Protein composite identifying method based on graph model
A protein complex and recognition method technology, applied in the field of first-order neighbor nodes to identify protein complexes, can solve the problems of sensitive input parameters and unsatisfactory recognition results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0033] Below in conjunction with specific embodiment, further illustrate the present invention. It should be understood that these examples are only used to illustrate the present invention and are not intended to limit the scope of the present invention. In addition, it should be understood that after reading the teachings of the present invention, those skilled in the art can make various changes or modifications to the present invention, and these equivalent forms also fall within the scope defined by the appended claims of the present application.
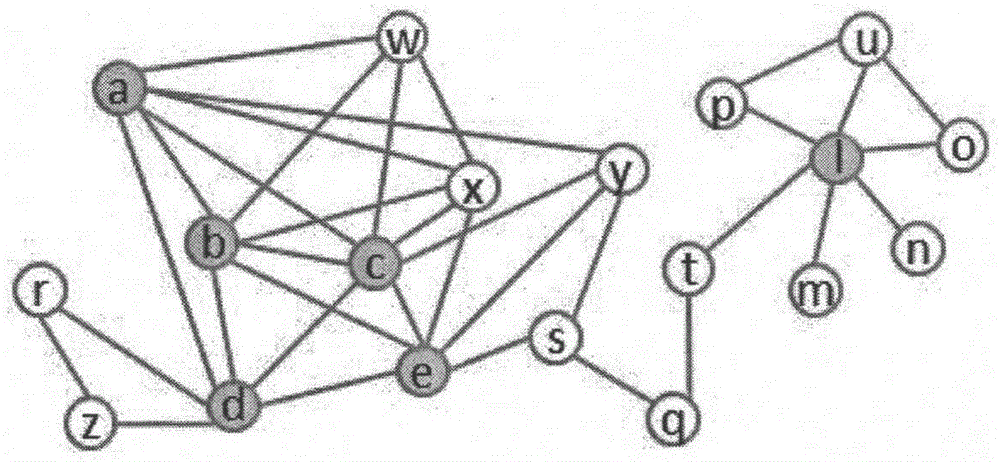
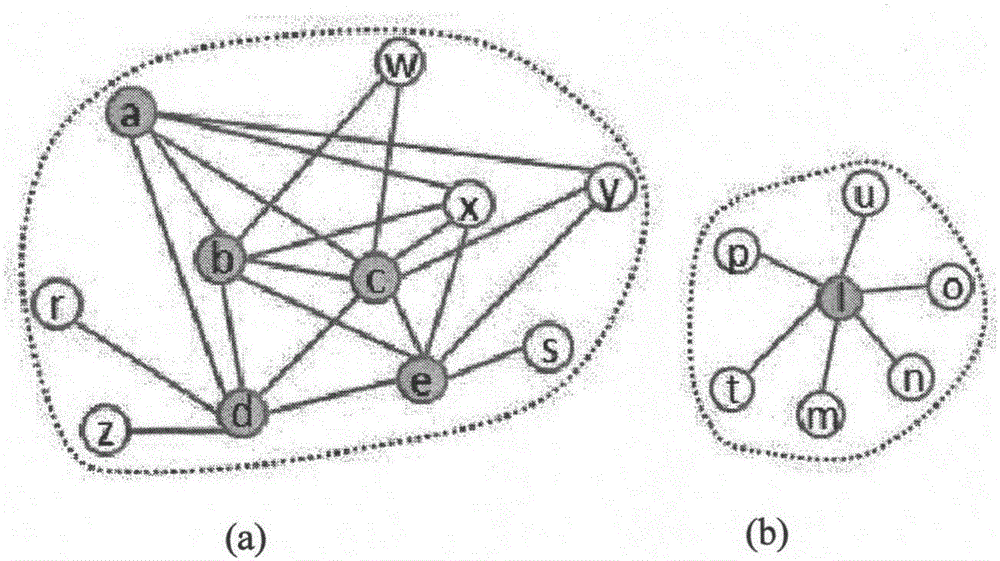
[0034] In this example, the protein interaction network of a given species can be regarded as a network graph G=(V, E), V is a protein node, E is a set of protein interaction edges, and the network is removed from the set of all edges Self-connected edges and repeated edges in . In order to discover the defined protein complexes from the protein interaction network G, some concepts are defined first to prepare for the identifi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com