Protein biological network motif identification method integrating topological attributes and functions
A biological network and motif recognition technology, applied in the field of systems biology, can solve the problems of false positives in protein interaction data and incomplete protein interaction data, and achieve simple results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
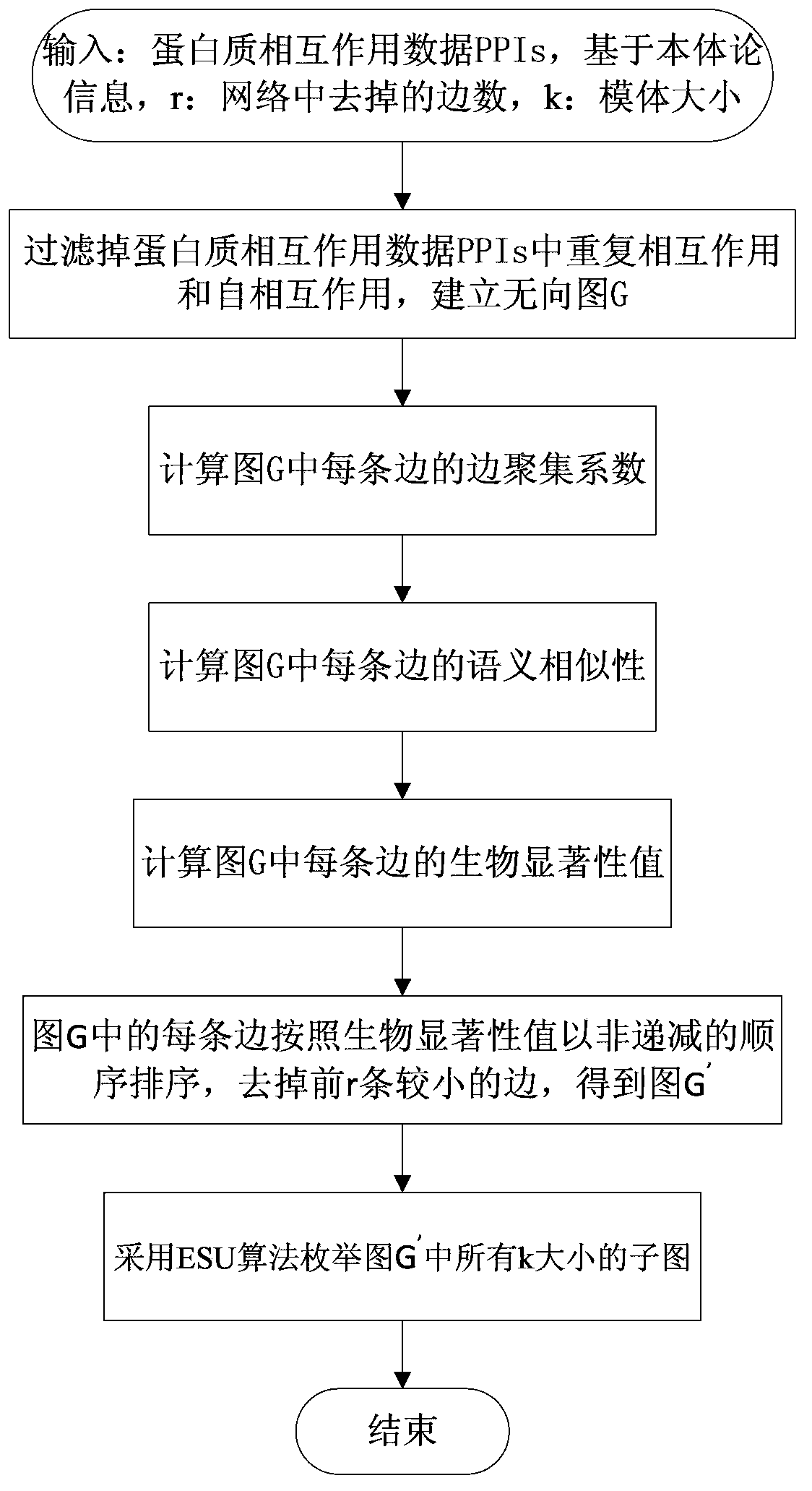
[0035] 1. Motif recognition model of protein biological network based on integrated topological properties and functions
[0036] The present invention defines the biological network motif as: comprehensively assessing the biological significance of the protein pair by integrating the edge aggregation coefficient and the semantic similarity of the GO phrase, and then removing the biologically non-significant edge according to the size of the value. The small-scale connected subgraphs found in the graph.
[0037] In order to clearly describe the protein biological network motif recognition model based on the integration of topological properties and functions, the inventors define the model as follows:
[0038] A measure of the biological significance value of protein pairs is proposed here p c , its expression is as follows:
[0039] p c ( u , v ) = ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com