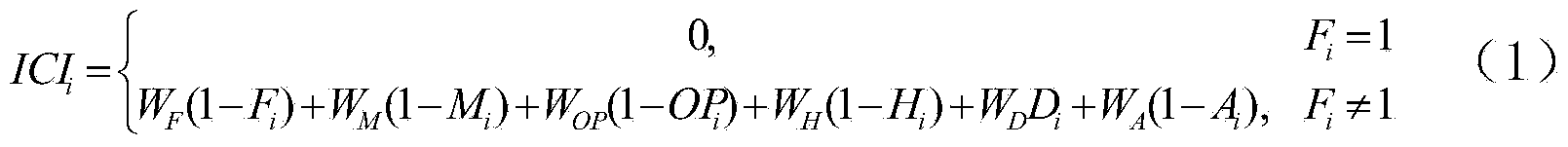Patents
Literature
82 results about "Biological significance" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Biological (Clinical) Significance: Biological significance is the significance of the difference between outcomes in the clinical situation and must be determined by the clinician with respect to the patient. Biological (clinical) significance is unrelated to statistical significance.
Methods for microdispensing patterened layers
InactiveUS6306594B1Efficient couplingShort response timeMicrobiological testing/measurementBurettes/pipettesAnalyteClinical settings
An efficient method for the microfabrication of electronic devices which have been adapted for the analyses of biologically significant analyte species is described. The techniques of the present invention allow for close control over the dimensional features of the various components and layers established on a suitable substrate. Such control extends to those parts of the devices which incorporate the biological components which enable these devices to function as biological sensors. The materials and methods disclosed herein thus provide an effective means for the mass production of uniform wholly microfabricated biosensors. Various embodiments of the devices themselves are described herein which are especially suited for real time analyses of biological samples in a clinical setting. In particular, the present invention describes assays which can be performed using certain ligand / ligand receptor-based biosensor embodiments. The present invention also discloses a novel method for the electrochemical detection of particular analyte species of biological and physiological significance using an substrate / label signal generating pair which produces a change in the concentration of electroactive species selected from the group consisting of dioxygen and hydrogen peroxide.
Owner:I STAT CORP
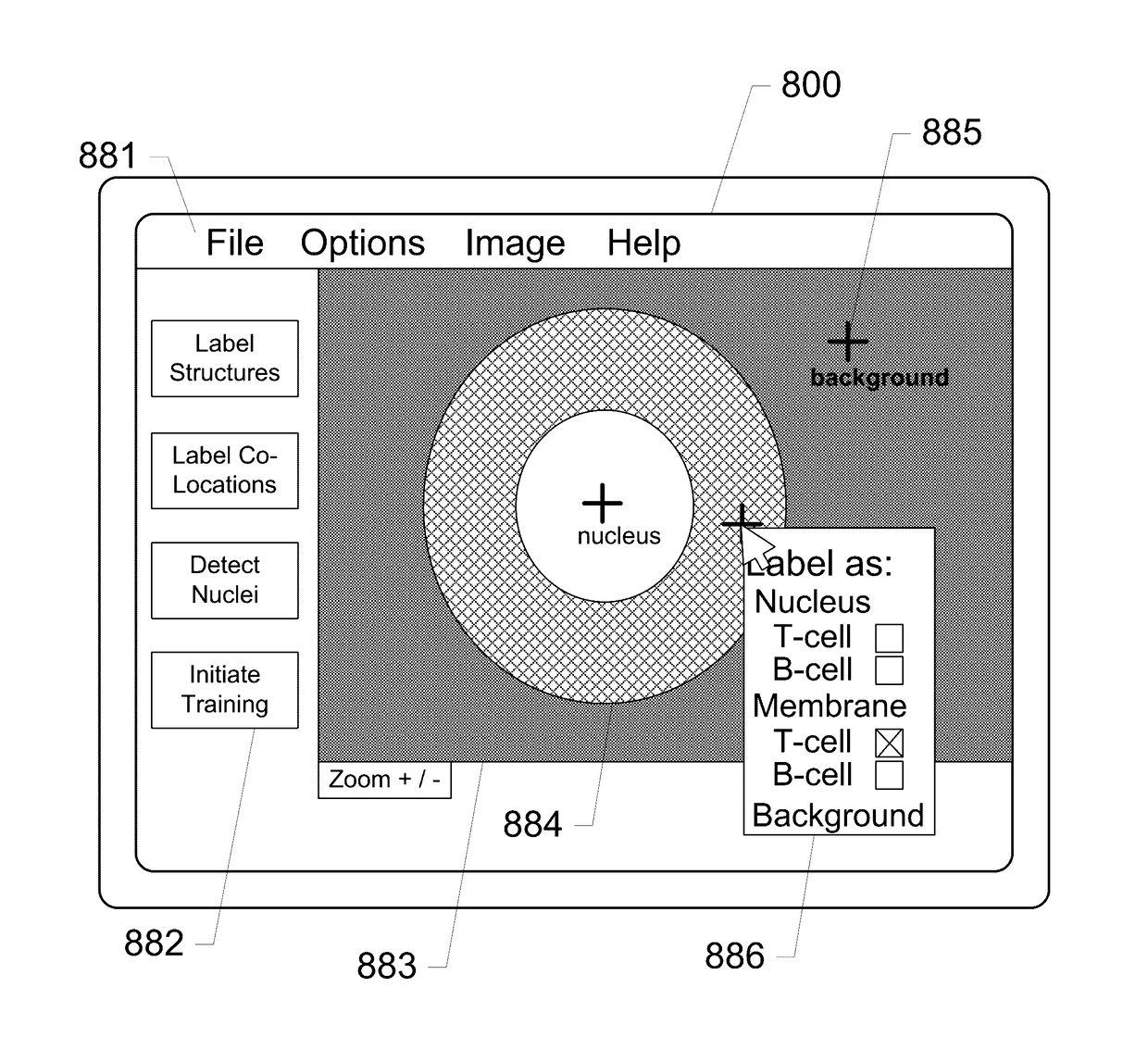
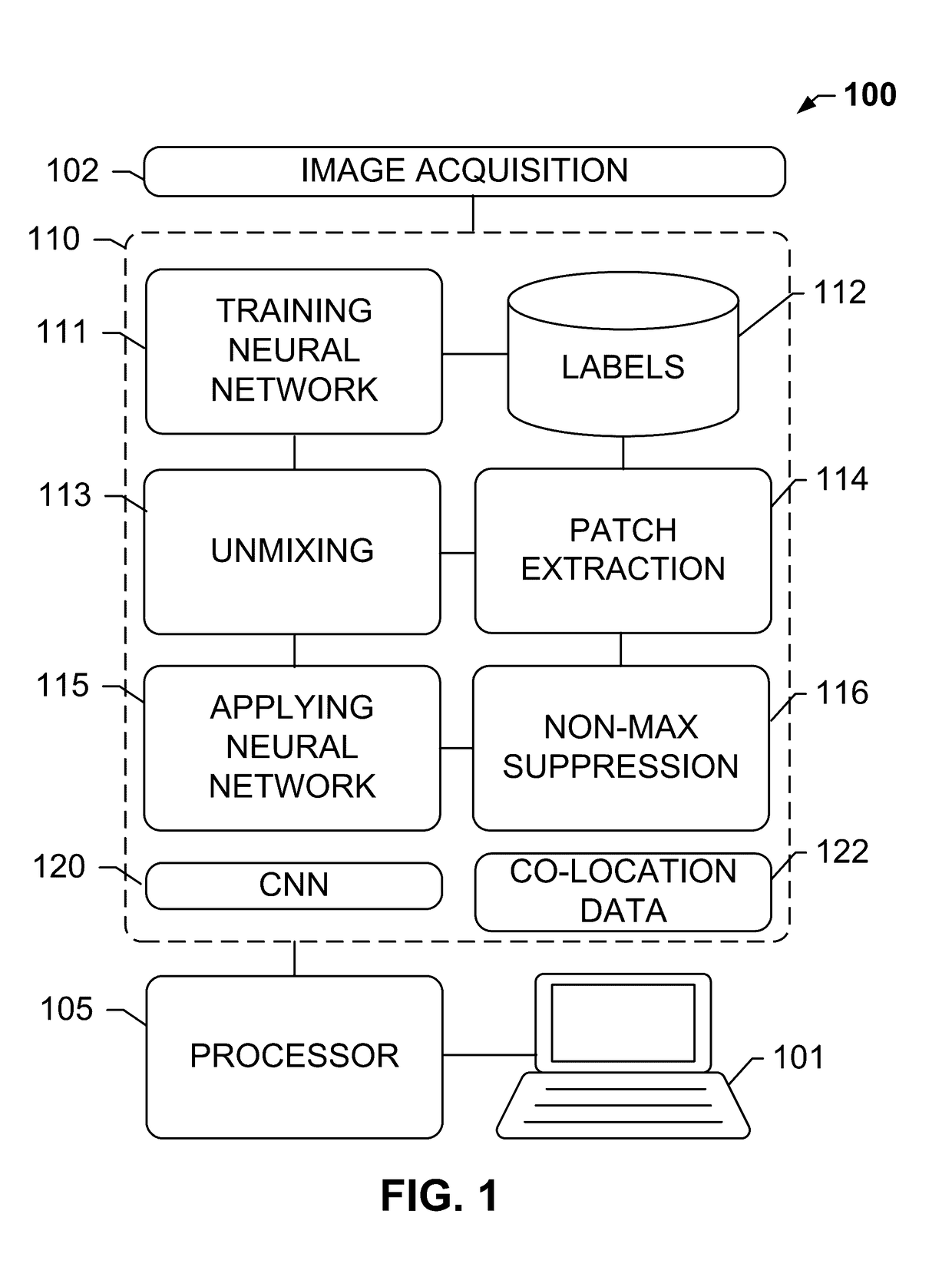
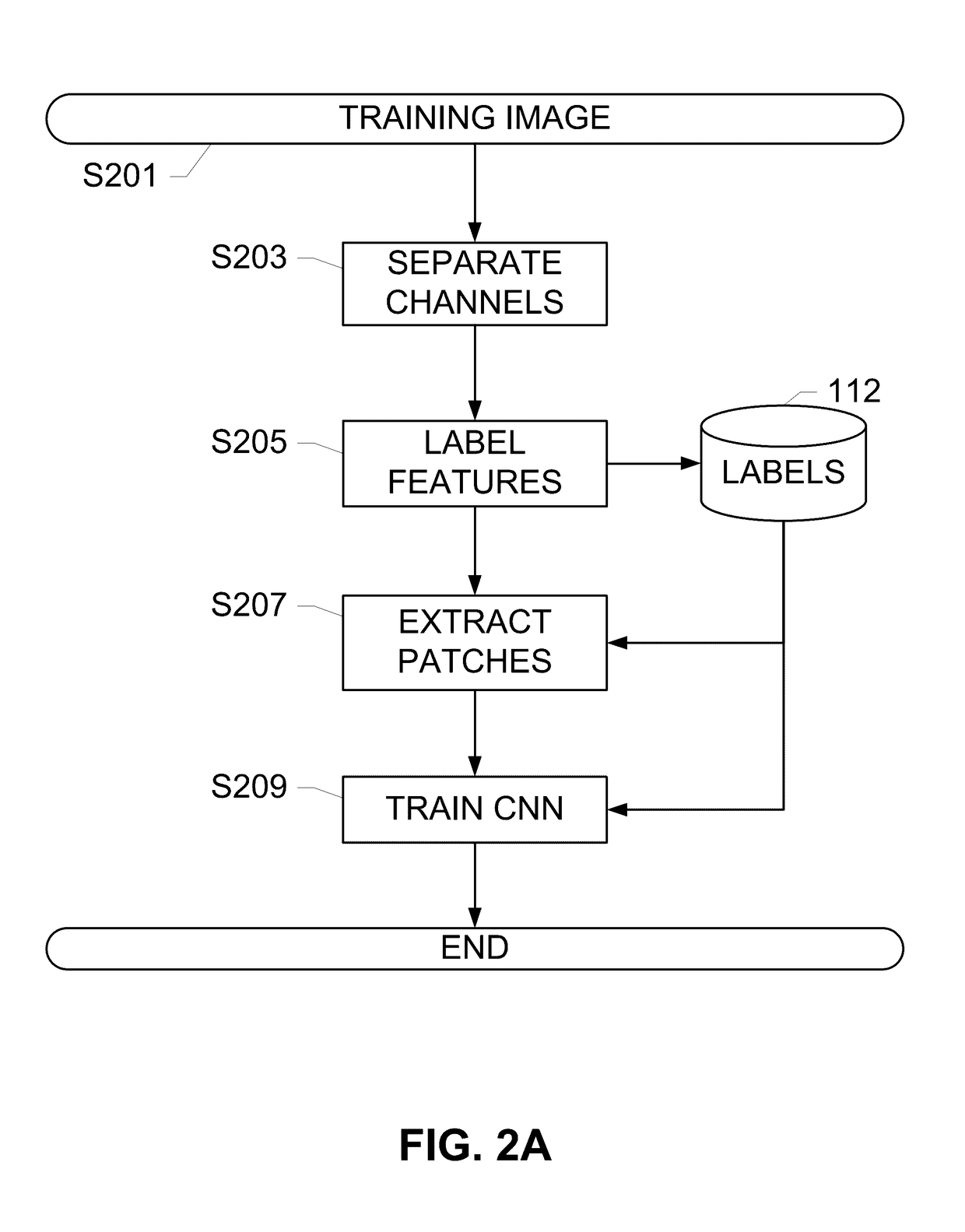
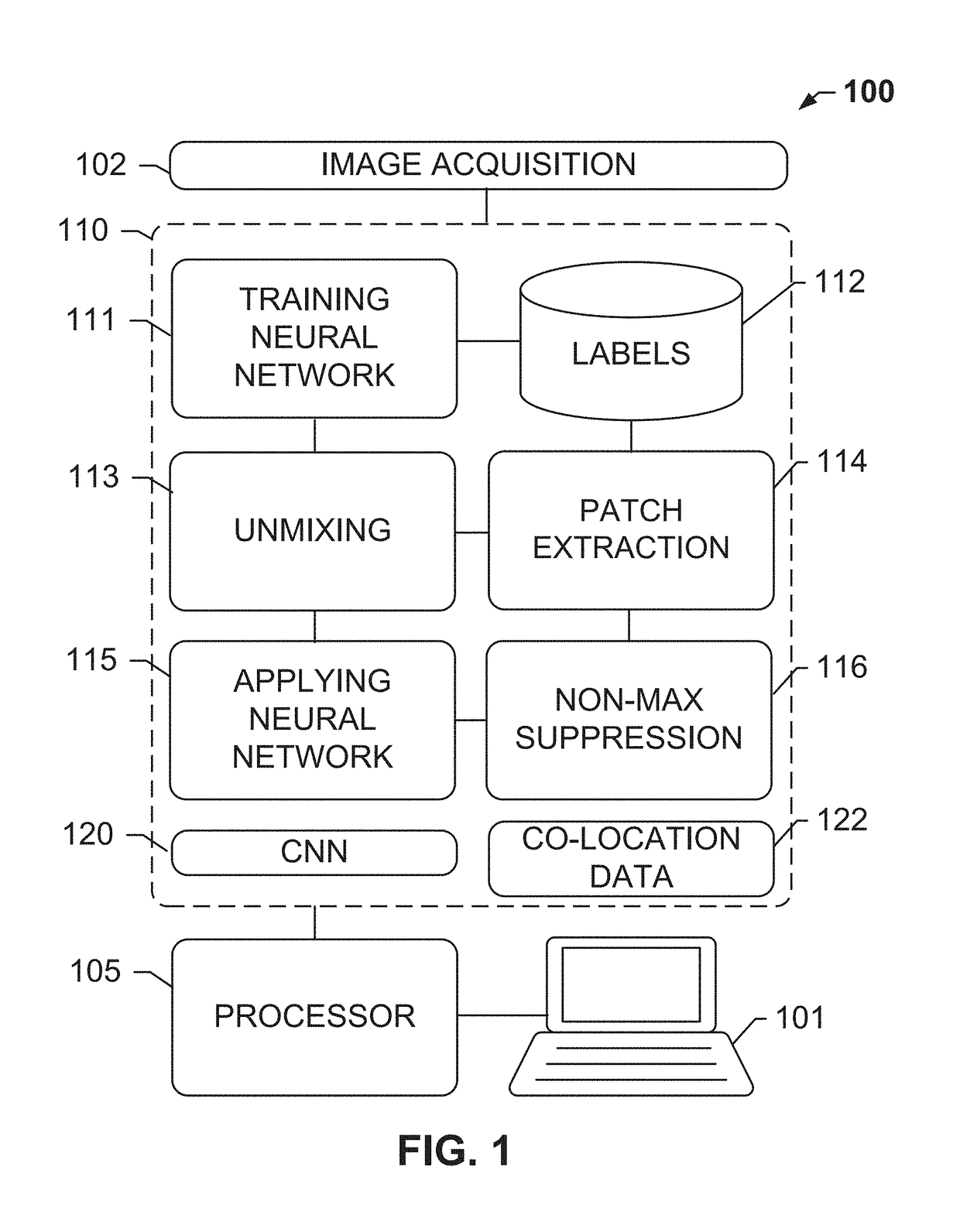
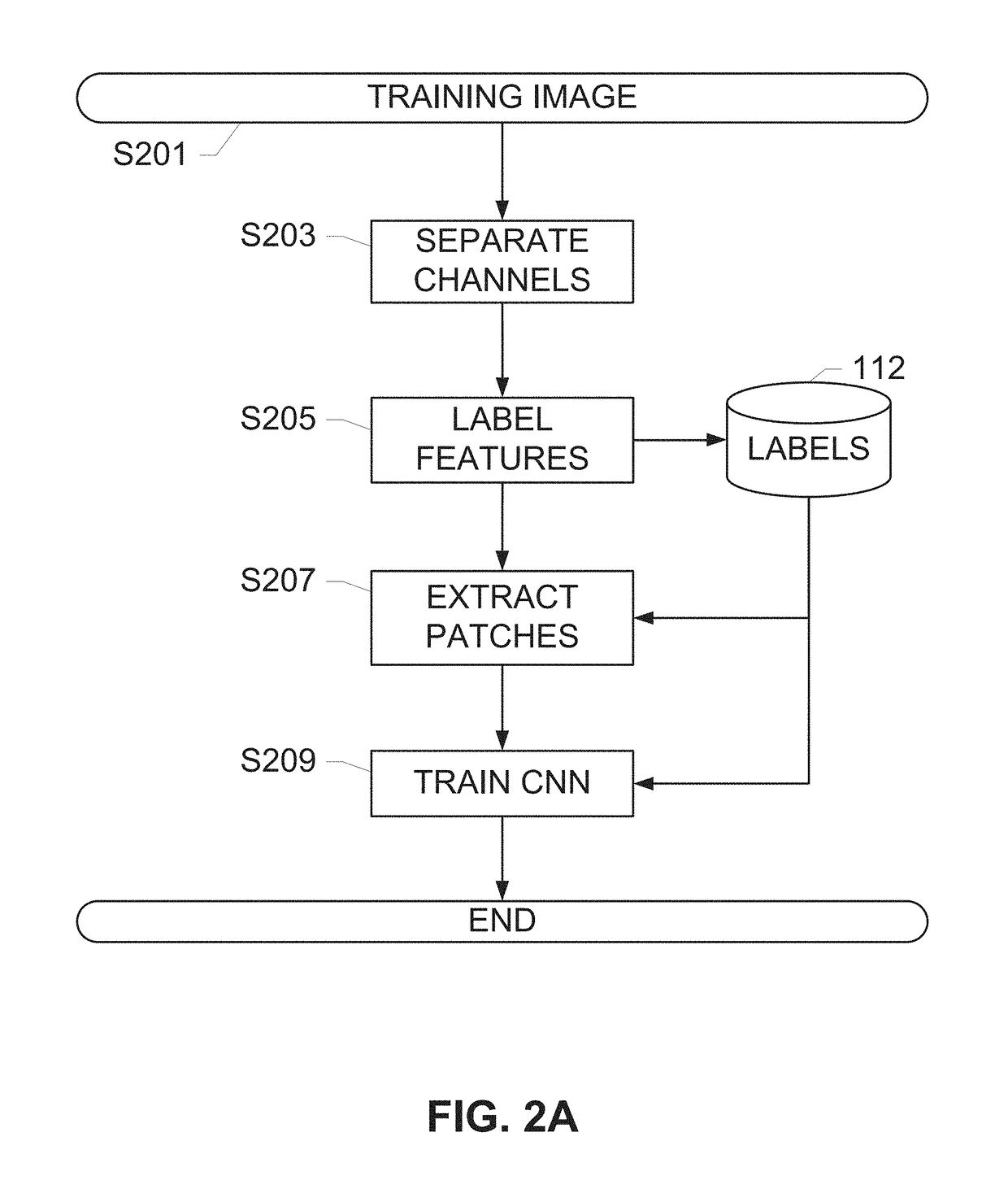
Systems and methods for detection of structures and/or patterns in images
ActiveUS20170169567A1Lighten the computational burdenImprove stabilityImage enhancementImage analysisRgb imageNon maximum suppression
The subject disclosure presents systems and computer-implemented methods for automatic immune cell detection that is of assistance in clinical immune profile studies. The automatic immune cell detection method involves retrieving a plurality of image channels from a multi-channel image such as an RGB image or biologically meaningful unmixed image. A cell detector is trained to identify the immune cells by a convolutional neural network in one or multiple image channels. Further, the automatic immune cell detection algorithm involves utilizing a non-maximum suppression algorithm to obtain the immune cell coordinates from a probability map of immune cell presence possibility generated from the convolutional neural network classifier.
Owner:VENTANA MEDICAL SYST INC
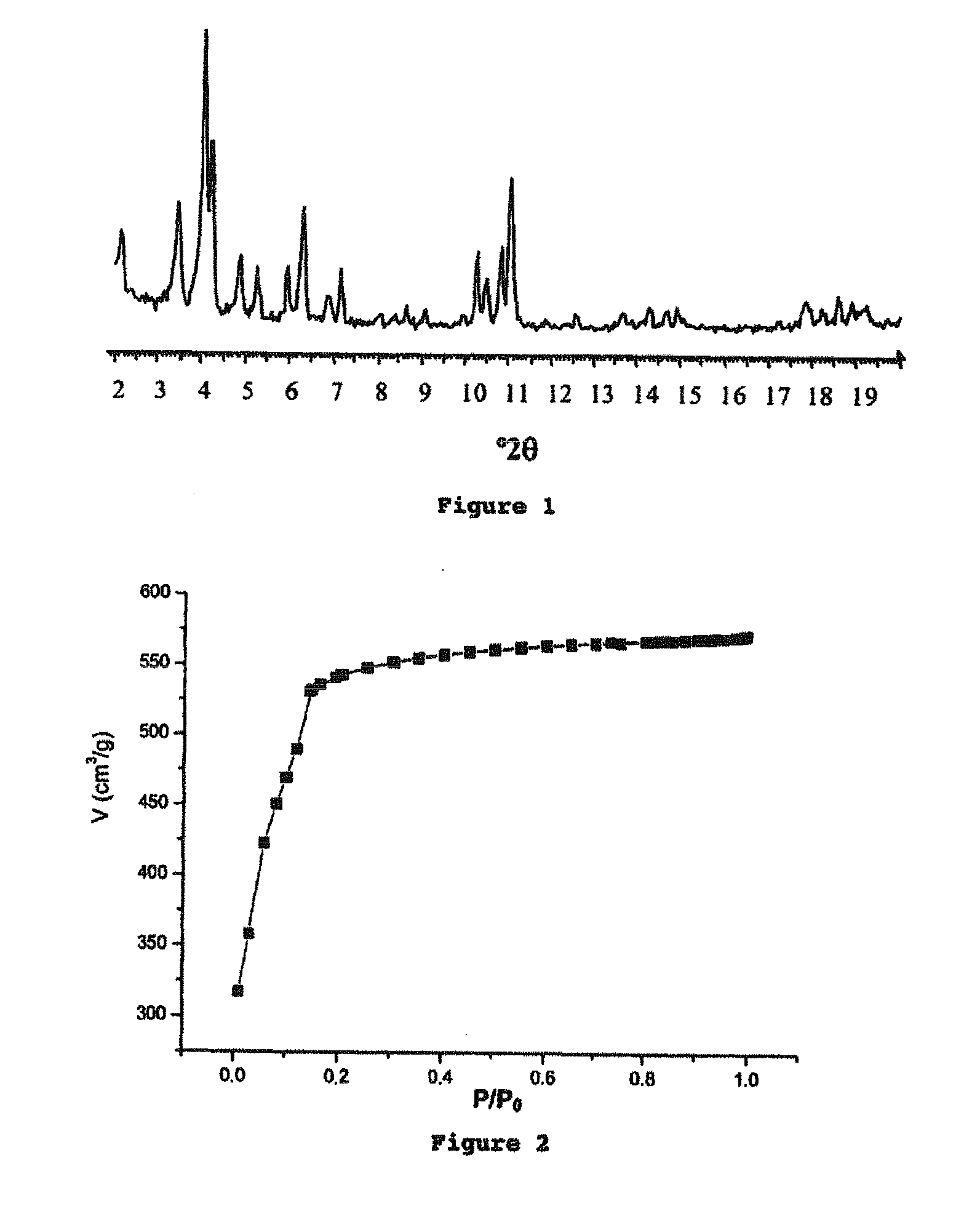
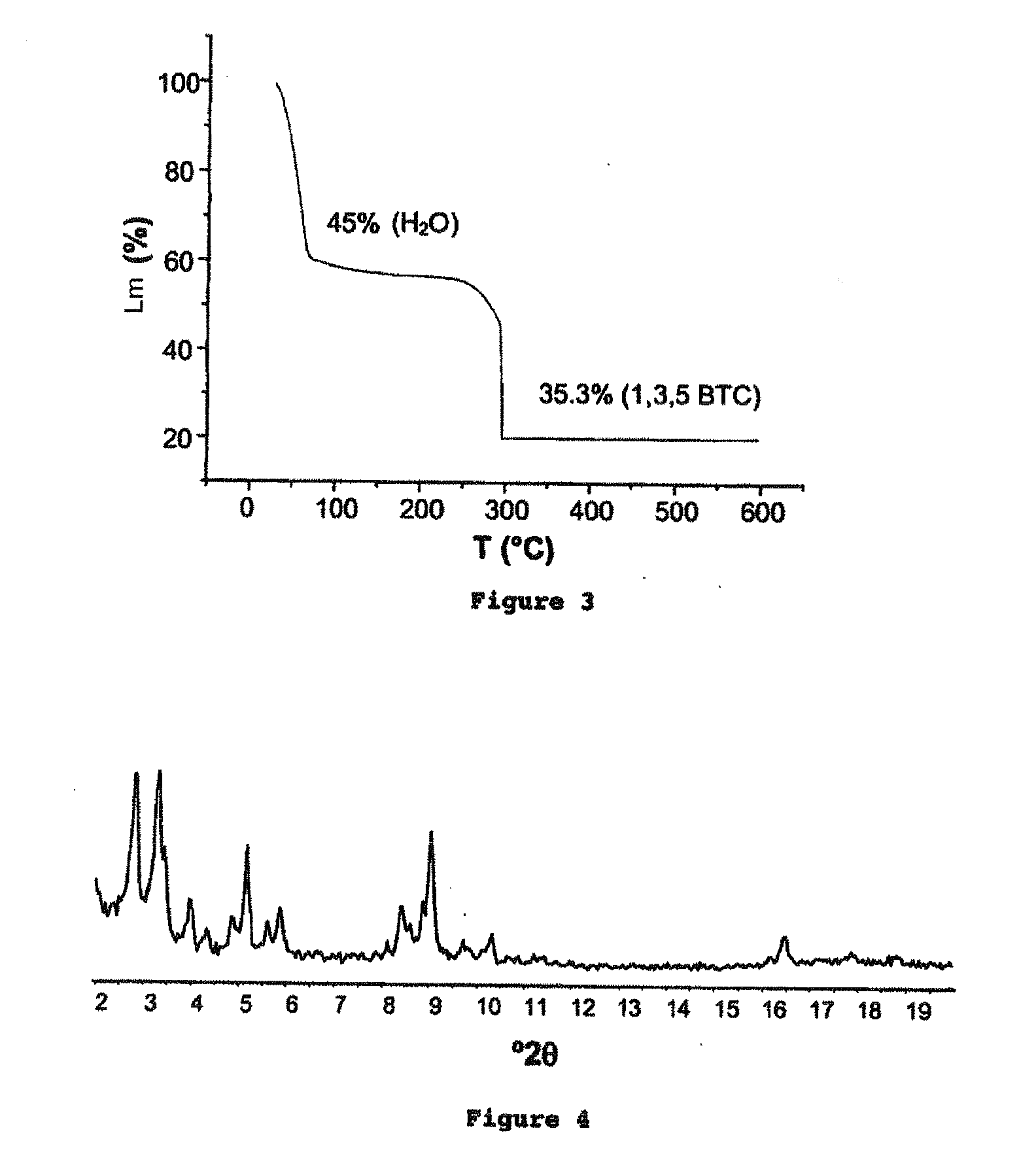
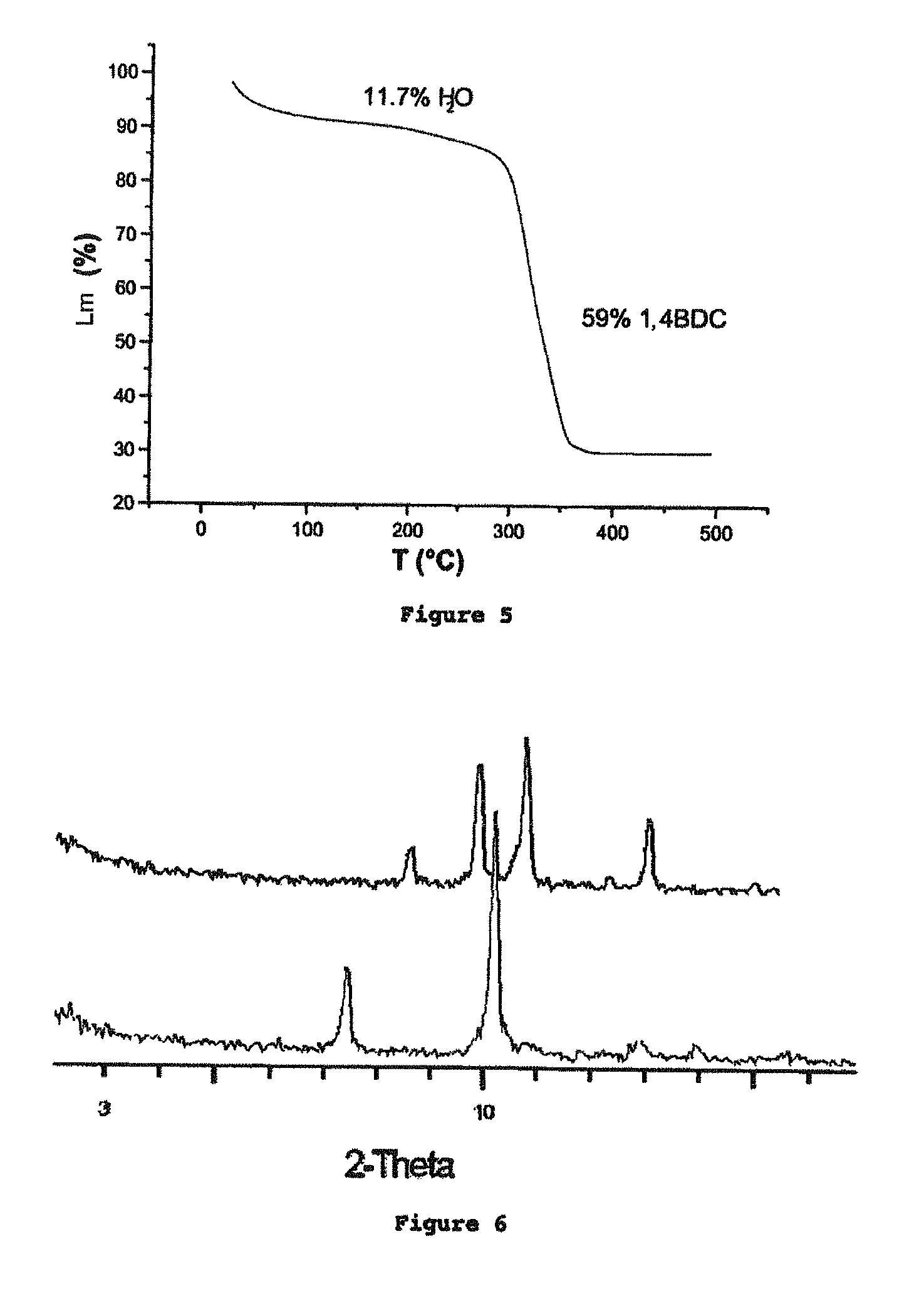
Porous crystalline hybrid solid for adsorbing and releasing gas of biological interest
The invention relates to solids made of a porous crystalline metal-organic framework (MOF) loaded with at least one gas of biological interest, and to a method for preparing the same. The MOF solids of the present invention are capable of adsorbing and releasing in a controlled manner gases having a biological interest. They can be used in the pharmaceutical field and / or for applications in the cosmetic field. They can also be used in the food industry.
Owner:CENT NAT DE LA RECHERCHE SCI +3
Phosphorus-containing Schiff base structured flame retardant and preparation method thereof
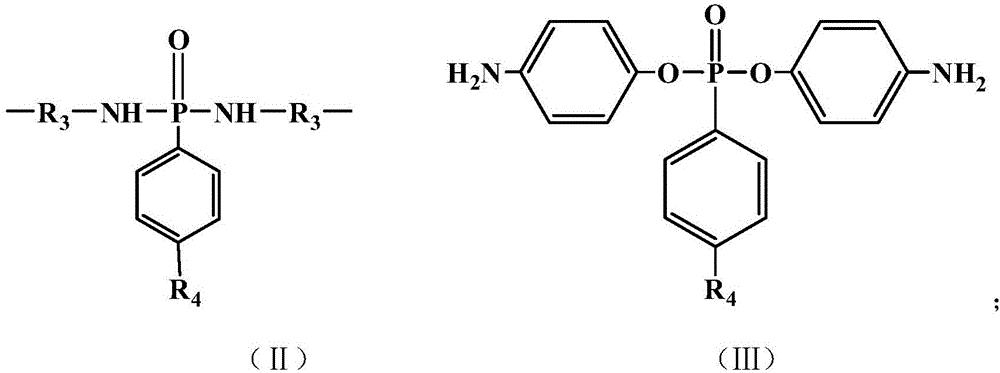
The invention discloses a phosphorus-containing Schiff base structured flame retardant and a preparation method thereof. The phosphorus-containing Schiff base structured flame retardant consists of hyperbranched macromolecules; a hyperbranched repeat unit is shown in the formula (I) as shown in the specification; in the formula, R1 is a phosphorus-containing structure, and R2 is a trifunctional structure; the molecular weight of the flame retardant is 500-100000. The flame retardant disclosed by the invention consists of hyperbranched macromolecules and is relatively large in molecular weight; when the flame retardant is used in polyurethane foams, the carbon residue amount of polyurethane can be greatly increased; the flame retardant disclosed by the invention is free of halogen and is a green and environment-friendly flame retardant; the flame retardant contains substances of a Schiff base structure, and due to existence of a C=N structure, very important chemical and biological significances can be achieved due to lone pair electrons on N atoms.
Owner:UNIV OF SCI & TECH OF CHINA
Systems and methods for detection of structures and/or patterns in images
ActiveUS10109052B2Easy to trainImprove stabilityImage enhancementImage analysisRgb imageNon maximum suppression
Owner:VENTANA MEDICAL SYST INC
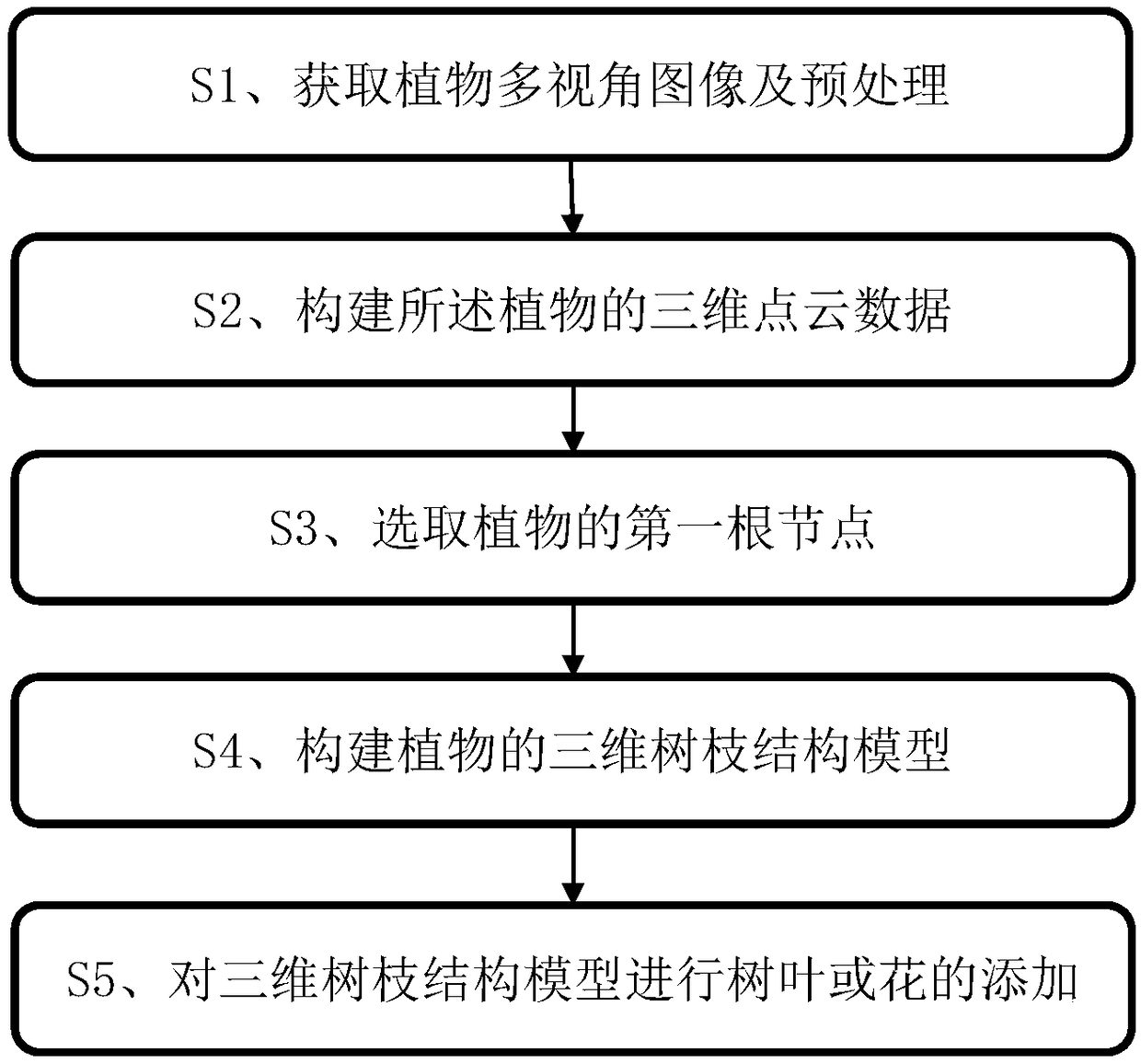
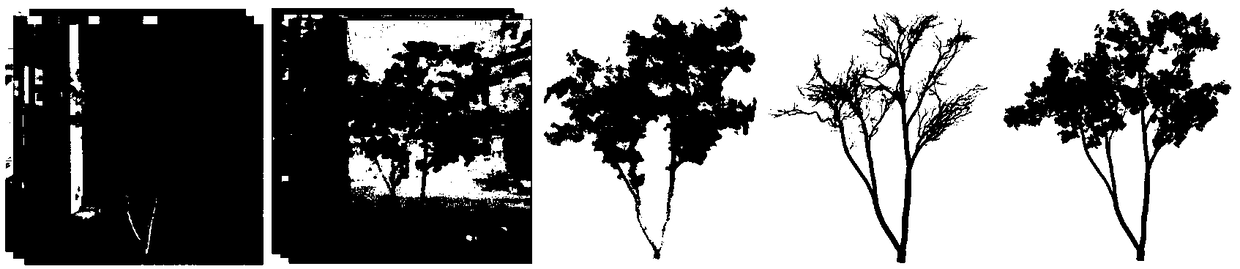
Process-type plant model reconstruction method based on multi-view images
ActiveCN109285217AImprove realismDetails involving processing stepsCharacter and pattern recognitionGeometric consistencyReconstruction method
The invention belongs to the technical field of plant modeling and computer graphics processing, in particular to a process-type plant model reconstruction method based on multi-view images, aiming atsolving the problems of large error and low precision between the plant model constructed by the prior method and the real data. The method comprises the following steps: acquiring the plant multi-view picture information; performing dense depth estimation based on color and geometric consistency; acquiring 3D point cloud data based on multi-view depth information fusion; a three-dimensional treebranch structure model of the plant is constructed by using a pre-constructed branch structure growth parameter representation model and the three-dimensional point cloud data to restrict plant growth. The invention provides a solution for reconstructing a complete tree model from a multi-view image. The obtained point clouds and models have a high degree of consistency with the original image, which skillfully combines plant process modeling and data-driven plant reconstruction method, not only to ensure the accuracy of the model, but also to maintain the biological significance of plants.
Owner:INST OF AUTOMATION CHINESE ACAD OF SCI
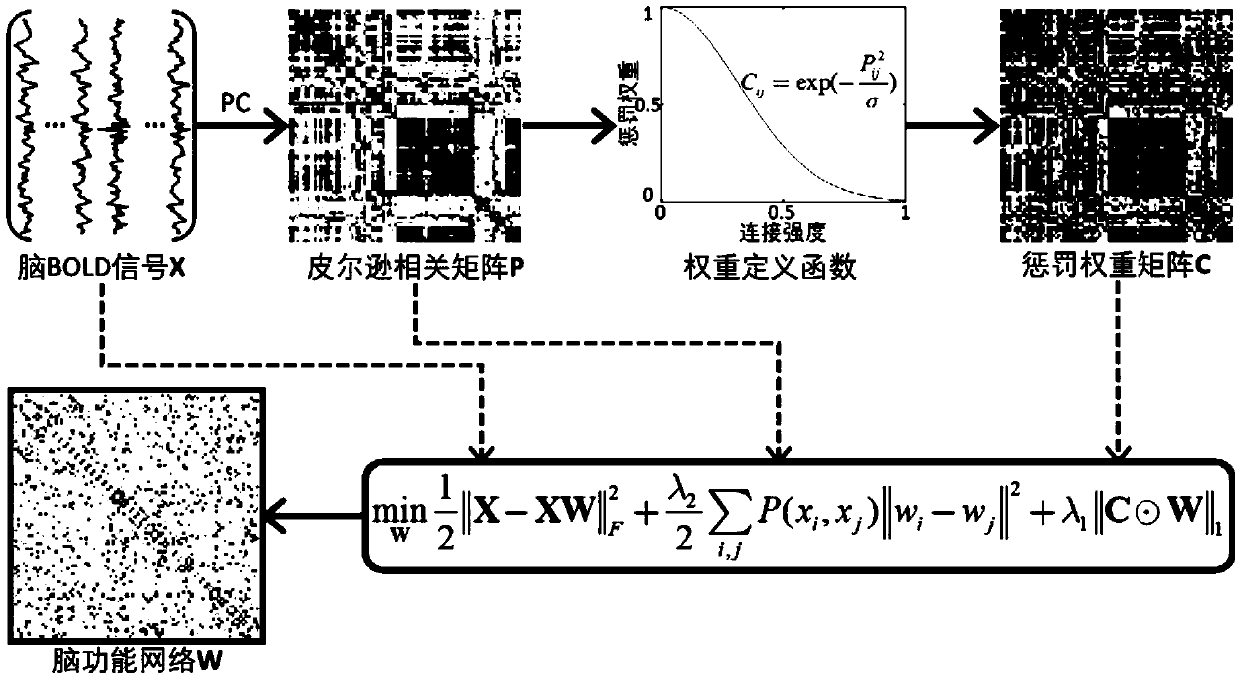
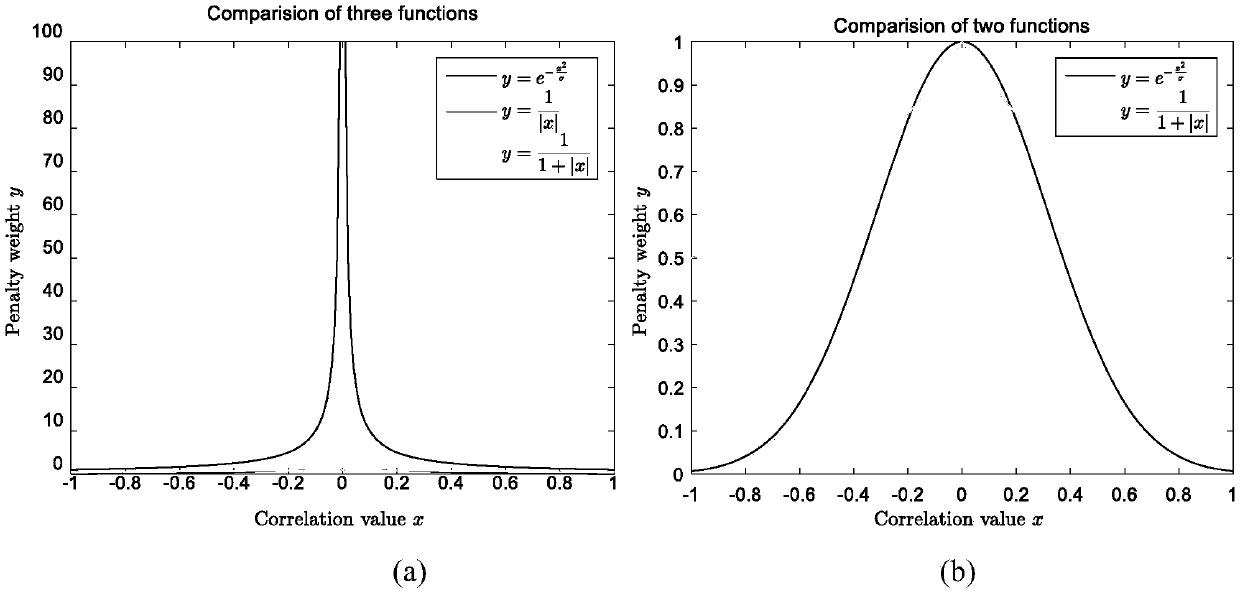
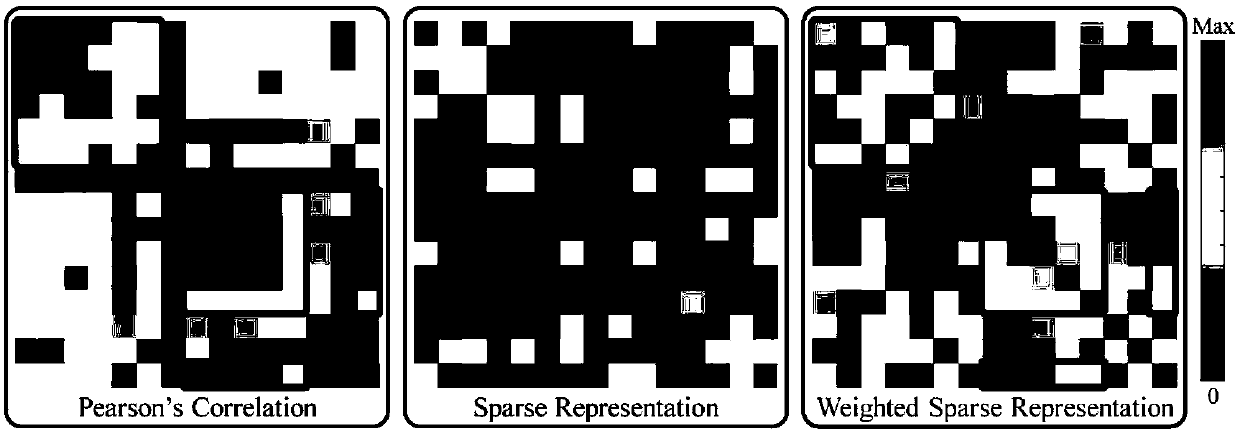
Weighted graph regularization sparse brain network construction method
PendingCN109065128ASolve the problem that the solution does not convergeImprove classification accuracyImage enhancementImage analysisDiseaseFunctional connectivity
The invention discloses a weighted graph regularization sparse brain network construction method. According to the method, on the basis of analysis of keeping similar local manifold characteristics ofsimilar data in original data space after projection, constrained modeling is carried out on association between brain connection by using a graph regularization item; with consideration of correlation analysis, the similarity degree of essences in a time sequence signal sequence of a brain area can be measured, sparse modeling is constrained by using measured functional connection strength priorinformation, and a weighted sparse regularization item is established; and then combined modeling is carried out on the whole brain function network and a brain network with the biological significance is constructed. According to the invention, on the basis of integration of correlation analysis, sparsity and graph regularization constraint into the unified modeling frame, a brain function network is constructed effectively by utilizing the similarity and locality of functional magnetic resonance brain image data fully; and the method is also used for neurological disease identification andbiological marker analysis.
Owner:ZHENGZHOU UNIV
Method for inducing gynogenesis of cyprinus carpio L. by using megalobrama amblycephala sperms
InactiveCN102919185ABreed fastAccelerated homozygosityClimate change adaptationPisciculture and aquariaCyprinusEmbryo
The invention discloses a method for inducing gynogenesis of cyprinus carpio L. by using megalobrama amblycephala sperms. The method comprises the following steps of: firstly, selecting a female gynogenesis of cyprinus carpio L. and a male megalobrama amblycephala as parent fishes, secondly, performing artificial induced spawning to obtain megalobrama amblycephala sperms and cyprinus carpio L. spawns; and finally, uniformly mixing ultraviolet-inactivated megalobrama amblycephala seminal sperms and the cyprinus carpio L. spawns, scattering the mixture on a mesh which is flatly paved in water, after 5 minutes, placing activated spawns in cold water of 5 DEG C to perform cold shock treatment for 18 to 25 minutes, and incubating embryos after the cold shock treatment at 21 to 23 DEG C in water to obtain diploid gynogenesis cyprinus carpio L. According to the method, the measures of activating the gynogenesis of the cyprinus carpio L. spawns by using the inactivated megalobrama amblycephala seminal sperms and doubling genomes are used, so that the induced descendants only contain genes of female parents, the homozygosis of genes can be accelerated, and the method has important biological significance in the aspects of color inheritance law study and genetic breeding of the cyprinus carpio L.
Owner:BEIJING FISHERIES RES INST
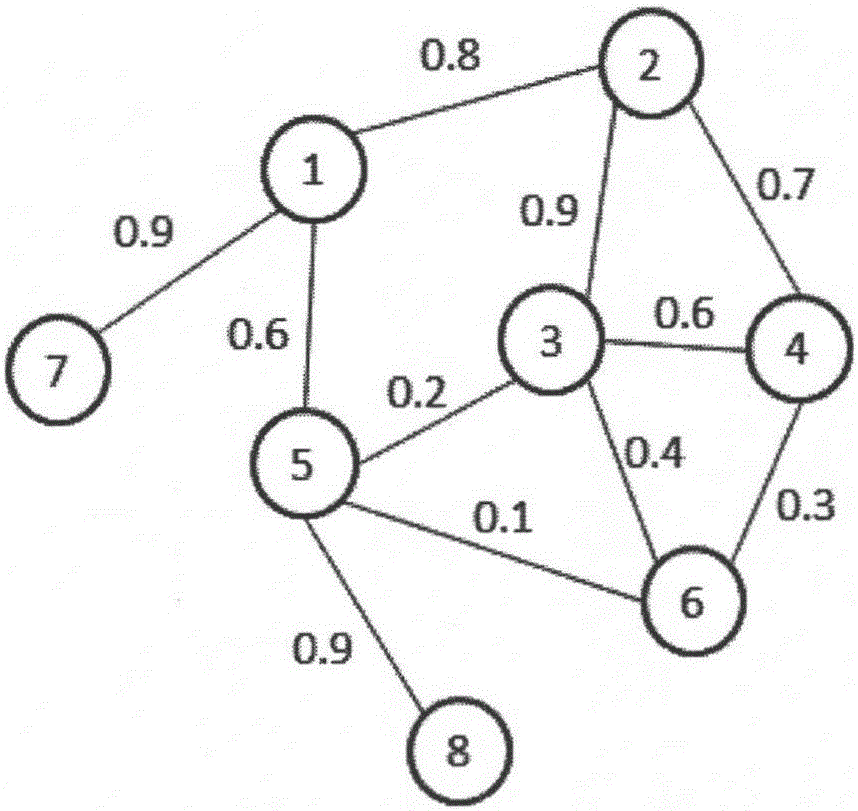
Protein complex recognizing method based on range estimation
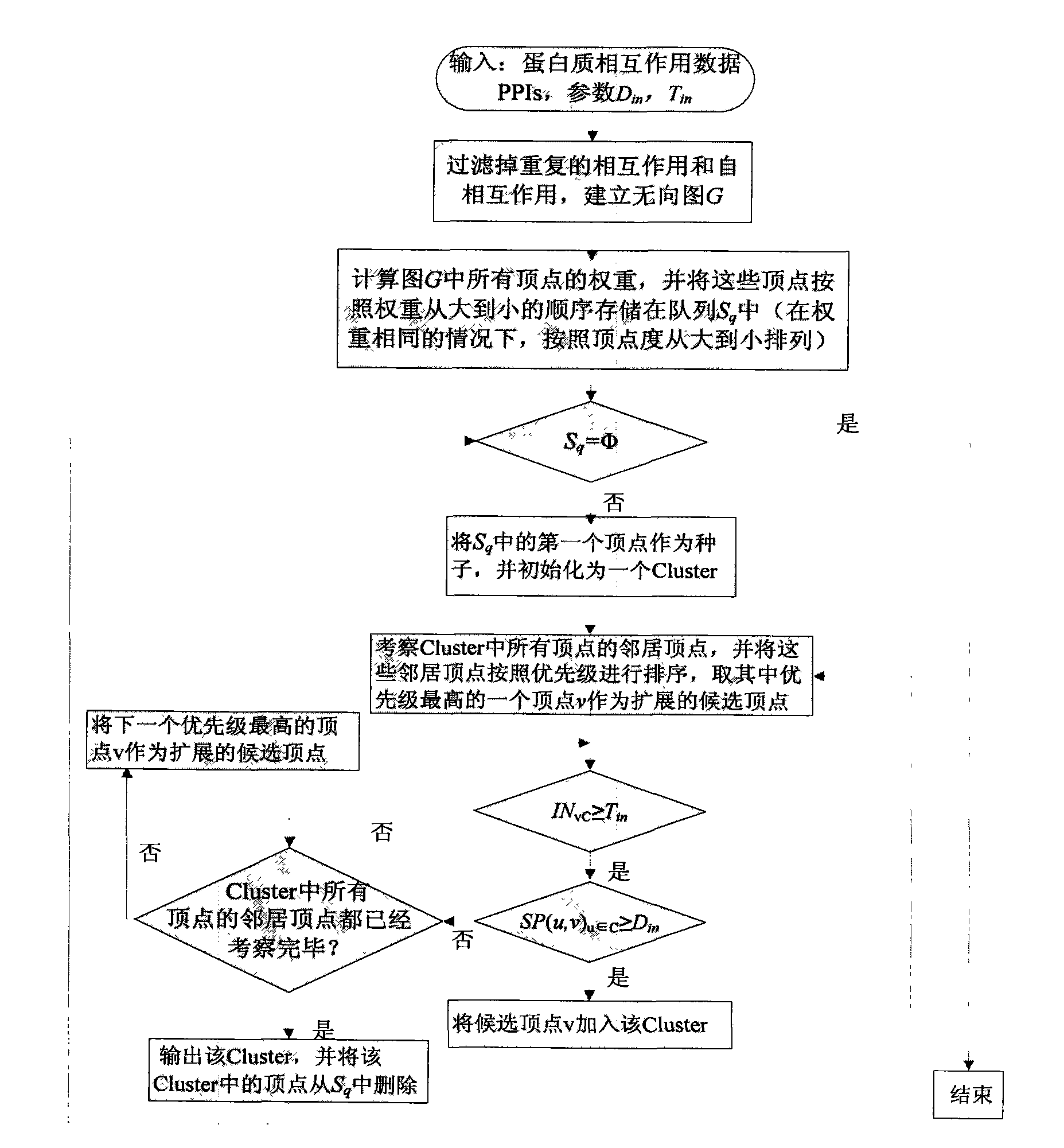
InactiveCN101246520AEfficient identificationImprove robustnessSpecial data processing applicationsShortest distanceProtein-protein complex
The invention discloses a protein compound recognition method based on the range estimation, which takes the most short distance between protein apexes as a key parameter to recognize the protein compound, and controls the dense degree of the recognized the protein compound by the function probability between the protein apexes and the protein compound based on finding that the most short distance between the protein apexes in the known protein compound generally does not surpass 2. The realization of the invention is simply; a plurality of known proteins compound with the biological significance can be recognized through the protein interaction network, the invention has the very good toughness for the high proportion false positive and false negative which universally exist in the protein interaction large-scale data, and solves the chemical experiment cost to be expensive and the biology difficult problems that the quantity of single recognition is small and the dynamic compound is very difficult to recognized effectively.
Owner:CENT SOUTH UNIV
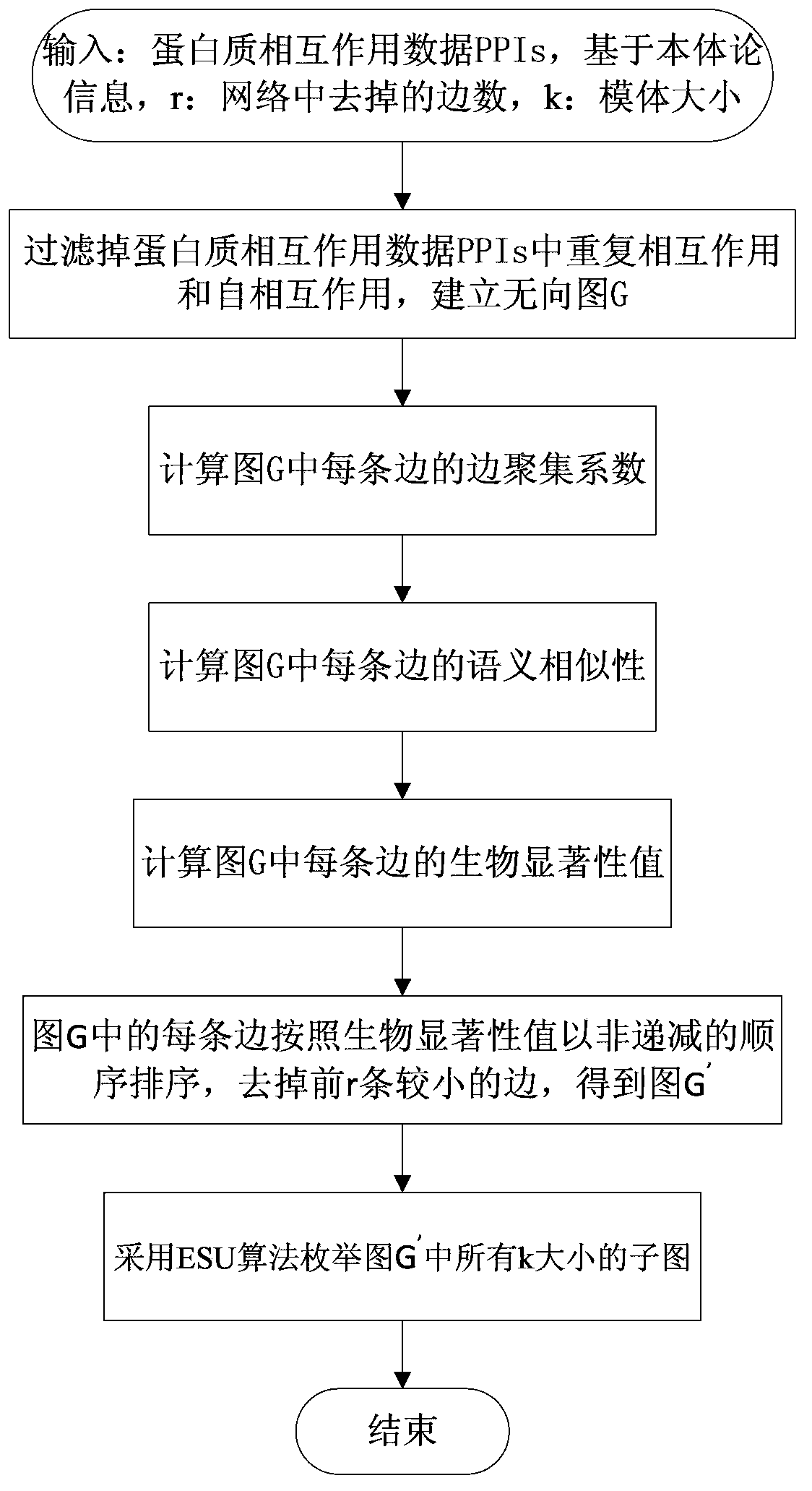
Protein biological network motif identification method integrating topological attributes and functions
ActiveCN103514381AAccurate identificationEasy to integrateSpecial data processing applicationsGene ontologyLarge scale data
The invention discloses a protein biological network motif identification method integrating topological attributes and functions. The protein biological network motif identification method integrating the topological attributes and functions (Ecc-GOSS) is based on the biology significance of a motif, and comprehensively evaluates the biological significance of protein-protein interaction by integrating edge clustering coefficients and semantic similarity of GO phrases. The protein biological network motif identification method integrating the topological attributes and functions is easy to implement, a great amount of network motifs with biological significance can be identified accurately according to PPI information and gene ontology information, and the protein biological network motif identification method integrating the topological attributes and functions has good robustness on high-percentage false positive prevailing among the large-scale data of the protein-protein interaction.
Owner:HUNAN UNIV
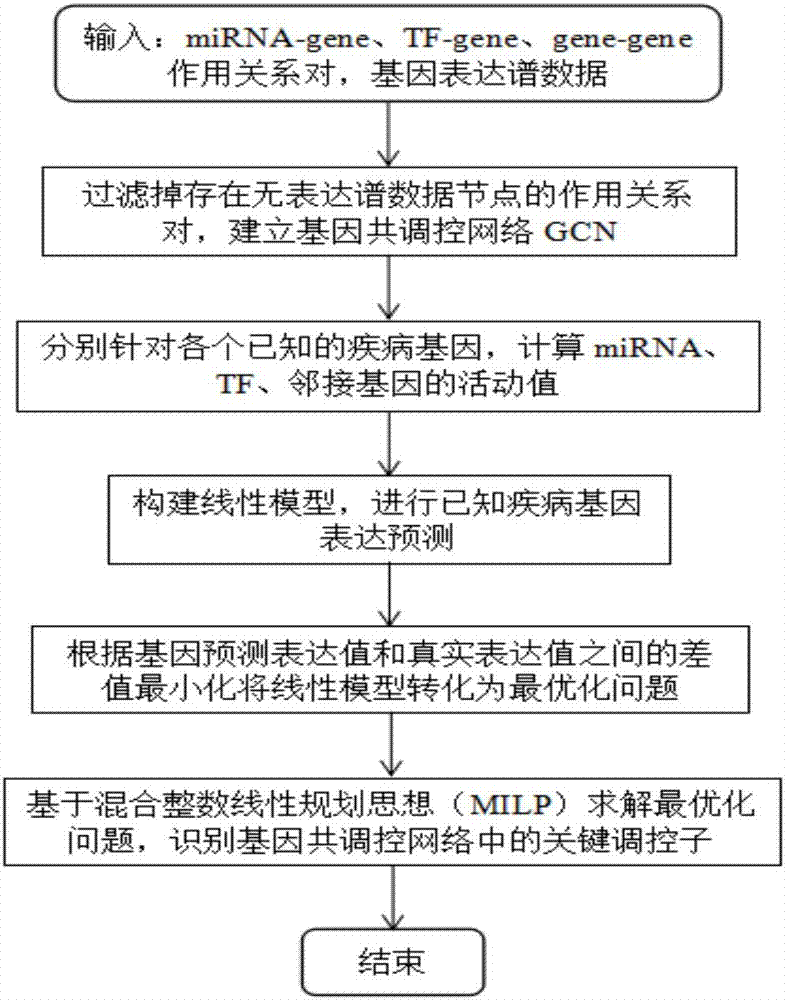
Method for recognizing key regulator in gene co-regulatory network based on linear model
ActiveCN106874704AHigh precisionAccurate identificationBiostatisticsSystems biologyLinear modelGene expression profiling
The invention discloses a method for recognizing a key regulator in a gene co-regulatory network based on a linear model. The gene expression profile data and the gene regulation relation data are utilized to forecast the expression of the known disease gene in the manner of establishing the linear model, so as to complete the recognition for the key regulator in the gene co-regulatory network. The method provided by the invention is simply realized; the key regulator in the gene co-regulatory network can be accurately recognized only according to the gene expression profile data and the gene regulation relation; tests prove that the recognized regulator has an important biological significance and has important theoretical significance and practical value in researching the disease mechanism.
Owner:HUNAN UNIV
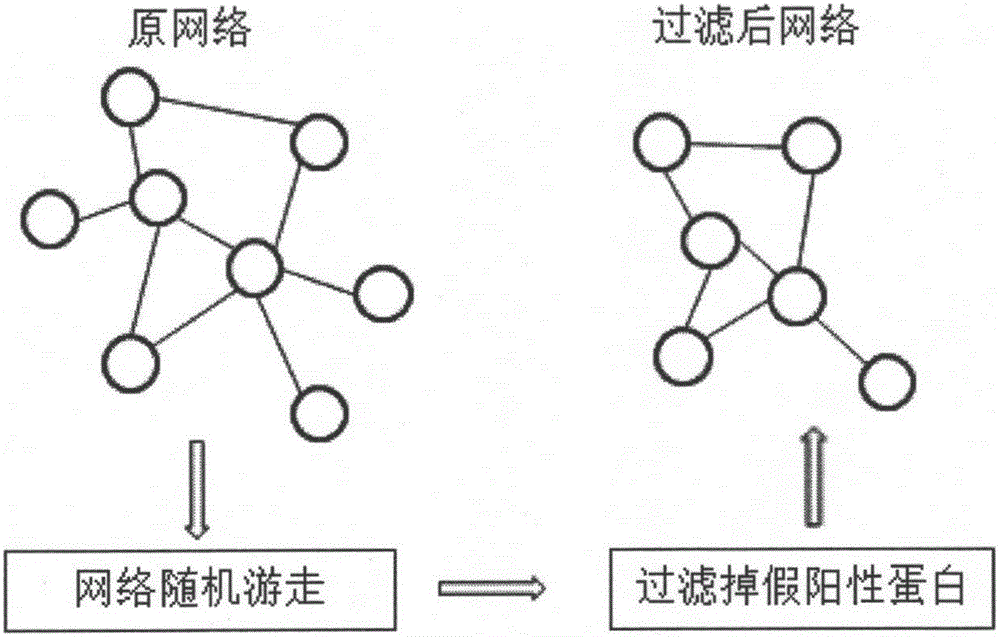
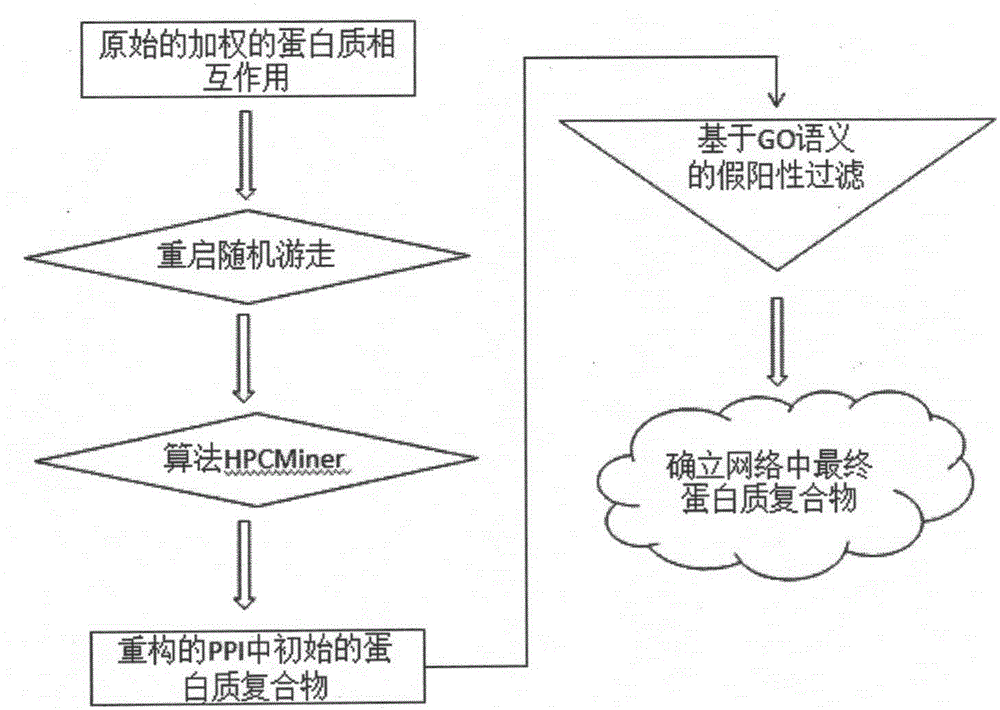
Protein composite identification method based on random walking model
InactiveCN106355044ASolve the noiseOvercome limitationsProteomicsGenomicsExperimental validationProtein insertion
The invention provides a protein composite identification method based on a random walking model. Interaction data and false-negative or false-positive noisy data truly existing on a protein network are forecasted through the random walking algorithm. On the protein interaction network obtained after false-negative data and false-positive noisy data are removed, protein composites with the biological significance are identified through a H-index graph model, the semantic similarity between the protein composites is calculated according to a GO body, and the identified protein composites are finally determined. According to the protein composite identification method based on the random walking model, the algorithm is insensitive to input parameters, and the effectiveness of the provided algorithm is verified through experiments.
Owner:SHANGHAI DIANJI UNIV
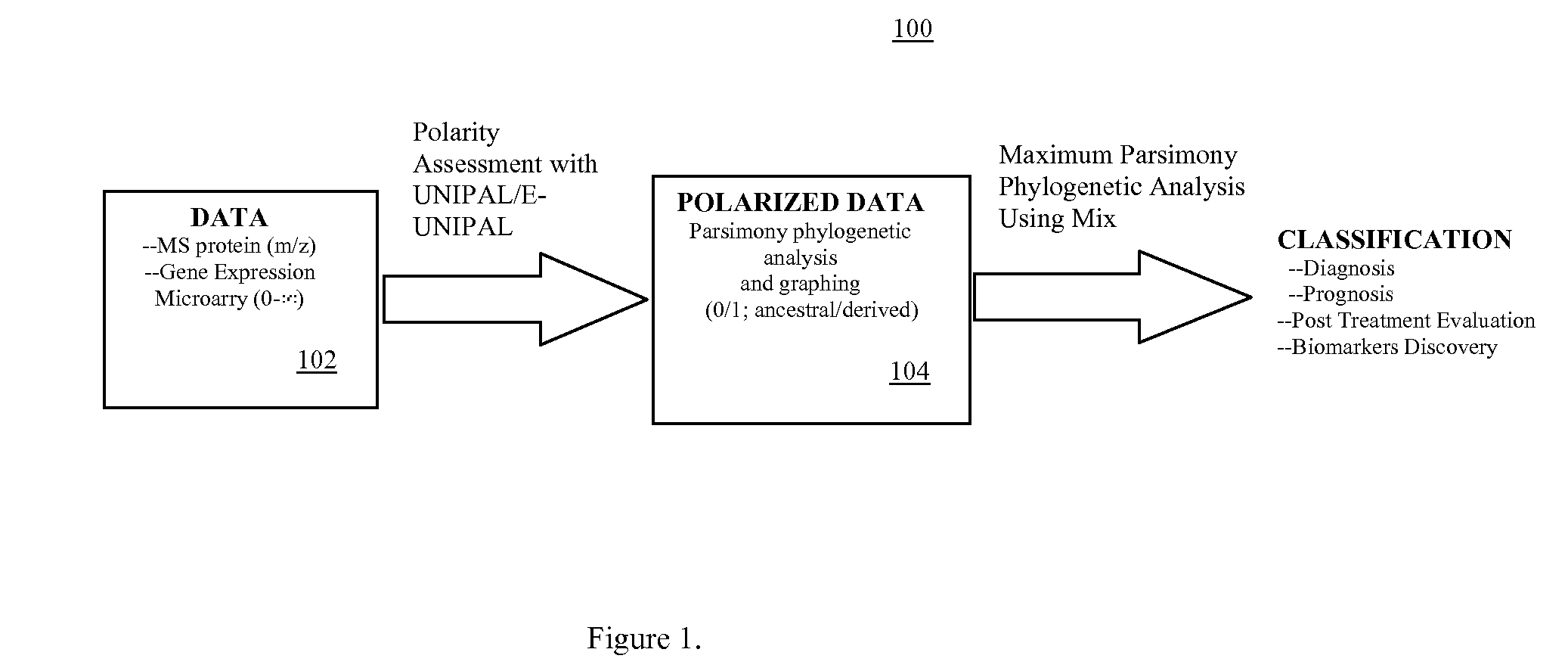
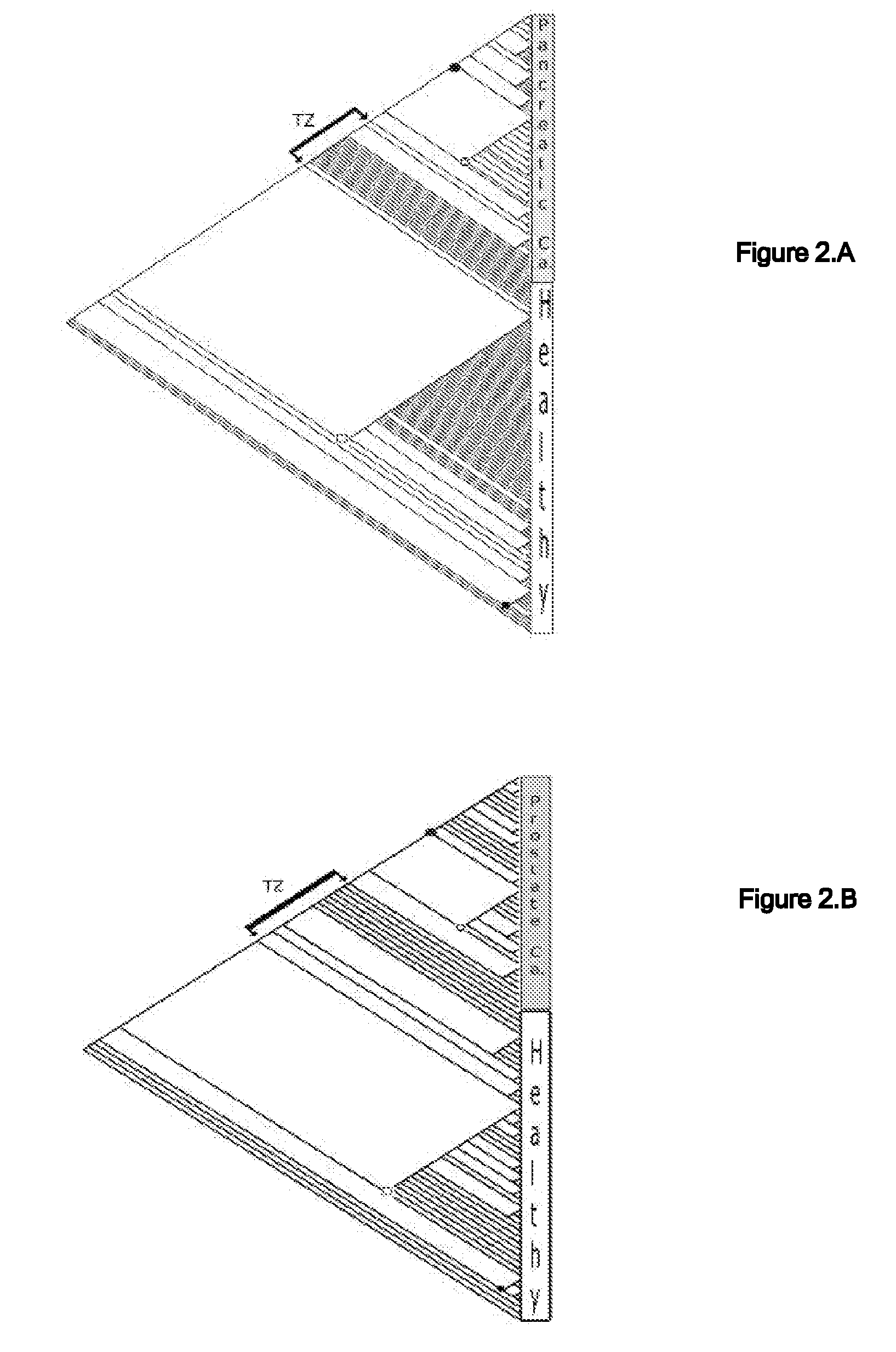
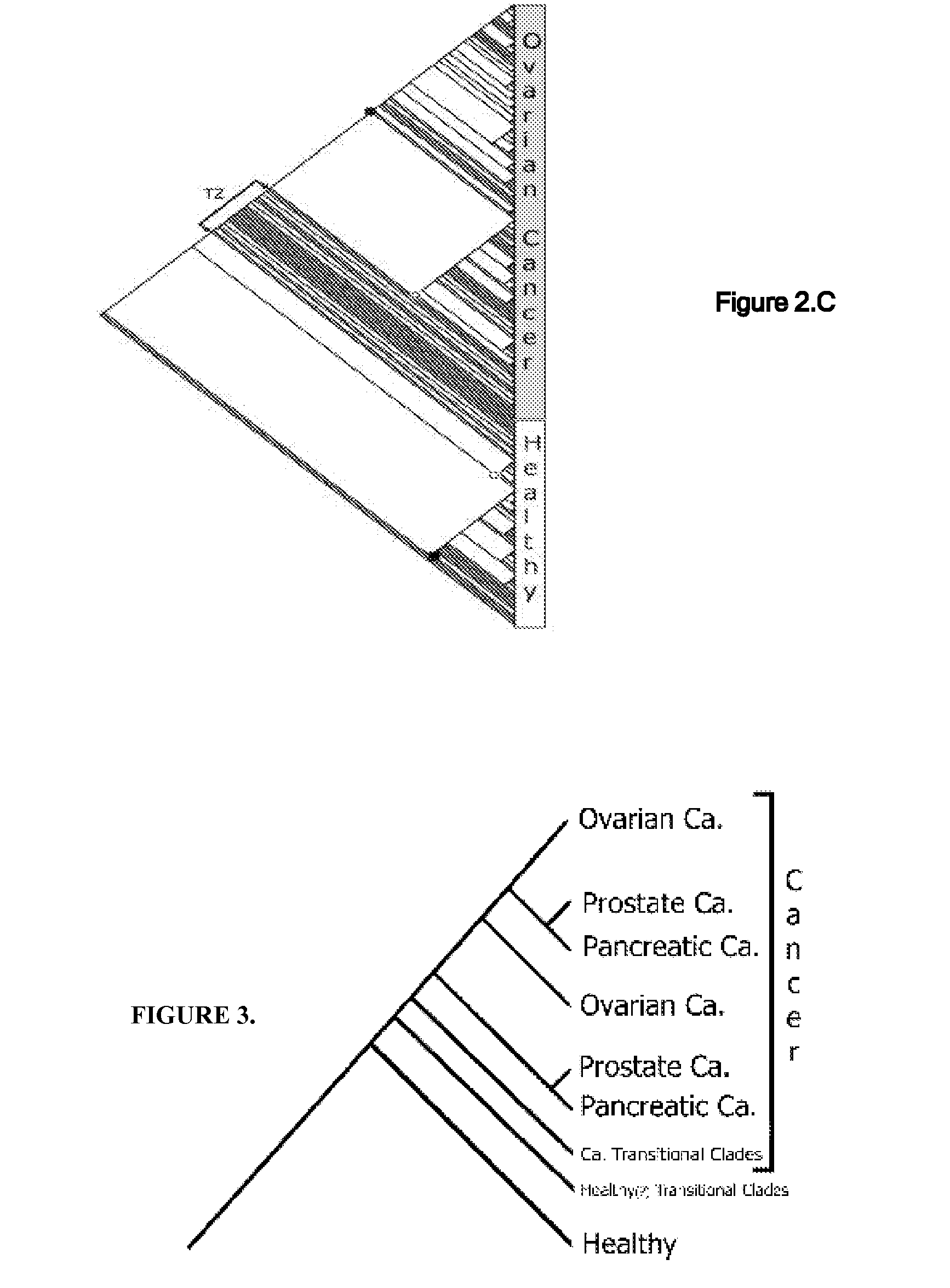
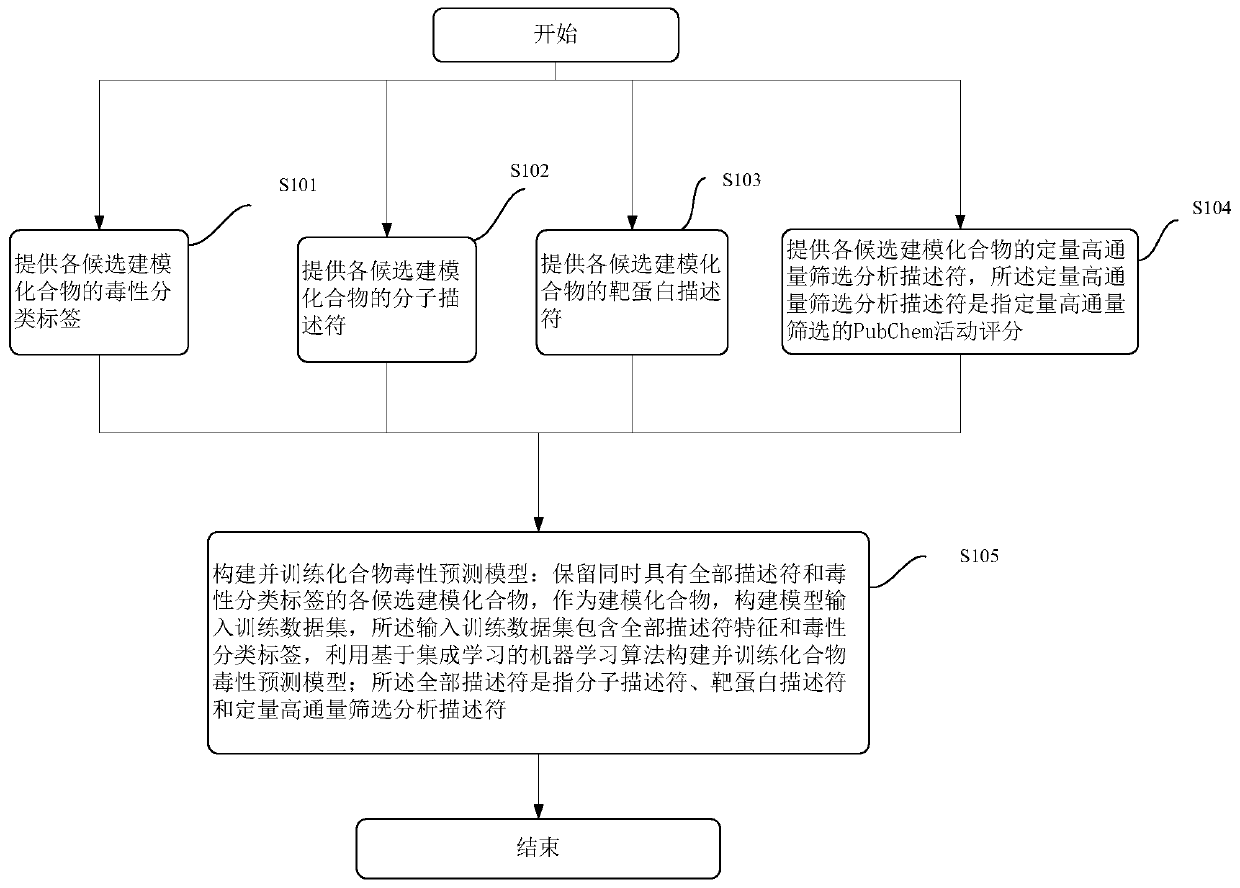
Phylogenetic Analysis of Mass Spectrometry or Gene Array Data for the Diagnosis of Physiological Conditions
ActiveUS20070259363A1Facilitates interplatform comparabilityMedical simulationMedical data miningDiseasePotential biomarkers
A universal data-mining platform capable of analyzing mass spectrometry (MS) serum proteomic profiles and / or gene array data to produce biologically meaningful classification; i.e., group together biologically related specimens into clades. This platform utilizes the principles of phylogenetics, such as parsimony, to reveal susceptibility to cancer development (or other physiological or pathophysiological conditions), diagnosis and typing of cancer, identifying stages of cancer, as well as post-treatment evaluation. To place specimens into their corresponding clade(s), the invention utilizes two algorithms: a new data-mining parsing algorithm, and a publicly available phylogenetic algorithm (MIX). By outgroup comparison (i.e., using a normal set as the standard reference), the parsing algorithm identifies under and / or overexpressed gene values or in the case of sera, (i) novel or (ii) vanished MS peaks, and peaks signifying (iii) up or (iv) down regulated proteins, and scores the variations as either derived (do not exit in the outgroup set) or ancestral (exist in the outgroup set); the derived is given a score of “1”, and the ancestral a score of “0”—these are called the polarized values. Furthermore, the shared derived characters that it identifies are potential biomarkers for cancers and other conditions and their subclasses.
Owner:AMRI HAKIMA +2
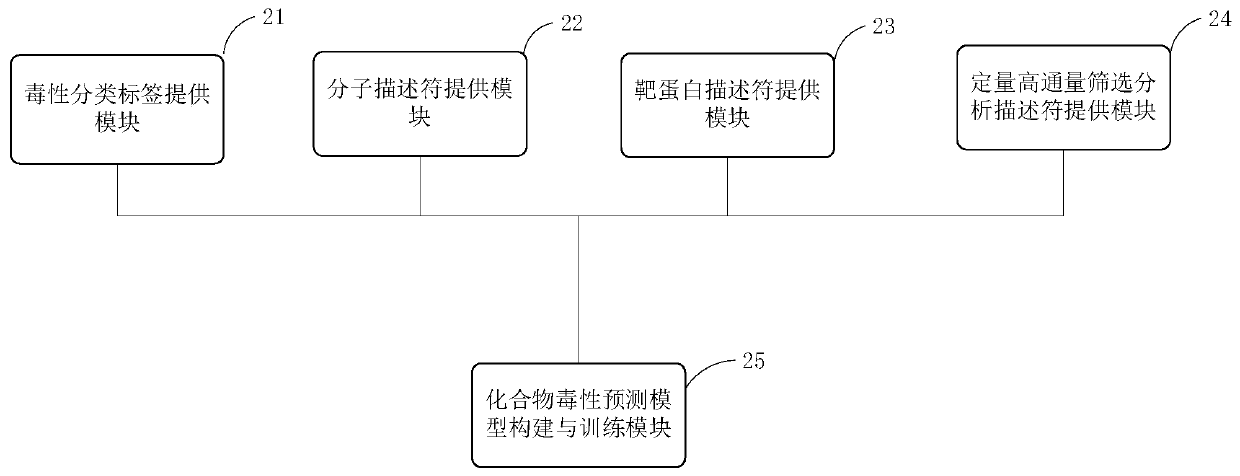
Modeling method and device of compound toxicity prediction model and application of compound toxicity prediction model
InactiveCN110890137AReliable Toxicity Prediction ResultsReduce false positive rateMolecular designSystems biologyProtein targetDrug biological activity
The invention provides a modeling method of a compound toxicity prediction model. The modeling method at least comprises the following steps of: S101, providing toxicity classification labels of candidate modeling compounds; S102, providing a molecular descriptor of each candidate modeling compound; S103, providing a target protein descriptor of each candidate modeling compound; S104, providing aquantitative high-throughput screening analysis descriptor of each candidate modeling compound, wherein the quantitative high-throughput screening analysis descriptor is a PubChem activity score of aspecified amount of high-throughput screening; and S105, constructing and training a compound toxicity prediction model. According to the method, physicochemical properties, biological activity and target protein action properties of drug candidate compounds can be fully utilized, and a drug toxicity prediction system is constructed by utilizing statistical modeling advantages of a machine learning algorithm based on ensemble learning, so that the model has interpretability and prediction performance, and has the better physicochemical and biological significance and research value.
Owner:上海尔云信息科技有限公司
Cellular arrays comprising encoded cells
InactiveUS20050158702A1Determine effectBioreactor/fermenter combinationsBiological substance pretreatmentsSensor arrayFiber
A biosensor, sensor array, sensing method and sensing apparatus are provided in which individual cells or randomly mixed populations of cells, having unique response characteristics to chemical and biological materials, are deployed in a plurality of discrete sites on a substrate. In a preferred embodiment, the discrete sites comprise microwells formed at the distal end of individual fibers within a fiber optic array. The biosensor array utilizes an optically interrogatable encoding scheme for determining the identity and location of each cell type in the array and provides for simultaneous measurements of large number of individual cell respnses to target analyses. The sensing method utilizes the unique ability of cell populations to respond to biologically significant compounds in a characteristic and detectable manner. The biosensor array and measurement method may be employed in the study of biologically active materials in situ environmental monitoring, monitoring of a variety of bioprocesses, and for high throughput screening of large combinatorial chemical libraries.
Owner:ILLUMINA INC
Methods of Identifying Microsatellite Instability
ActiveUS20190194731A1Improve the level ofMicrobiological testing/measurementTumor SampleWilms' tumor
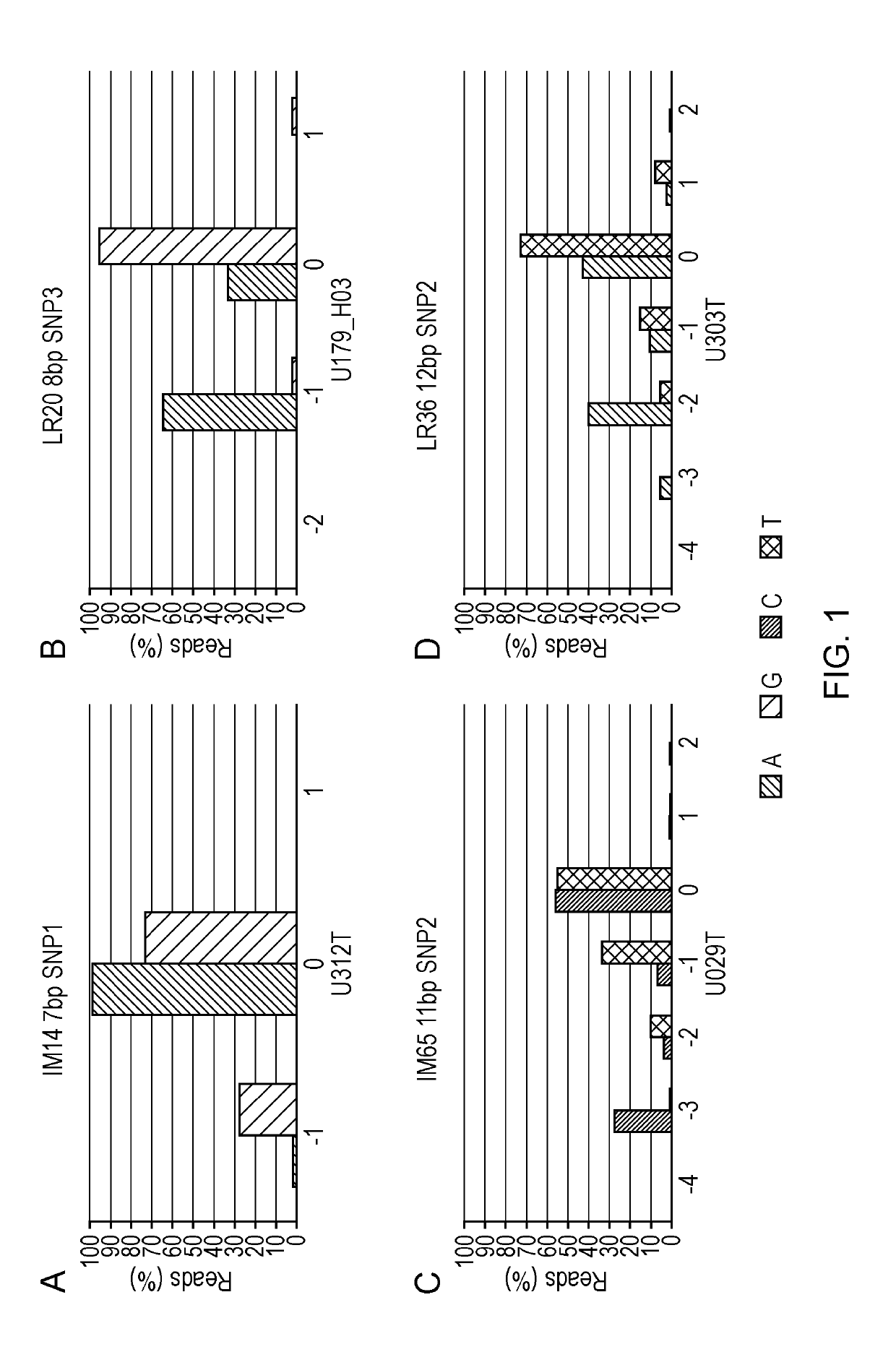
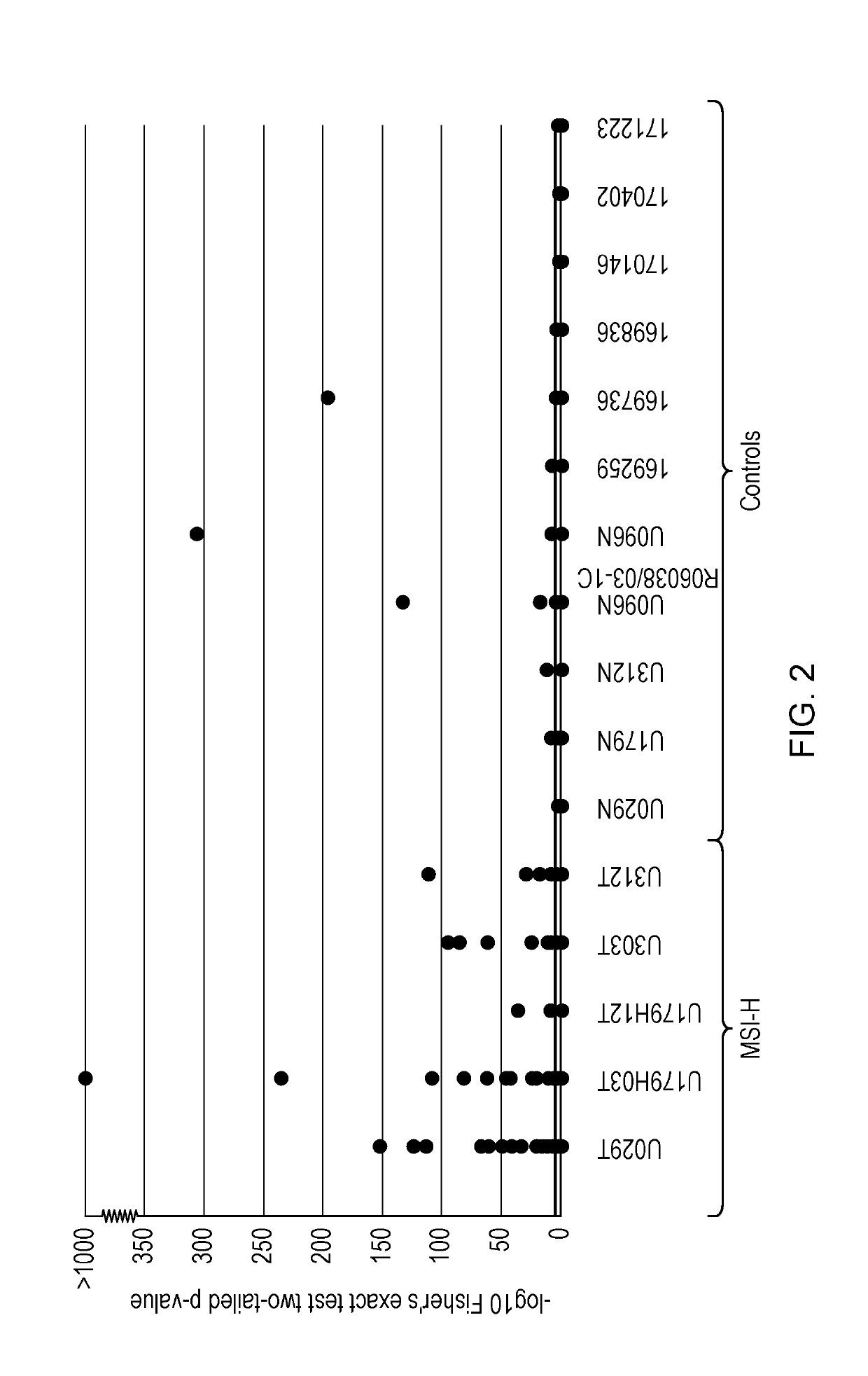
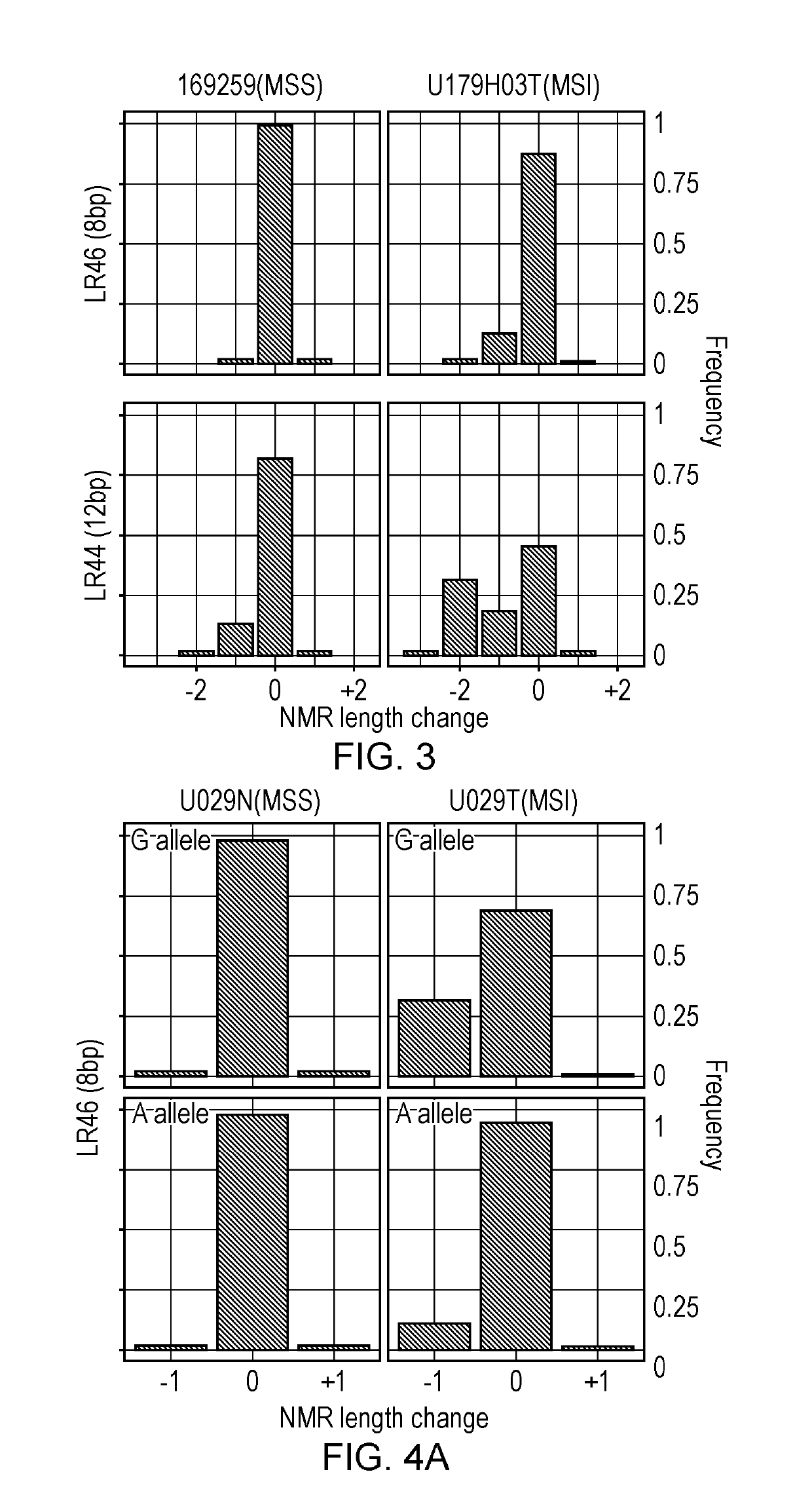
The present invention relates to methods and kits for identifying microsatellite instability (MSI) in a sample. In particular it relates to identifying microsatellite instability in a tumor sample, which may be from a subject suspected of having colorectal cancer or Lynch syndrome. The methods and kits can be used to identify mismatch repair defects. More particularly the invention relates to a panel of markers for a sequencing based MSI test, that can differentiate between MSI-H and MSS CRCs. The invention also allows for determination of biological significance, differentiating between PCR and sequencing errors and MSI induced indels / mutations.
Owner:CANCER RES TECH LTD
Protein composite identifying method based on graph model
The invention provides a protein composite identifying method based on a graph model; a protein interactive network of a given species is regarded as a network drawing =(V, E), wherein V is combining point of protein, and E is set of protein-protein interaction side; the self-connecting side and the repeat side in a network are removed from the set of all sides; the method includes steps of firstly acquiring a nuclear protein vertex set of the protein composite, and then expanding a first-order neighbor of the edge combining point thereof, and forming a graph model; judging its connectivity according to characteristics of the graph model, and finding out all dense subgraphs, namely, protein composite. The method provided by the invention can regard the graph model as the nucleus of the protein composite; through investigating and expanding the first-order neighbor combining point of the graph model, the protein composite is identified; an algorithm provided by the invention is applied to the known yeast protein network; the experimental result indicates that the algorithm can identify multiple protein composites with biological significance, and the algorithm is not sensitive to the input parameter.
Owner:SHANGHAI DIANJI UNIV
Method for distant hybridization between subfamilies of red crucian carps and xenocypris davidi bleekers
ActiveCN101669448ALess internal organsMiscellaneousClimate change adaptationPisciculture and aquariaSubfamilyCarp
The invention discloses a method for the distant hybridization between subfamilies of red crucian carps and xenocypri davidi bleekers, comprising the following steps: firstly, selecting red crucian carps and xenocypri davidi bleekers with maturated gonads and good body features to be respectively used as female parent fishes and male parent fishes for hybridization, and carrying out artificial induced spawning on the female and male parent fishes in a breeding season; secondly, selecting the male and the female parent fishes with good effect of induced spawning for carrying out artificial dry-method insemination, and carrying out automatic hatching in running water after the insemination is finished; thirdly, breeding hatched fries, and detecting and screening the bred fries to obtain redcrucian carp and xenocypri davidi bleeker hybridized triploid fishes, red crucian carp and xenocypri davidi bleeker hybridized tetraploid fishes and natural gynogenesis red crucian carps. In the invention, the characteristics of the bred hybridized generation are improved, and the insemination rate and the hatching rate of distant hybridization are remarkably improved; in addition, the method is anew way for generating the tetraploid fishes and the natural gynogenesis red crucian carps, and has important biological significance in the aspects of the biological evolution and the genetic breeding of fishes.
Owner:HUNAN NORMAL UNIVERSITY
Method for distant hybridization of cyprinuscarpiohaematopterus and megalobramaamblycephala subfamilies
ActiveCN106550909ASolve the problem of distant hybridizationImprove hatchabilityClimate change adaptationPisciculture and aquariaBroodstockCommon carp
The invention discloses a method for distant hybridization of cyprinuscarpiohaematopterus and megalobramaamblycephala subfamilies. The method comprises the following steps: taking cyprinuscarpiohaematopterus as female parent fish and megalobramaamblycephala as male parent fish, artificially inducing spawning of the female parent fish and the male parent fish in a breeding season, carrying out dry artificial insemination, incubating obtained fertilized roe in running water in an incubation tank to obtain hybrids of the cyprinuscarpiohaematopterus and the megalobramaamblycephala, and testing and screening the hybrids to obtain diploid cyprinuscarpiohaematopterus, diploid common-carp and crucian-carp hybrids and diploid crucian carp. The method for distant hybridization of the cyprinuscarpiohaematopterus and megalobramaamblycephala subfamilies integrates the excellent characters of the cyprinuscarpiohaematopterus and the megalobramaamblycephala, enriches the fish species on the market, lays a favorable foundation for selection and breeding of excellent species, provides precious resources for follow-up selection and breeding of novel polyploid fish and is of great biological significance in biological evolution and genetic breeding of fish.
Owner:湖南岳麓山水产育种科技有限公司
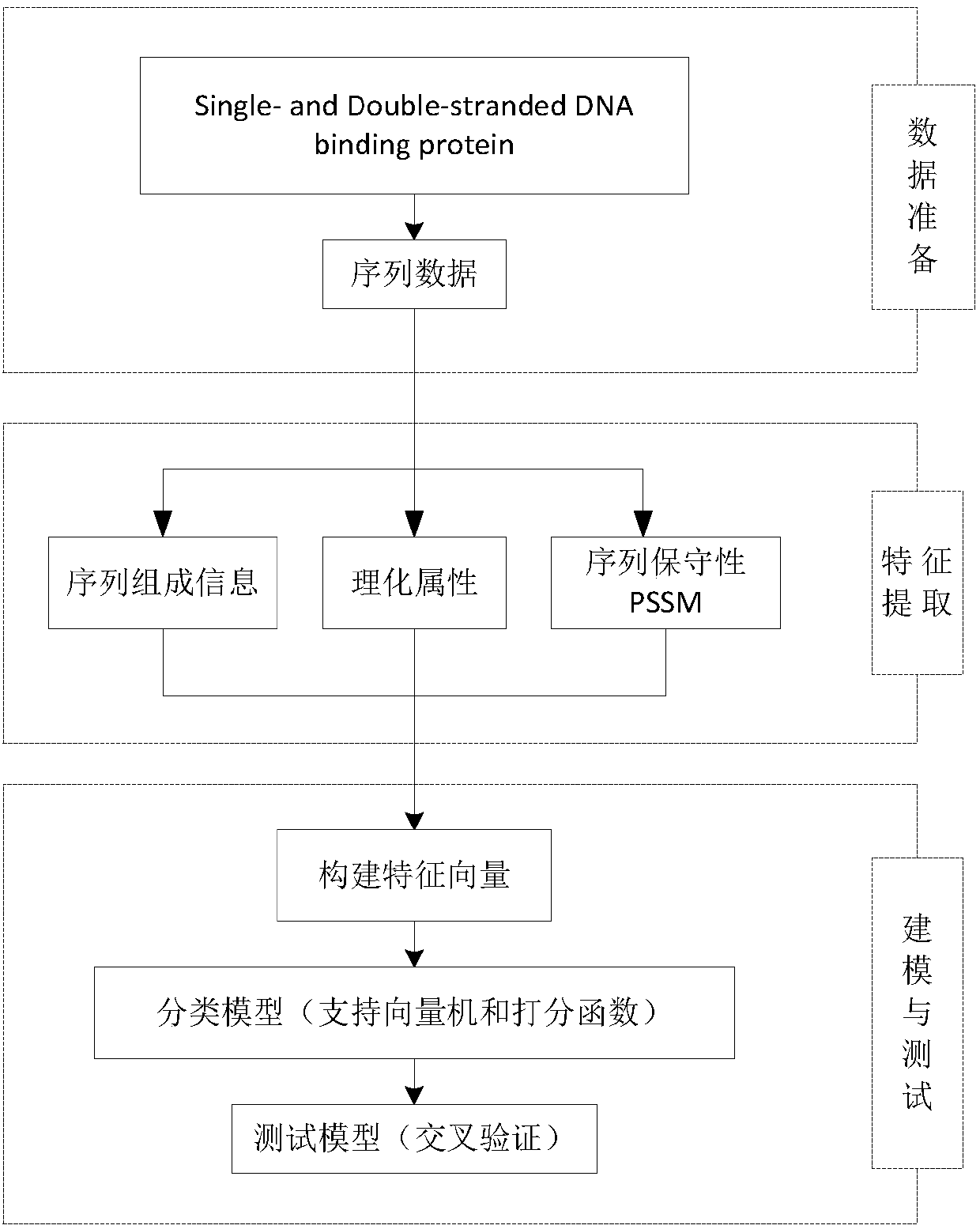
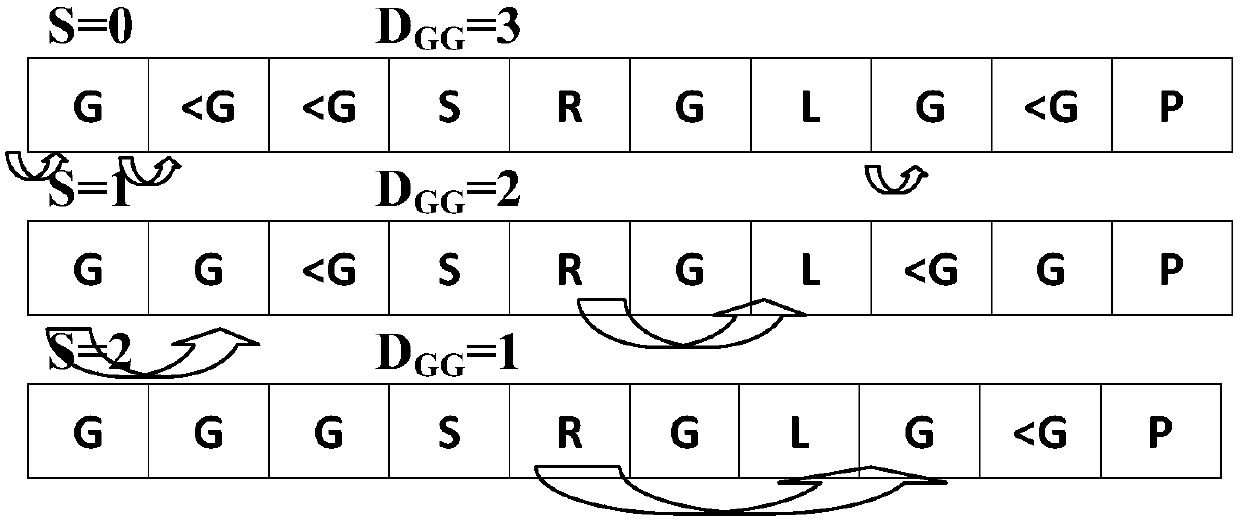
Characteristic extraction and classification method and device for DNA (Deoxyribo-Nucleic Acid) binding protein sequence information
InactiveCN108875310AImplement function annotationsAchieve classification goalsSpecial data processing applicationsData setProtein insertion
The invention relates to a characteristic extraction and classification method and device for DNA (Deoxyribo-Nucleic Acid) binding protein sequence information. The method comprises the following steps that: firstly, carrying out theory argumentation, and analyzing and arranging collected data to obtain a reliable dataset with a biological meaning and a statistical meaning; then, extracting effective protein sequence data characteristic parameters from a complex protein three-dimensional structure as a key link which is a way of converting sequence character information into digital characteristic information, designing a reasonable classification algorithm for extracted characteristic data, and screening the characteristics favorable for classification for realizing target classification;and finally, adopting a reasonable and fair evaluation system, including testing methods, checking means, evaluation index selection and the like, for classification performance. By use of the method, requirements on high-flux protein sequencing function annotation can be met, automated DNA binding protein sequence function annotation can be realized, and meanwhile, the characteristics which areput forward can assist biologists in carrying out experimental analysis and research on the DNA binding protein sequence.
Owner:HENAN NORMAL UNIV
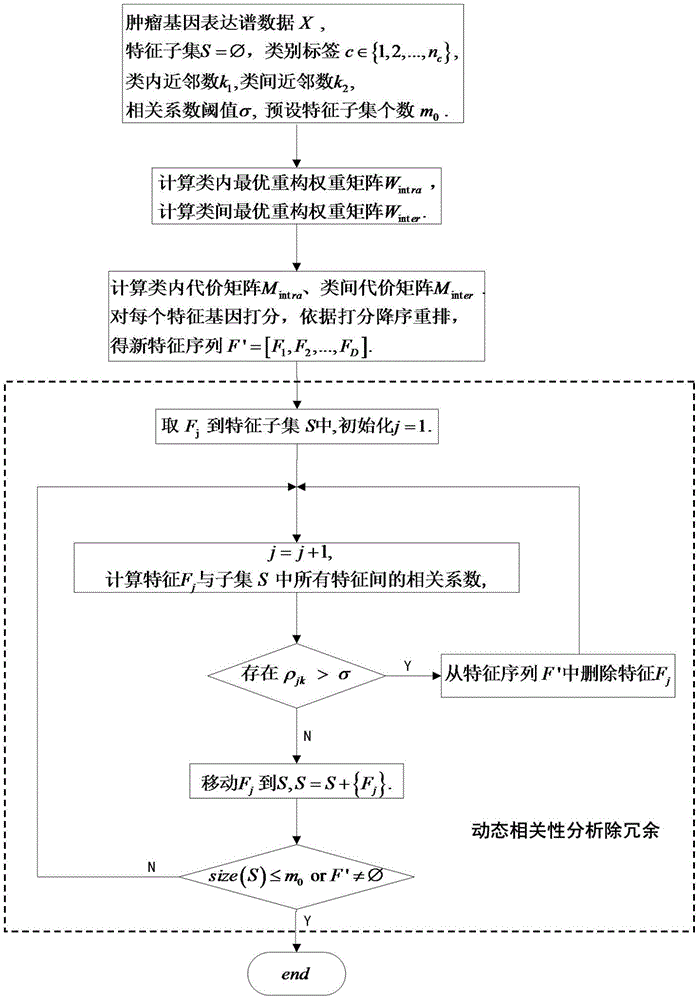
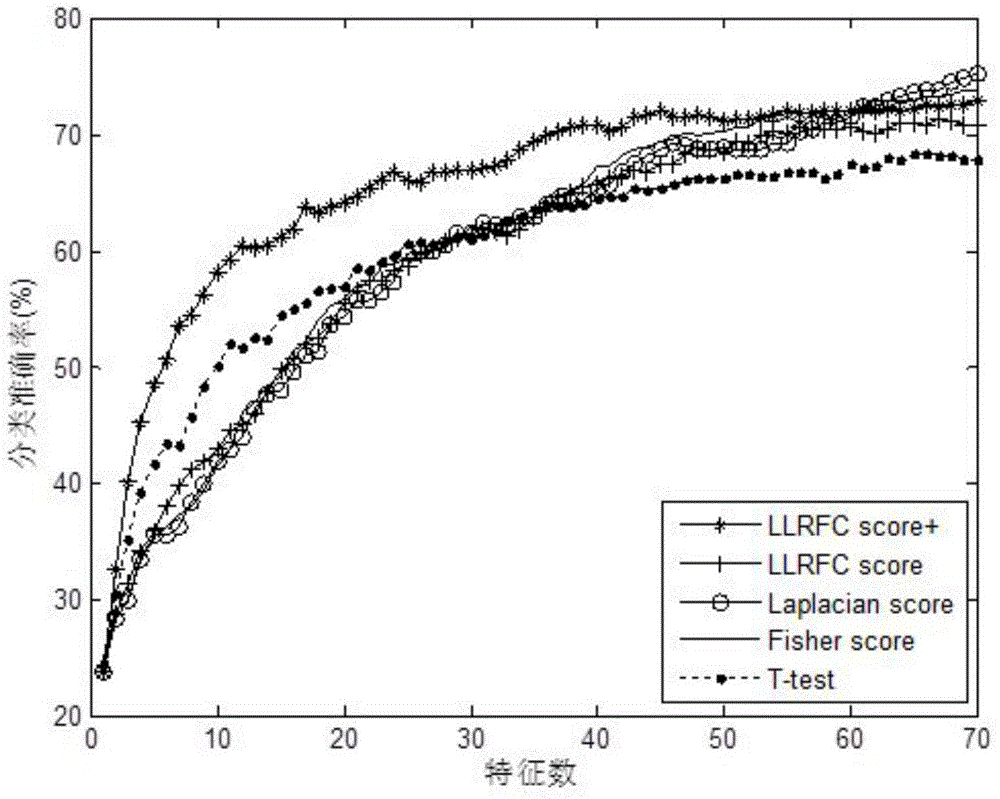
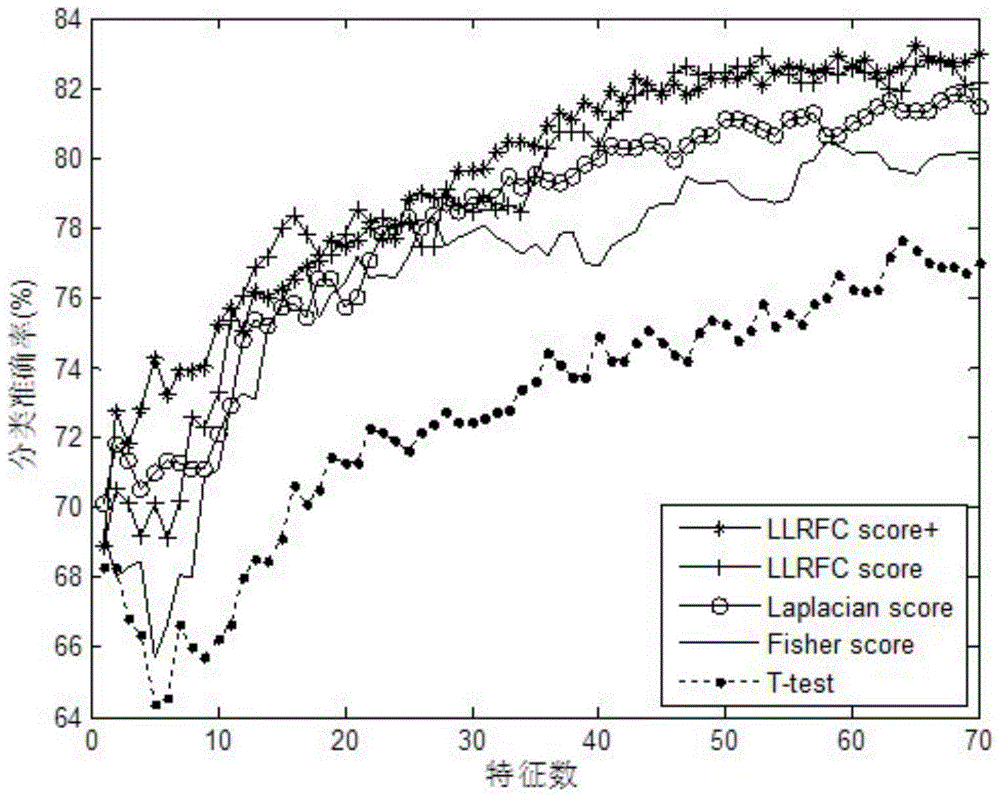
Redundancy removal feature selection method LLRFC score+ based on LLRFC and correlation analysis
InactiveCN105740653AOptimal trait gene subsetImprove classification accuracyBiostatisticsSpecial data processing applicationsDiseaseData set
The invention provides a redundancy removal feature selection method LLRFC (Locally Linear Representation Fisher Criterion) score+ based on LLRFC and correlation analysis. A DNA (Deoxyribonucleic Acid) microarray technology provides a new direction for clinic tumor diagnosis. Performance of gene expression data corresponding to different kinds of tumor is different; through the analysis on the tumor gene expression data, study personnel can realize the accurate recognition on the tumor and the tumor subtype in the molecular level; and important biological significance is realized on the diagnosis and the treatment of the tumor. The feature genes in LLRFC judging criterion descending sort gene expression data is used to be combined with the dynamic correlation analysis strategy for further eliminating redundant features; an LLRFC score+ algorithm is provided; and the optimum feature gene subset is selected. The feature selection method LLRFC score+ has the advantages that the classification precision of a classifier can be effectively improved; a sample data set does not need to meet the normal distribution; and the method is applicable to data in various distribution types. The feature selection method LLRFC score+ can help people to find the virulence gene of cancer, and the early-stage diagnosis, tumor staging and typing, prognosis treatment and the like of clinic tumor diseases are facilitated.
Owner:BEIJING UNIV OF TECH
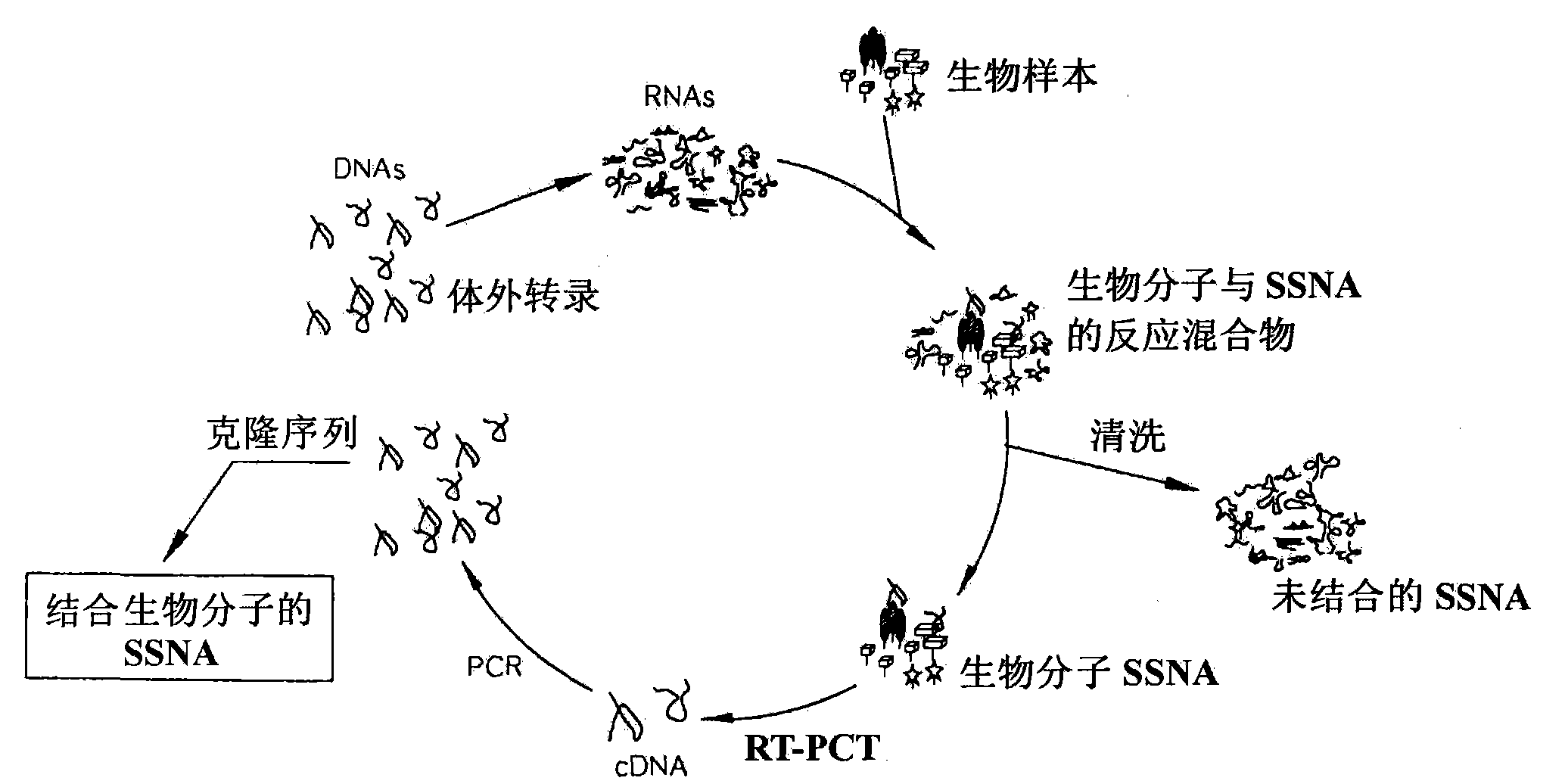
Nucleic acid chip for obtaining bind profile of single strand nucleic acid and unknown biomolecule, manufacturing method thereof and analysis method of unknown biomolecule using nucleic acid chip
InactiveCN101663406ALow costEasy to analyzeMicrobiological testing/measurementBiological testingNucleic acid detectionSingle strand
Disclosed are a nucleic acid chip for obtaining binding profiles between unknown biomolecules and single-stranded nucleic acids, a method for manufacturing the chip, and a method for analyzing the unknown biomolecules using the chip. The nucleic acid chip is used to analyze biological significance of the unknown biomolecule in the biospecimen. The nucleic acid chip can be manufactured by reactinga biospecimen containing an unknown biomolecule with random single-stranded nucleic acids having random base sequences to determine biomolecule-binding single stranded nucleic acids capable of bindingthe unknown biomolecule; and synthesizing capture single stranded nucleic acids composed of the determined biomolecule-binding single stranded nucleic acids and / or single stranded nucleic acids having base sequences complementary to those of said determined biomolecule-binding single stranded nucleic acids and affixing the capture single stranded nucleic acids on a substrate.
Owner:GENOPROT
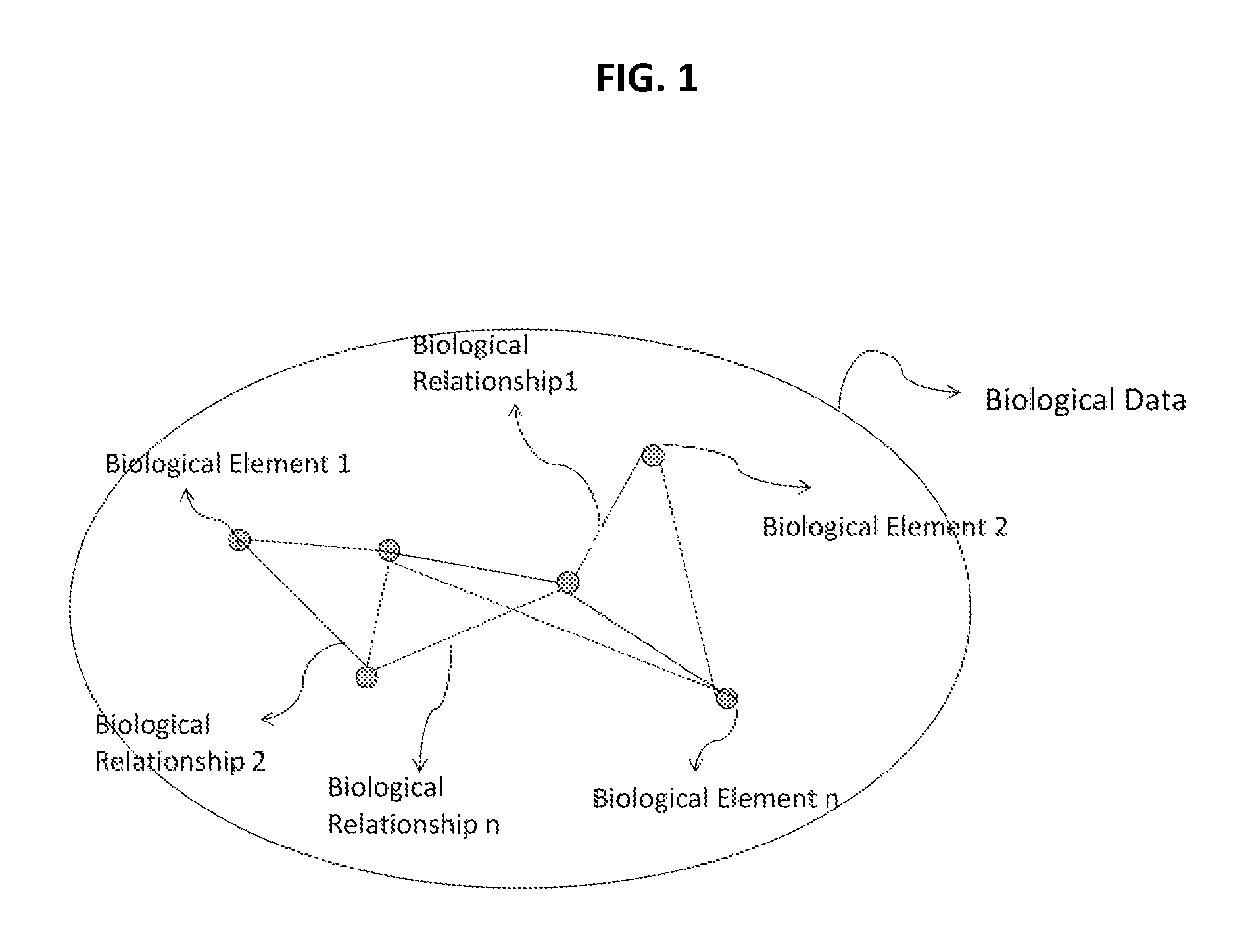
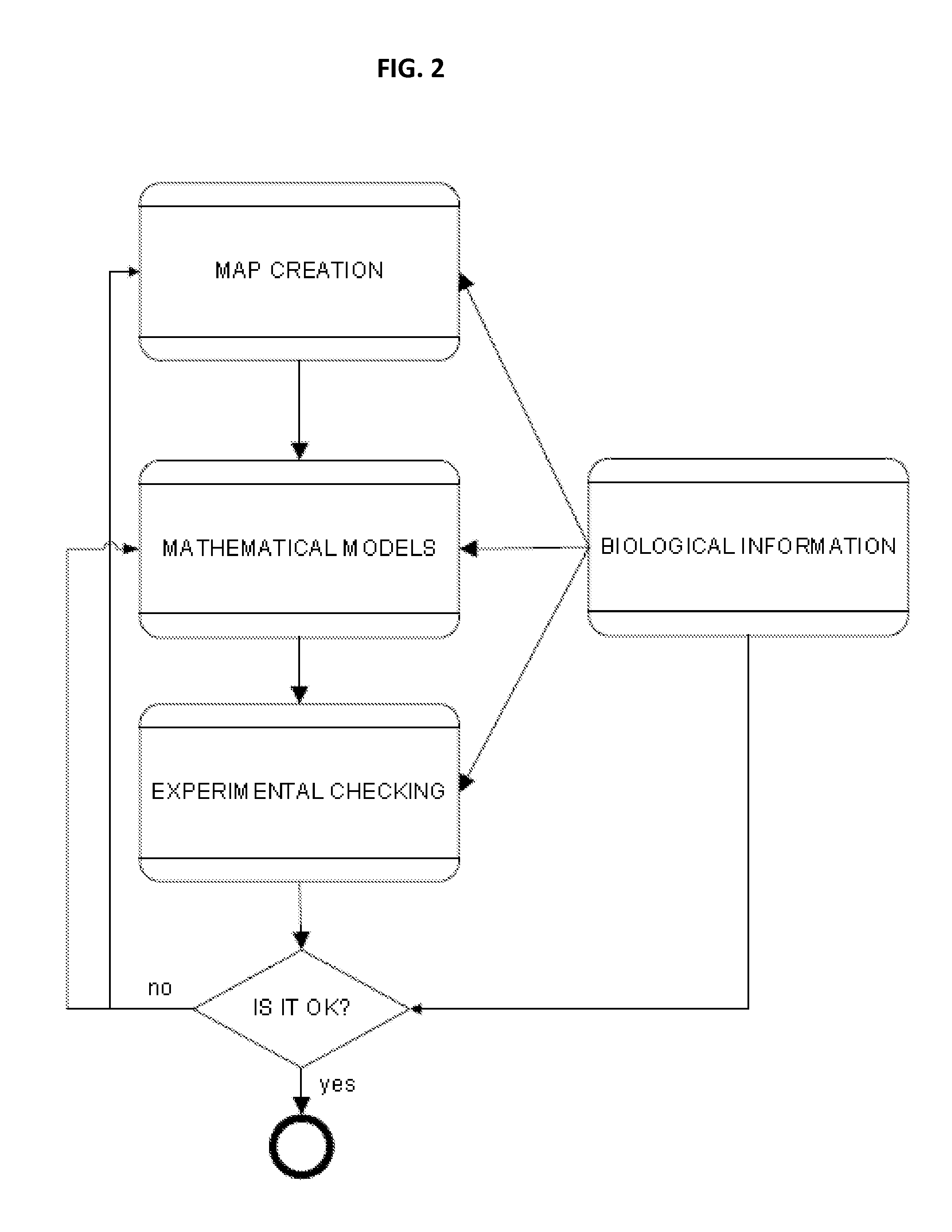
Methods and systems for identifying molecules or processes of biological interest by using knowledge discovery in biological data
InactiveUS20110098993A1Improve practicalityReduced activityAnalogue computers for chemical processesBiostatisticsMetaboliteAssociated organism
The present application relates to methods and systems of identifying molecules or processes of biological interest by using knowledge discovery in biological data. In particular, the present application describes new methods of creating a biological map, new methods of codifying such map, new methods of analyzing such map and new methods of identifying molecules and processes of biological interest. The present application provides methods and systems to identify new and useful direct or indirect therapeutic targets, molecular modulators, adverse events effectors, disease biomarkers, genetic biomarkers, safety-related biomarkers, diagnostic molecules, hormones, metabolites, or metabolic effectors of any type.
Owner:ANAXOMICS BIOTECH SL
Method for selecting natural secondary forest intermediate cutting object woods
InactiveCN103593563AOvercome the Difficulty of Selecting the Target Trees for Tendance and ThinningReasonable parenting planSpecial data processing applicationsSecondary forestHealth index
The invention discloses a method for selecting secondary forest intermediate cutting object woods. The method includes the steps of in cooperation with secondary forest characteristics and operation targets, selecting quantitative indexes, such as the freedom degree, the mixture degree, the health indexes, the purpose tree species characteristic indexes, the space density indexes and the open odds, which influence the probability that the woods in a secondary forest are determined as intermediate cutting objects, solving the weights of the six quantitative indexes with an analytic hierarchy process, analyzing correlations between the indexes which serve as independent variables and the intermediate cutting indexes which serve as dependent variables according to biological significances of the indexes for building the ICIi of the woods, and enabling the ICIi of the woods to be sort in a descending mode to determine the prior intermediate cutting object woods. The method is convenient to use, easy to operate and high in efficiency, and the accuracy can meet the requirement.
Owner:CENTRAL SOUTH UNIVERSITY OF FORESTRY AND TECHNOLOGY
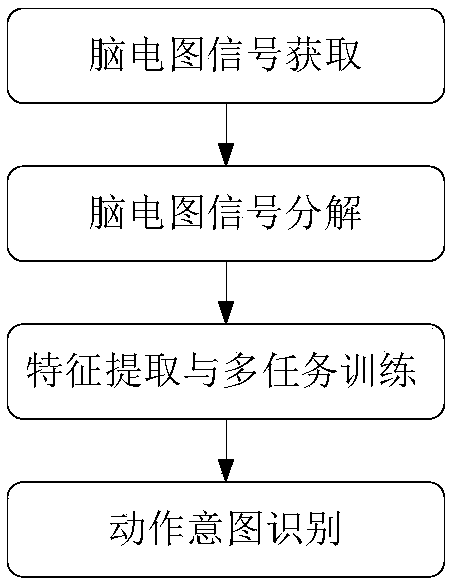
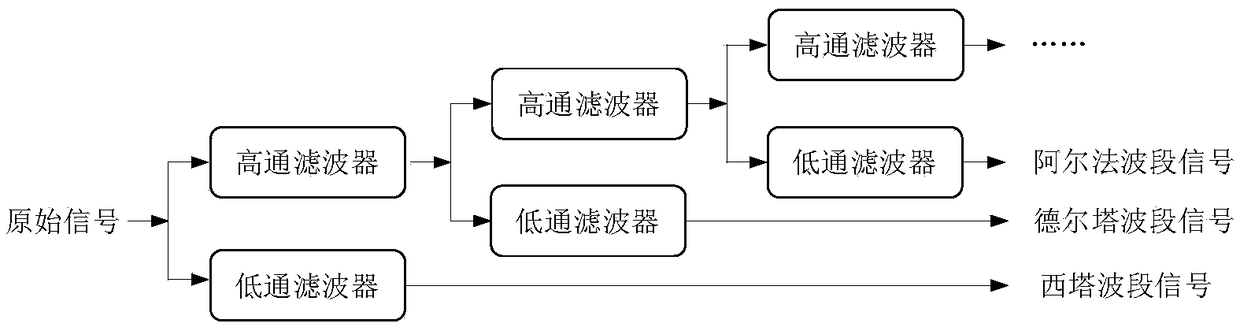
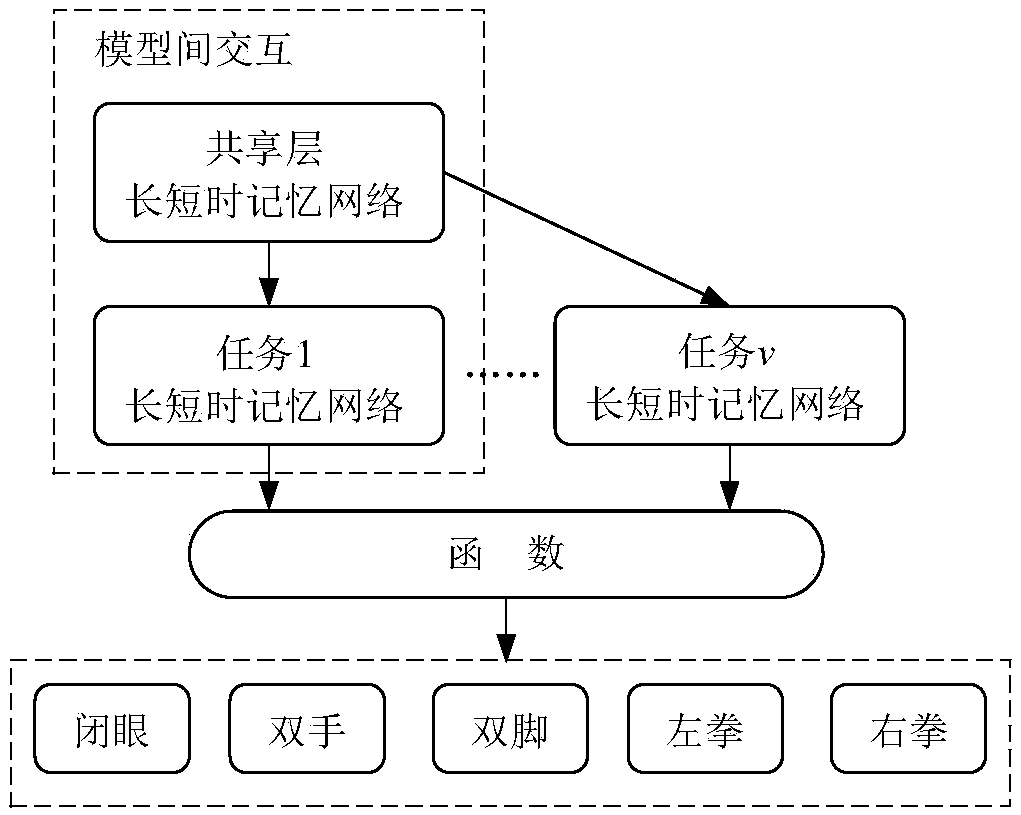
Motion intention recognition method of EEG signals based on multitask RNN model
ActiveCN109375776AImprove relevanceImproved binaryInput/output for user-computer interactionCharacter and pattern recognitionPattern recognitionData set
Multi-task RNN model based EEG signal action intention recognitioin method is disclosed. The invention relates to a method for identifying the action intention of EEG signals by a multi-task recurrentneural network, and solves the defect that the prior art cannot well process the signal noise and the time sequence information. The invention provides a novel framework for learning the unique characteristics from the EEG signals by applying the multi-task recurrent neural network. The correlation between different EEG frequency bands and biological significance is improved by learning the separated signals used for human motion intention recognition. Temporal correlation between different channels is also exploited. In this way, the identification of binary and multivariate intentions is improved. The invention performs extensive experiments on publicly available EEG signal reference datasets and compares our method with many of the most advanced algorithms. The experimental results show that the method proposed by the invention surpasses all comparison methods and achieves an accuracy rate of 97.8%.
Owner:NORTHEAST NORMAL UNIVERSITY
Single protein production in living cells facilitated by a messenger RNA interferase
InactiveUS20100035346A1Inhibit protein synthesisInhibit synthesisBacteriaHydrolasesEndoribonucleaseSingle-Stranded RNA
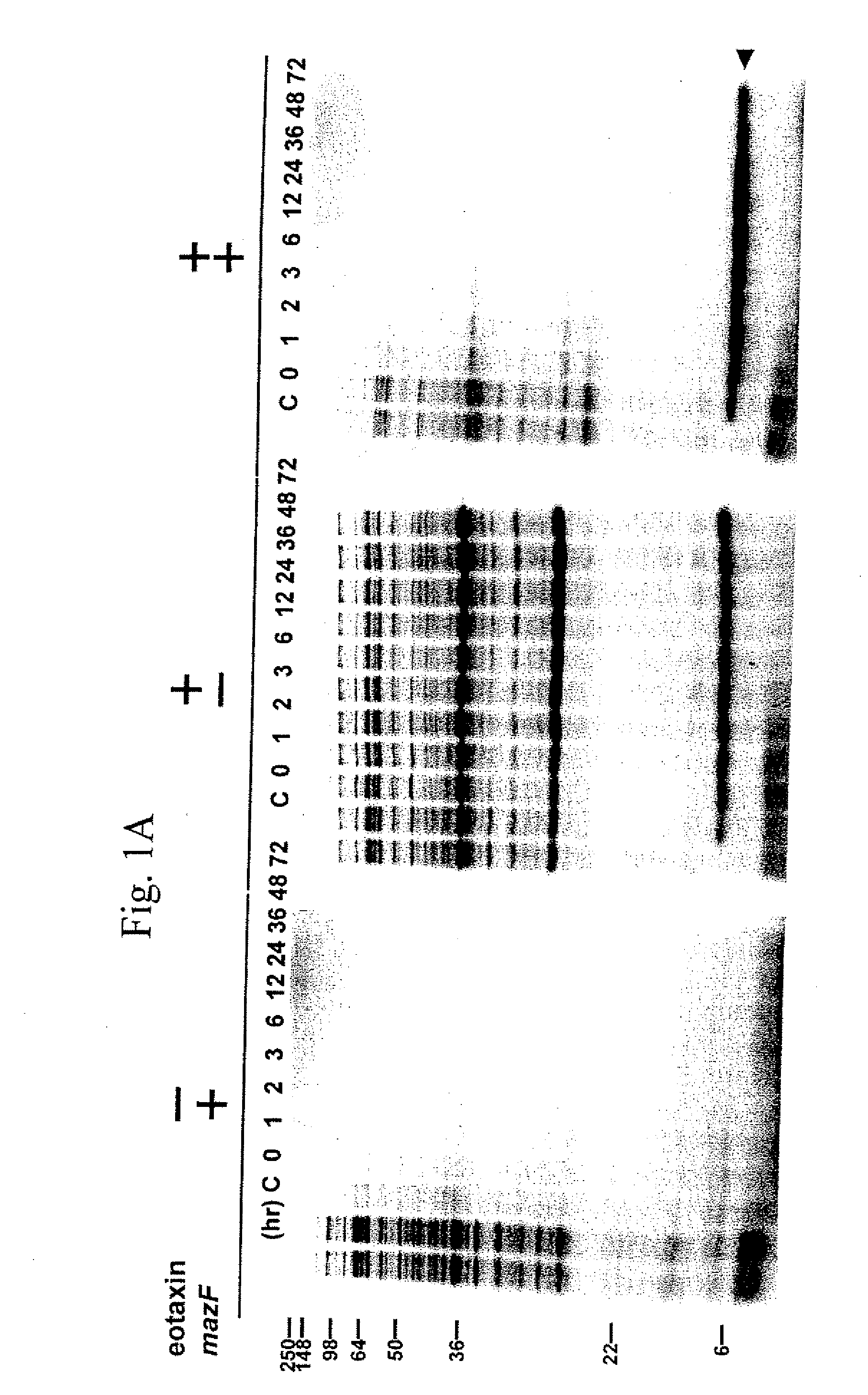
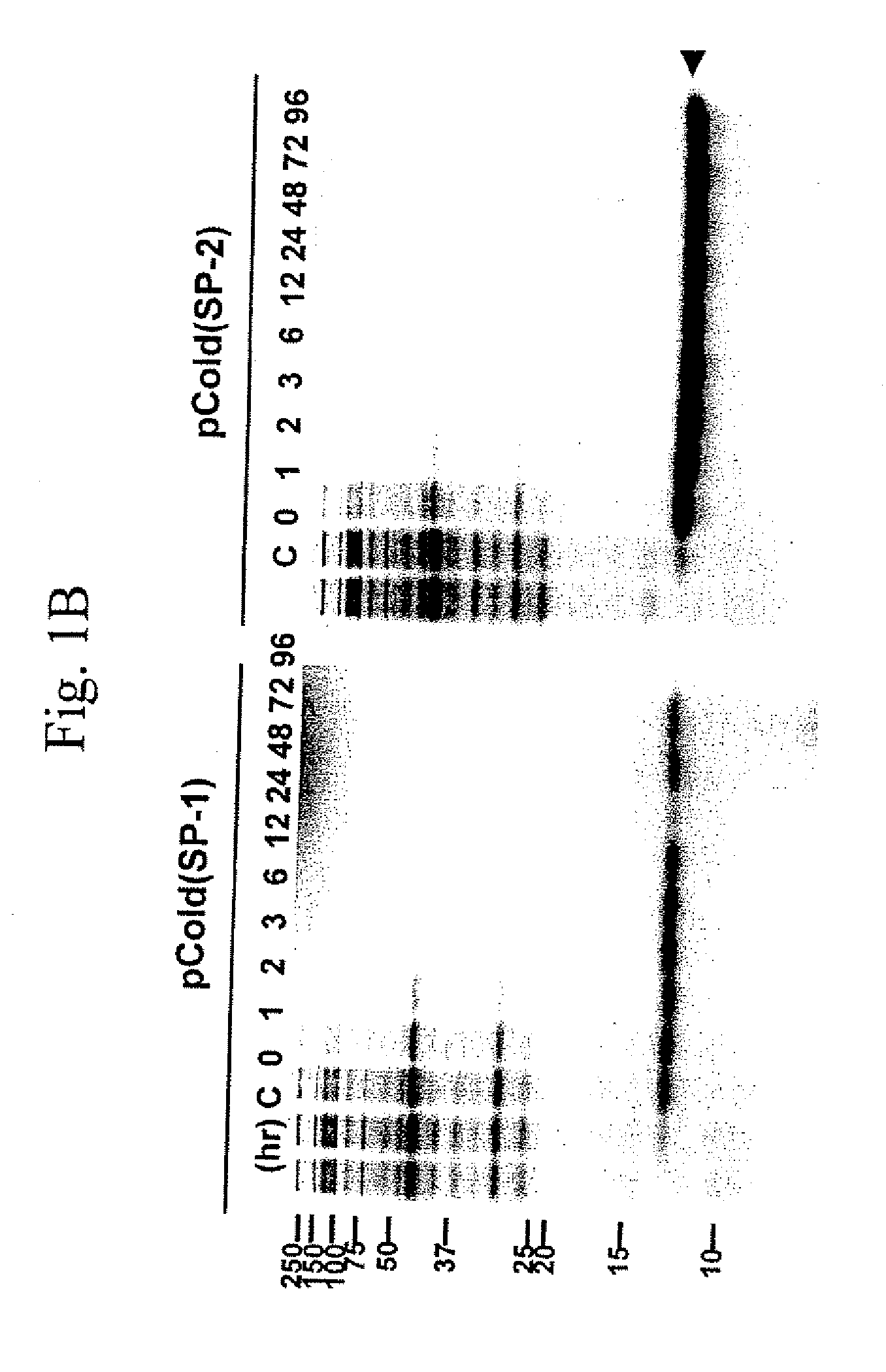
The present invention describes a single-protein production (SPP) system in living E. coli cells that exploits the unique properties of an mRNA interferase, for example, MazF, a bacterial toxin that is a single stranded RNA- and ACA-specific endoribonuclease, which efficiently and selectively degrades all cellular mRNAs in vivo, resulting in a precipitous drop in total protein synthesis. Concomitant expression of MazF and a target gene engineered to encode an ACA-less mRNA results in sustained and high-level (up to 90%) target expression in the virtual absence of background cellular protein synthesis. Remarkably, target synthesis continues for at least 4 days, indicating that cells retain transcriptional and translational competence despite their growth arrest. SPP technology works well for yeast and human proteins, even a bacterial integral membrane protein. This novel system enables unparalleled signal to noise ratios that should dramatically simplify structural and functional studies of previously intractable but biologically important proteins. The present invention also provides an optimized condensed single protein production system.
Owner:RUTGERS THE STATE UNIV
Comparative cancer survival models to assist physicians to choose optimal treatment
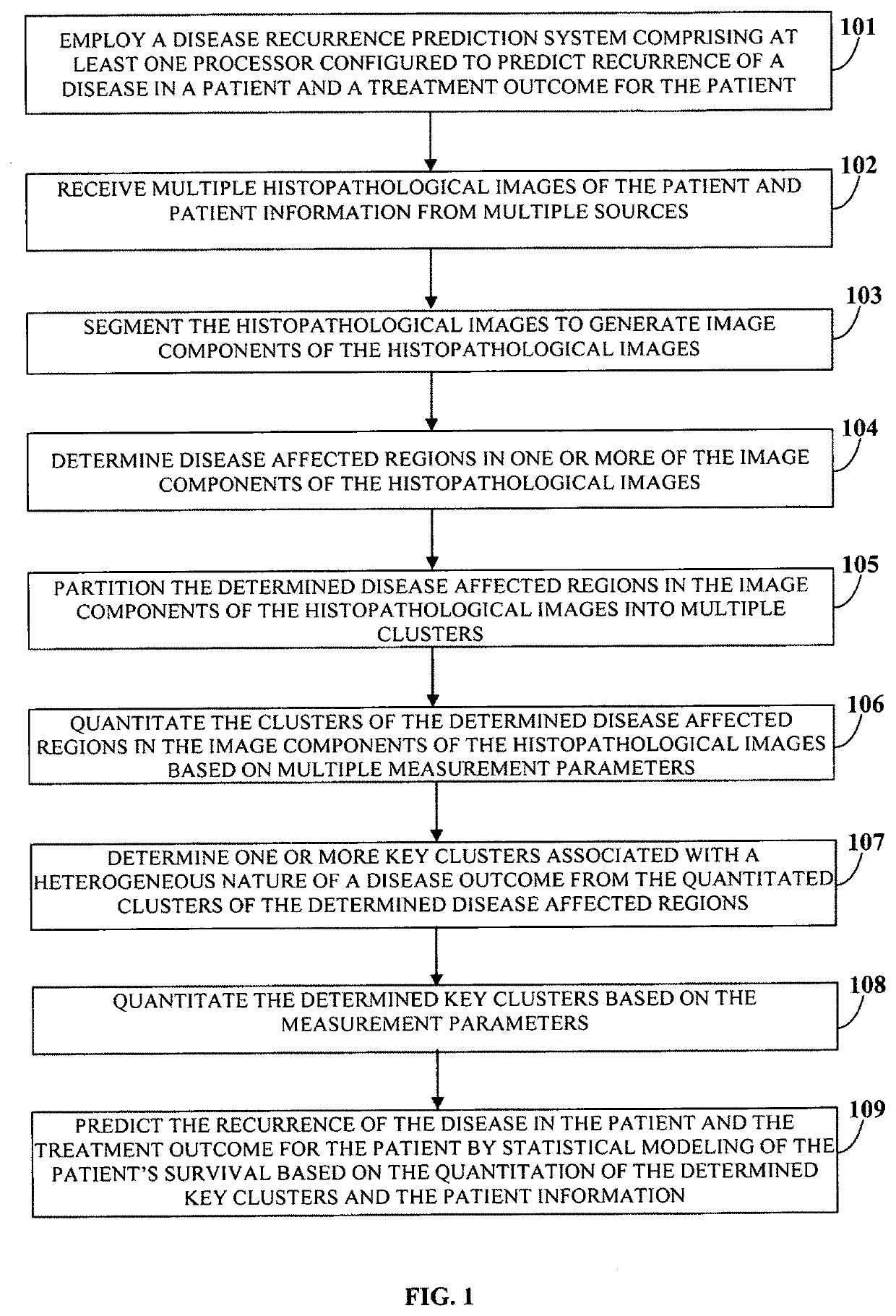
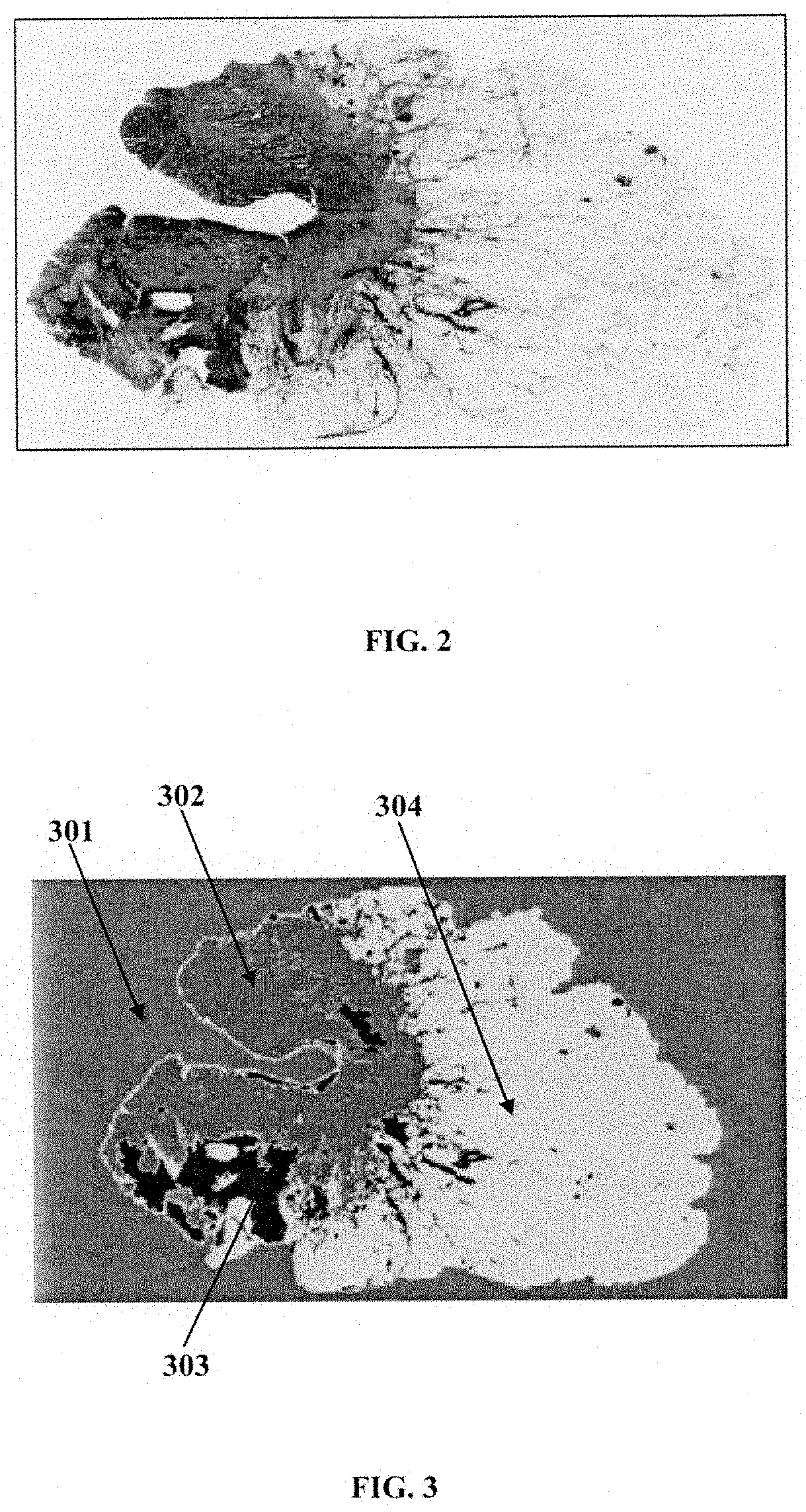
A computer implemented method and a system choosing optimal disease treatment among several possible treatment options for a patient are provided. The system computes cancer-free survival rates for each considered treatment based on predicting recurrence rate of a disease and / or cancer outcome for a particular patient. The treatment survival models use quantitative data from histopathological images of the patient, clinical data and other patient information. The system segments the histopathological images into biologically meaningful components; automatically determines disease-affected regions in one or more of the segmented image components. The system also partitions the disease-affected regions in each image into a number clusters. Those that are determined to be the most associated with the disease outcome are used as a source of the imaging information for the survival modeling. Optimal treatment is suggested as the treatment with probability of the cancer free survival within a certain time period is maximized.
Owner:TEVEROVSKIY MIKHAIL
Method of screening multiple sclerosis biomarkers
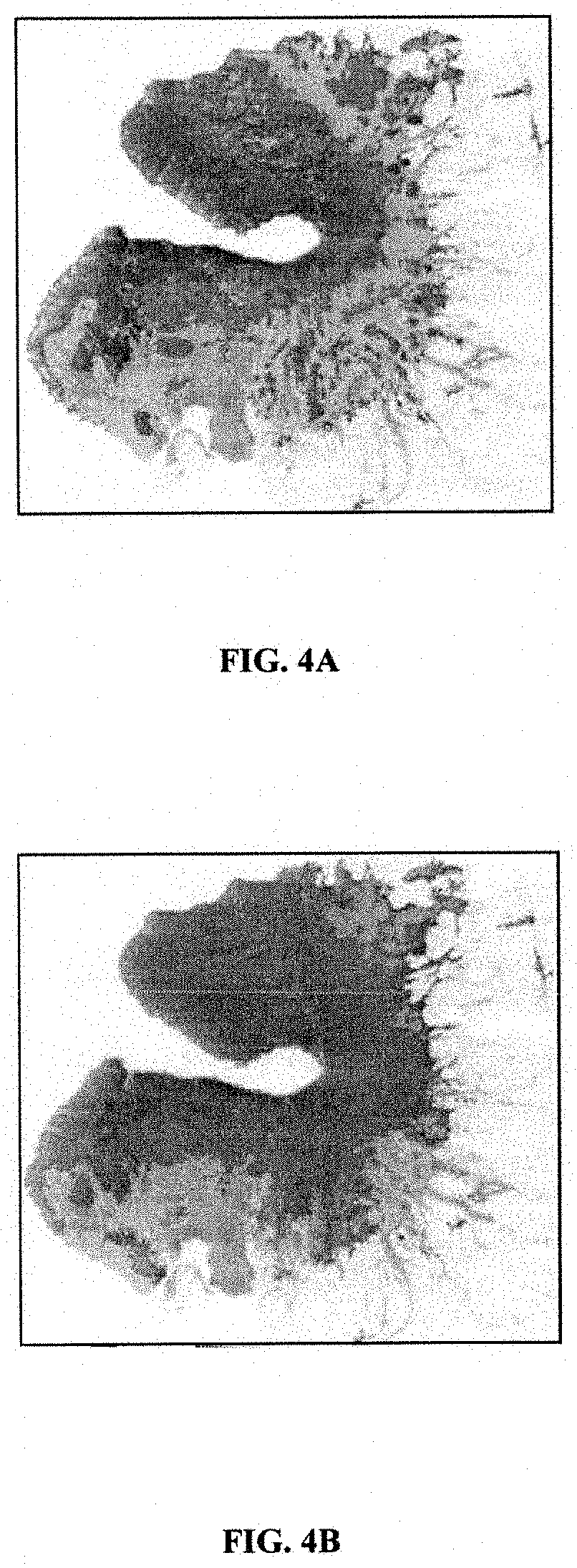
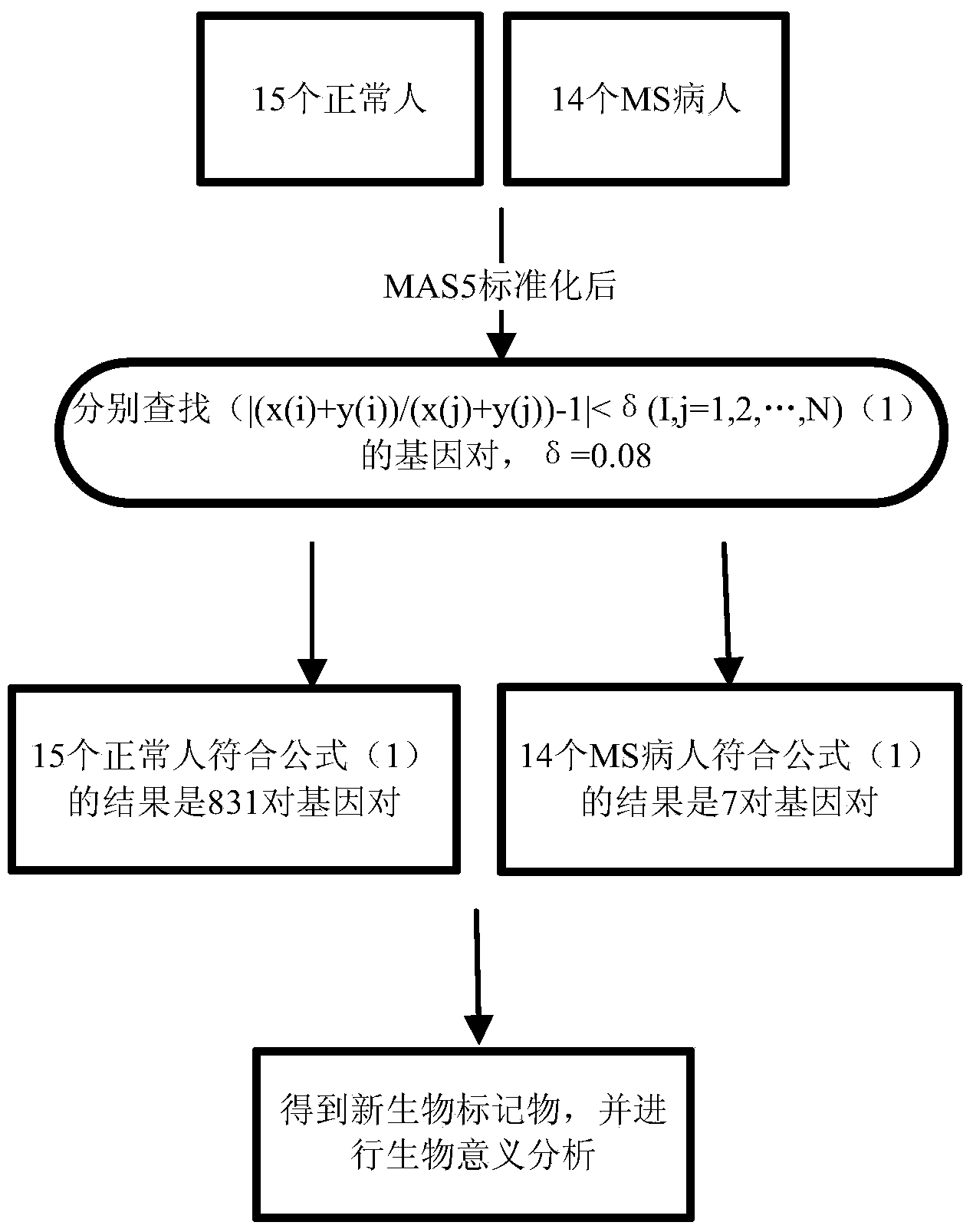
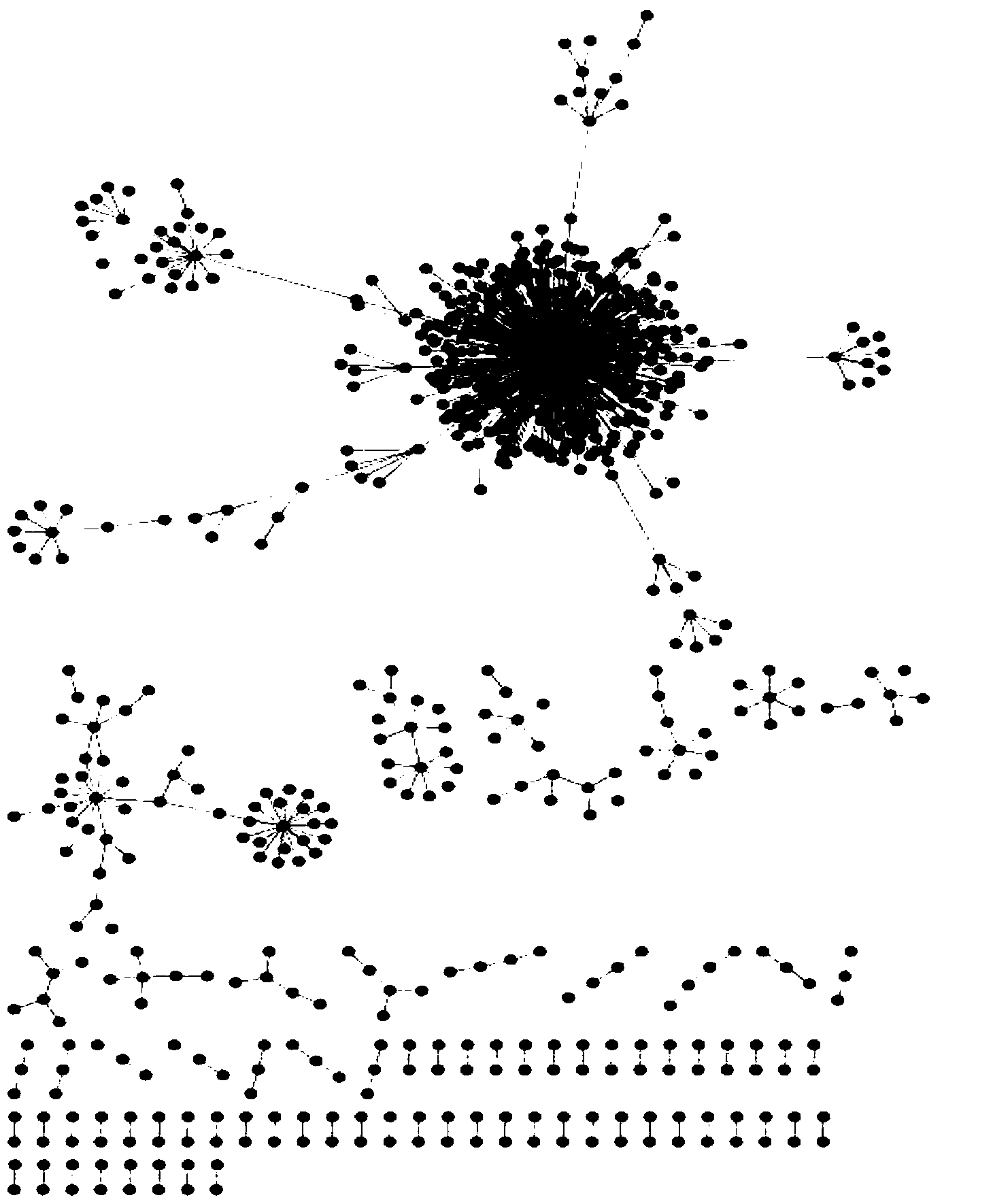
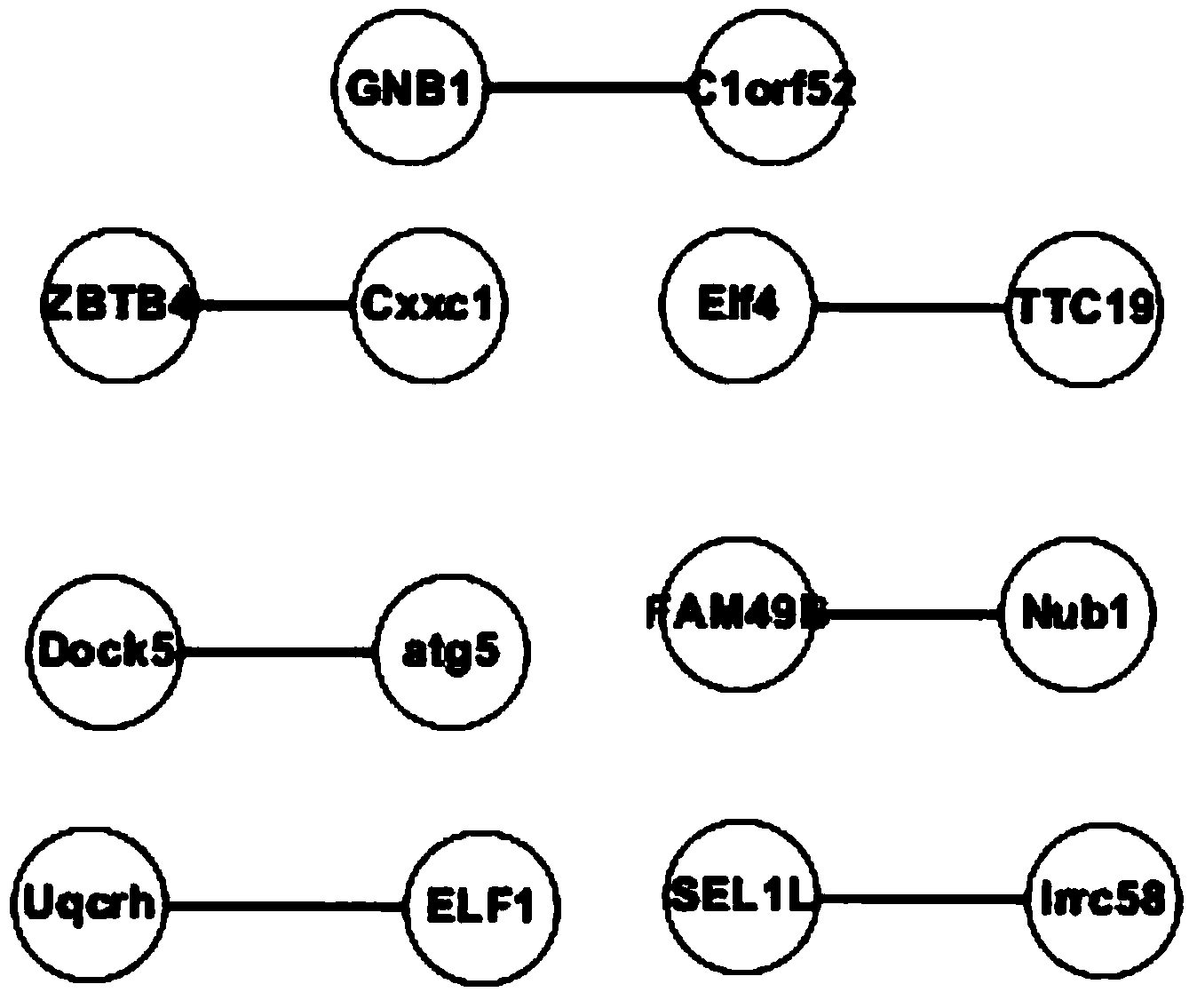
The invention belongs to the technical field of bioinformatics and particularly discloses a method of screening multiple sclerosis biomarkers. The method includes: acquiring a data set GSE21942 from an GEO database, and subjecting a sample to MAS5 standardization by a GPL570 platform; removing x or y gene expressions invariable according to a formula: (|(x(i)+y(i)) / (x(j)+y(j))-1|(delta(i, j=1, 2,..., N), and screening out mutually inhibiting gene pairs (x, y) to obtain a multiple sclerosis biomarker. Delta refers to degree of approximation between genes x and y, and is 0.08 in experiments; x(i) refers to gene expression quantity of the i-th sample of the gene x; y(i) refers to gene expression quantity of the i-th sample of the gene y. The mutually inhibiting gene pairs in mononuclear cells in peripheral blood of both normal people and MS patients to obtain the multiple sclerosis biomarker; by applying the screened multiple sclerosis biomarker in the study and analysis of biological significance, real conditions of the patients can be indicated.
Owner:SHENZHEN INST OF ADVANCED TECH
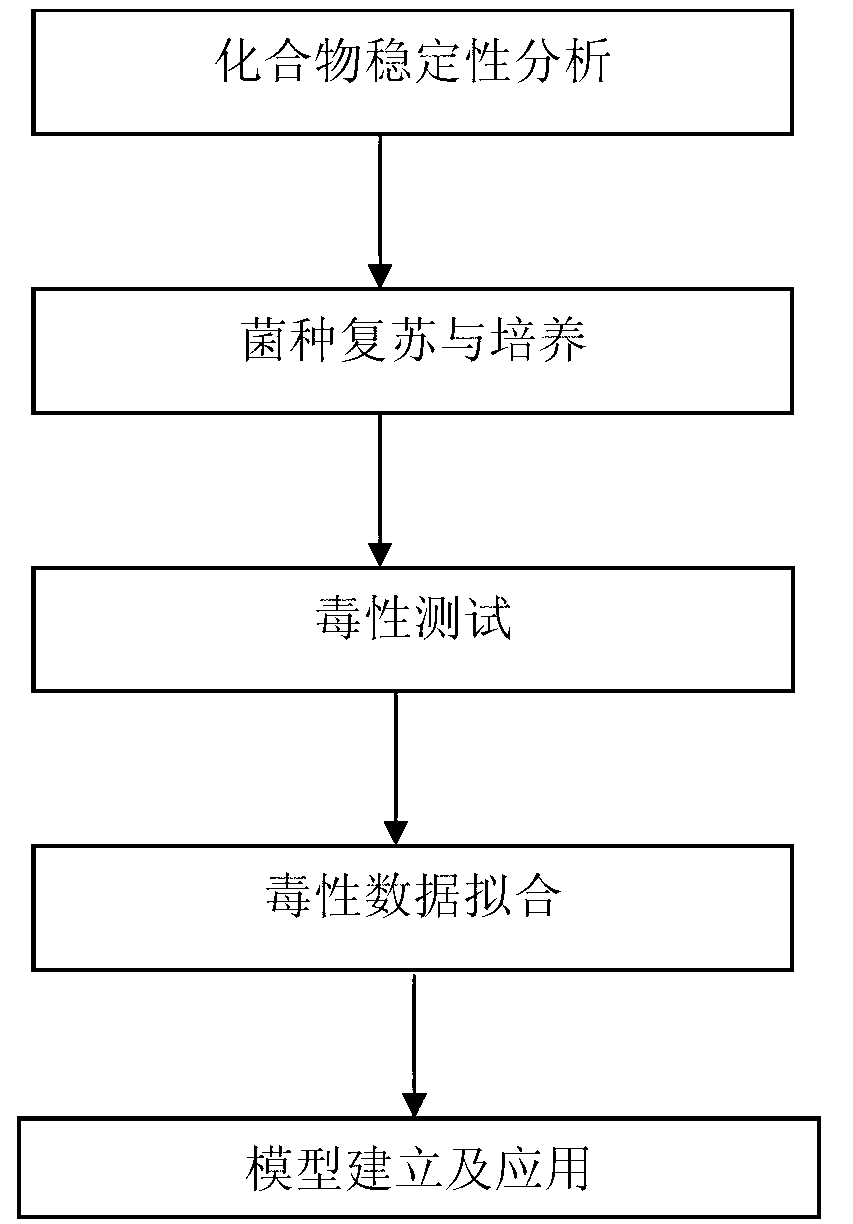
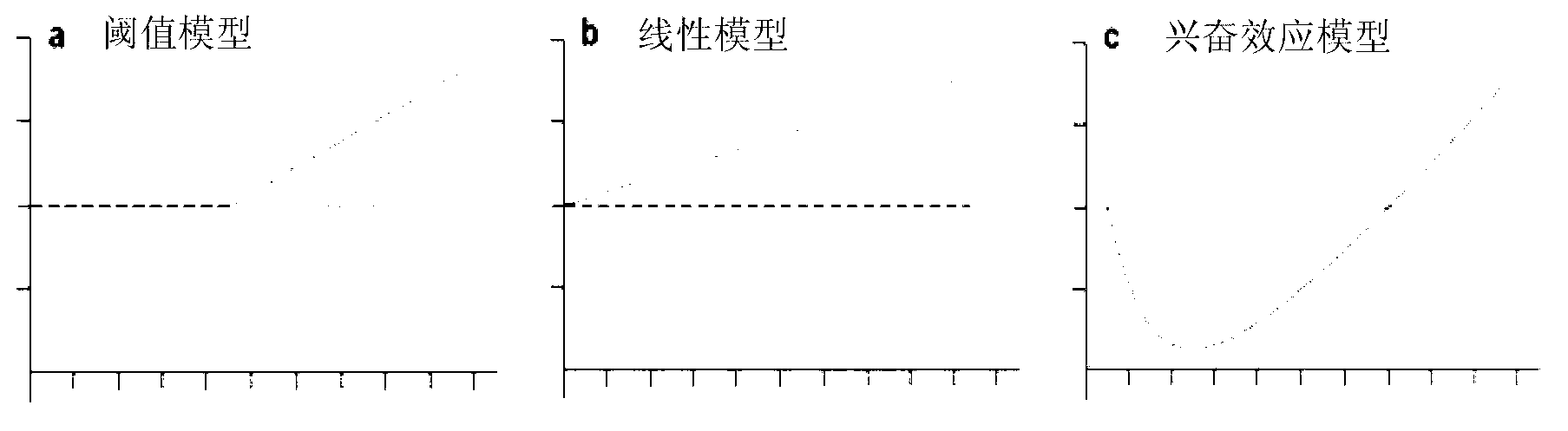
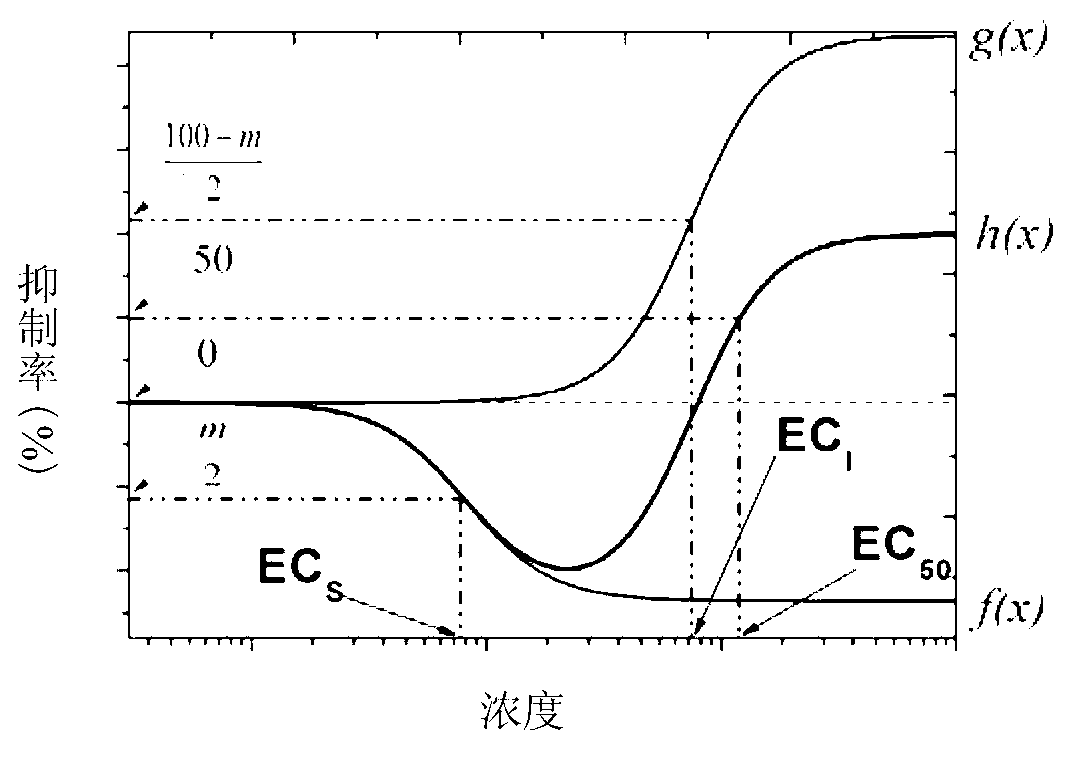
Method of establishing Hormesis dose-effect fitting model
InactiveCN103014116ASimple initializationEasy to initializeMicrobiological testing/measurementAcute toxicity testingHypothesis
The invention belongs to the field of environment pollution detection technology and relates to a method of establishing a Hormesis dose-effect fitting model, namely a Bilogistic model. The method comprises the following steps: (1) analyzing the stability of a compound; (2) recovering and culturing strains; (3) conducting toxicity test including acute toxicity test and chronic toxicity test; (4) calculating the inhibition ratio of the compound analyzed in the step (1) to the strains recovered and cultured in the step (2) by applying the experiment result obtained in the step (3), and conducting toxicity representation by taking inhibition ratio as an index; and (5) fitting the toxicity data to obtain the Bilogistic model. The data fitting is more convenient through the model; parameters in the model are initialized simply; the model is established based on a receptor mechanism hypothesis, thereby having great biological significance; good fitting can be conducted under the condition of high stimulatory effect; the Hormesis effect can be effectively guided to the application in the drug design.
Owner:TONGJI UNIV
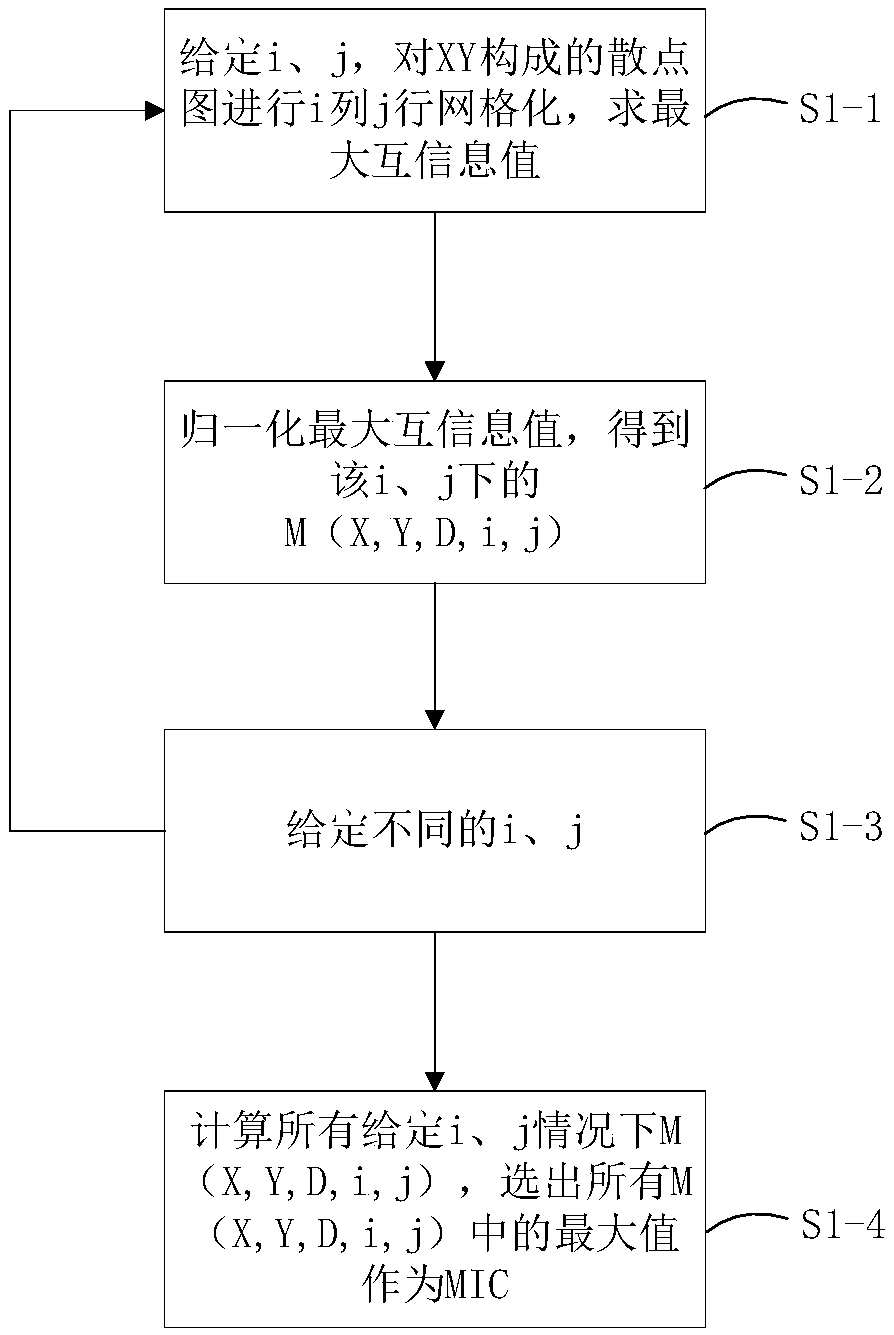
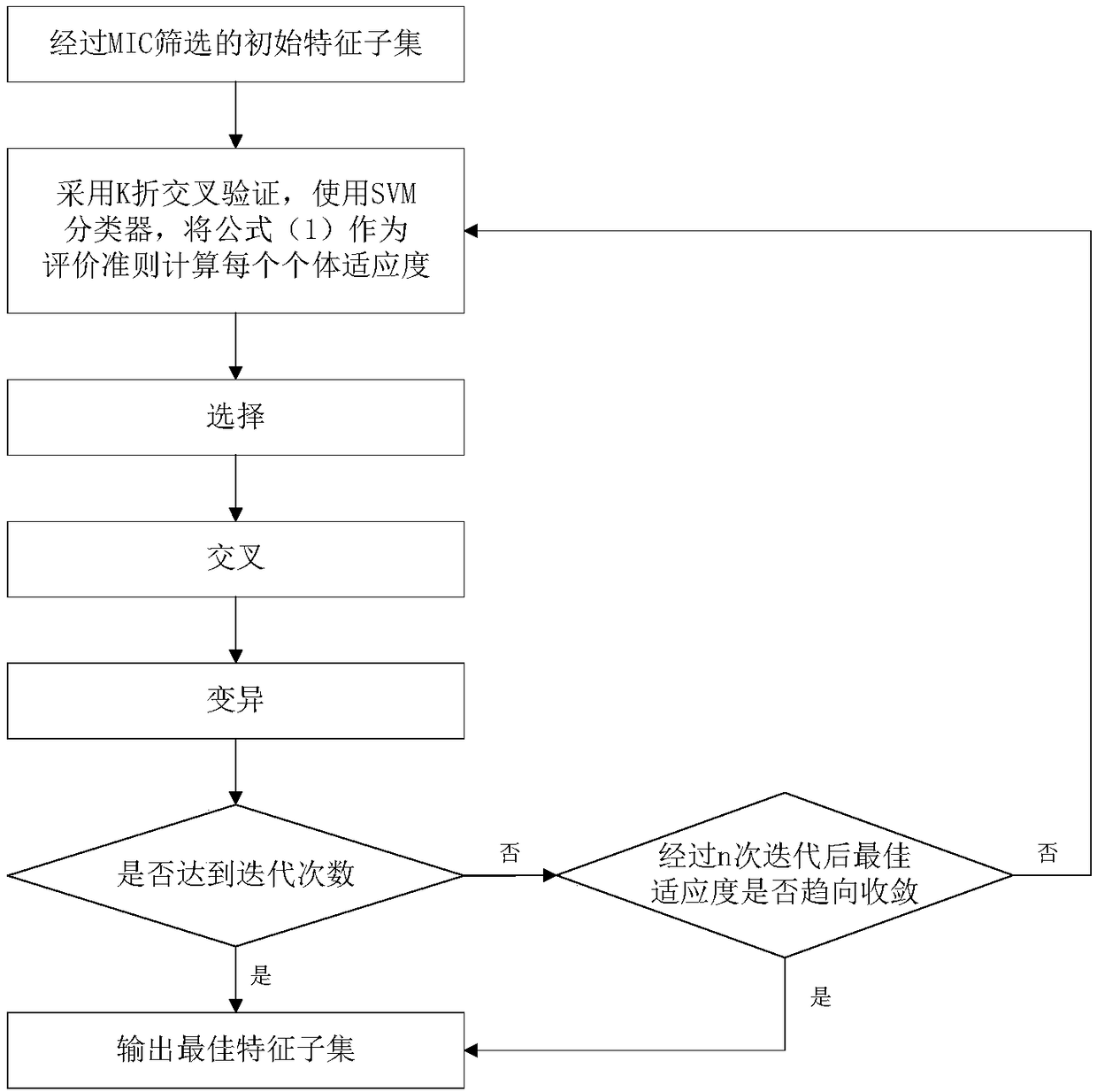
High-dimensional data feature selection method based on filtering method and genetic algorithm
InactiveCN108805159ASolve the problem of high time calculation overhead and unsuitable for high-dimensional dataSolve the problem of removing useful featuresCharacter and pattern recognitionGenetic algorithmsSmall sampleProbit
The invention discloses a high-dimensional data feature selection method based on a filtering method and a genetic algorithm. Traditional feature selection methods are not suitable for high-dimensional and small-sample data because they have the limitation of high probability of falling into local optimization and deleting useful features. According to the invention, firstly, the maximum information coefficient is adopted to calculate the correlation between features of input data and class marks; then, the features are sorted in descending order according to values of the correlation, a threshold value is set, and features with weak correlation are deleted; and finally, the remaining features with strong correlation are subjected to random searching optimization by the genetic algorithm to obtain an optimal feature subset. The invention can effectively carry out feature selection on high-dimensional data and realize dimension reduction. The result of feature selection has important significance for sample class judgment, and when the method is applied to gene expression profile data, the selected features also have important biological significance.
Owner:HANGZHOU DIANZI UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com