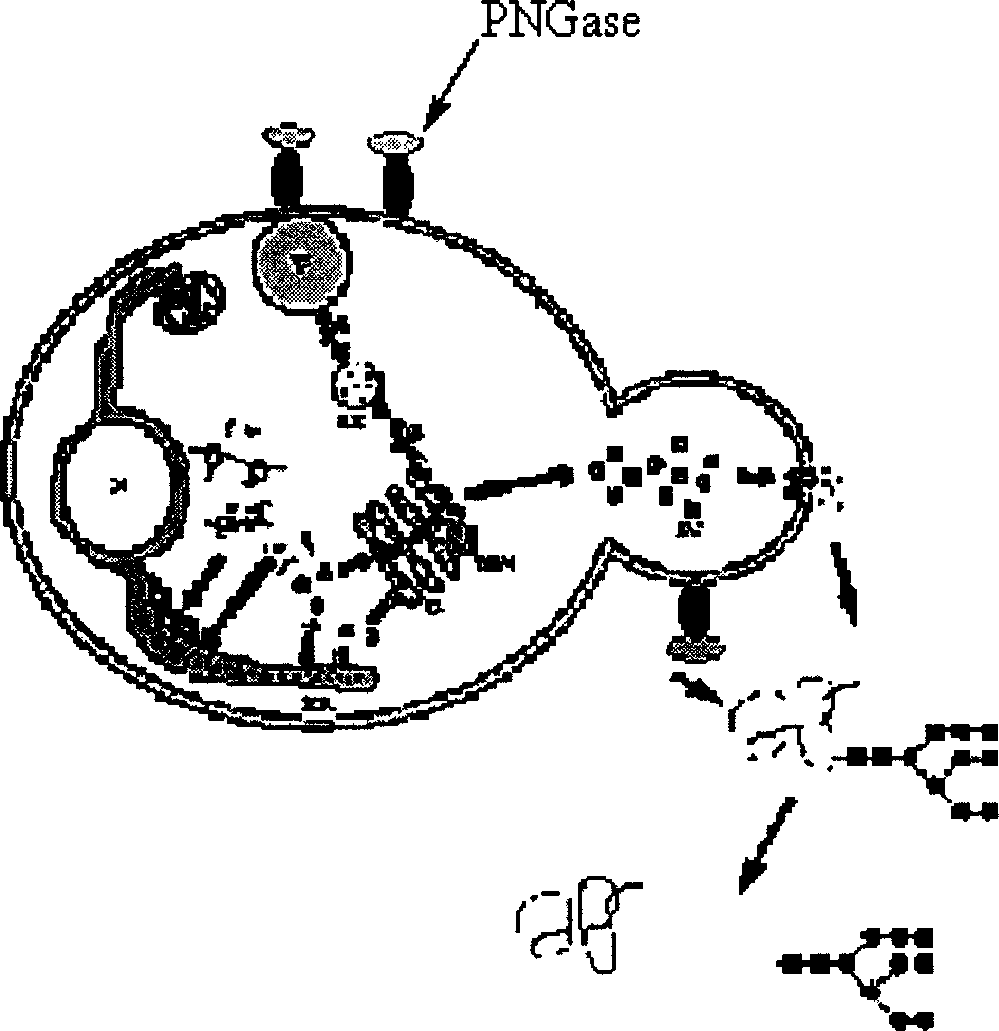
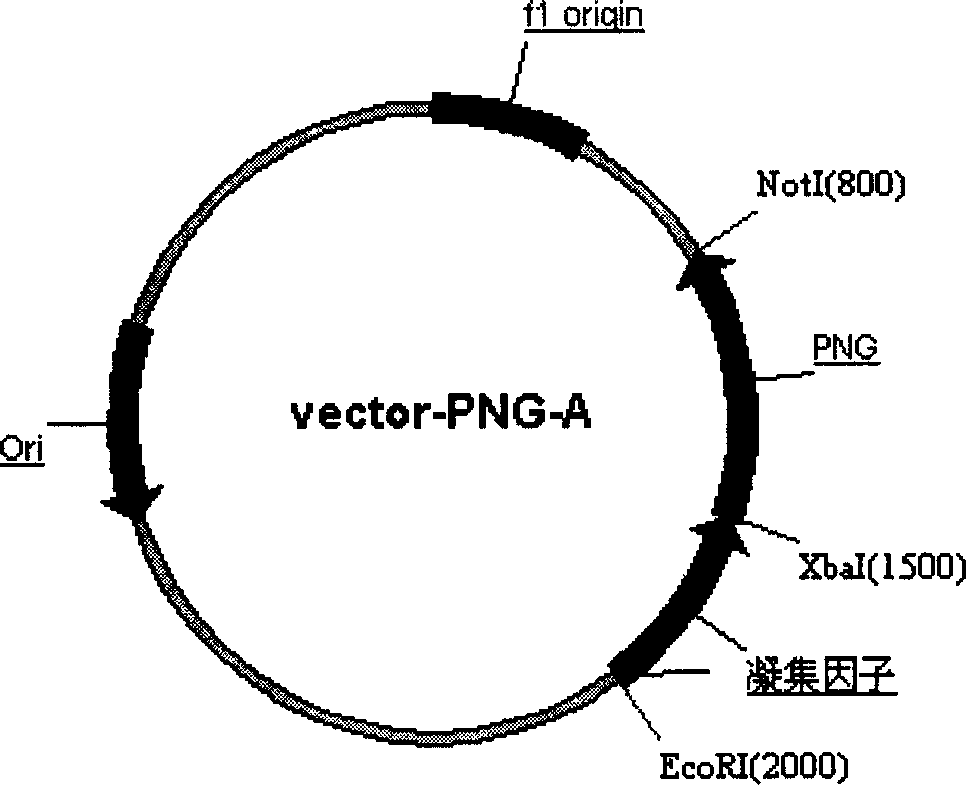
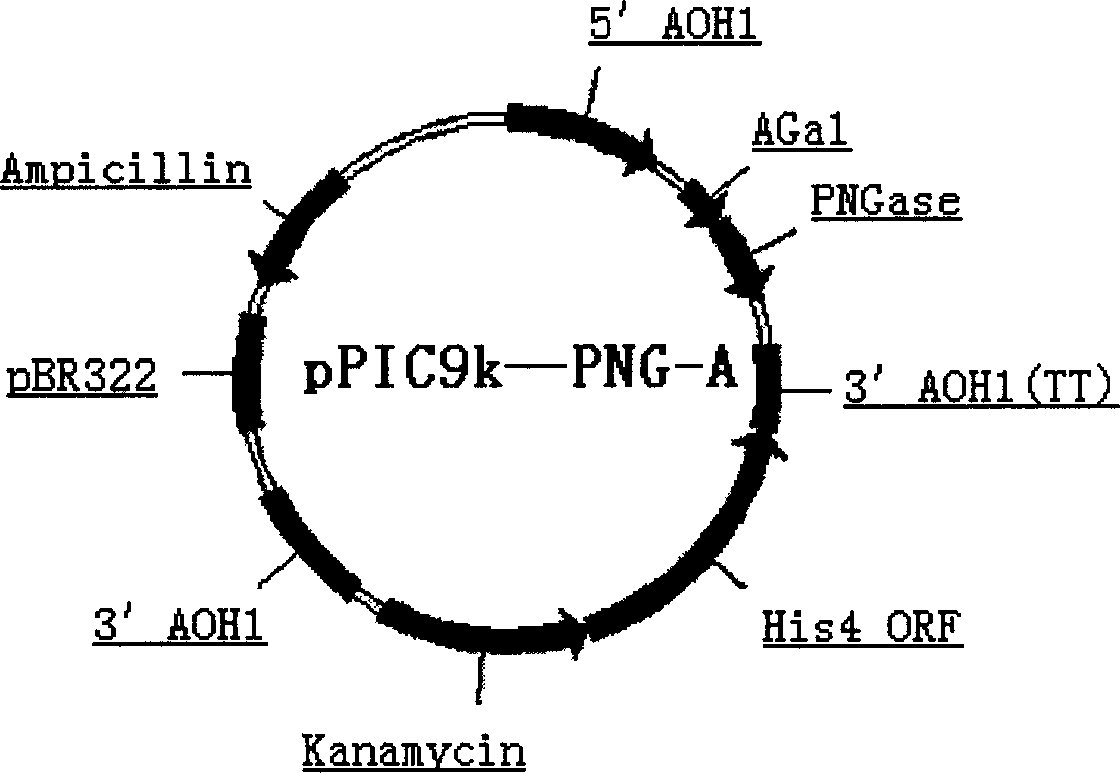
Production of Non-N glycosylated protein from yeast
A technology for glycosylated protein and glycoprotein, which is applied in the production field of non-N-glycosylated protein, can solve the problems of loss of enzymatic activity and reduced enzymatic activity, and achieves the effect of simple method and cost reduction.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Production of non-N-glycosylated erythropoietin (EPO) in Pichia pastoris strains
[0041] 1. Materials: EPO gene: Genbank [gi: 4503588], pPIC9K vector (Invitrogen Ltd), Pichia GS115 strain containing PNGaseF
[0042] 2. Method:
[0043] 1), the construction of surface expression N-glycoamidase yeast strain:
[0044] Using 5-ATATGAATTCGCTCCGCCAGATAATACCGT-3; 5-CACGTCTAGAGTTTGTAACTACCGGAG-3 as primers to clone the N-glycoamidase gene from Pseudomonas meningitidis ATCC 33958 (purchased from ATCC), PCR amplification conditions: 50 μl reaction system, 10×PCR buffer 5 μl , MgCl 2 (25mmol / L) 3μl, dNTP (25mmol / L) 1μl, primers 1 and 2 (20pmol / L) 1μl each, genomic DNA 1μl, Taq DNA polymerase 0.5U, after pre-denaturation at 94°C for 5min, according to the following parameters: 94 Denaturation at ℃ for 45s, annealing at 56℃ for 45s, extension at 72℃ for 2min, 28 cycles, and the last cycle of extension at 72℃ for 10min. The XbaI and NotI sites inserted into the pBluescriptSK(+) ...
Embodiment 2
[0050] Production of non-N-glycosylated interferons in yeast strains
[0051] 1. Materials: Interferon gene: Genbank [gi: 50593016], pPIC9K vector (Invitrogen Ltd), Pichia GS115 strain containing PNGase F
[0052] 2. Method:
[0053] 1), the construction of surface expression N-glycoamidase Pichia strain:
[0054] Using 5-ATATGAATTCGCTCCGCCAGATAATACCGT-3; 5-CACGTCTAGAGTTTGTAACTACCGGAG-3 as primers to clone the N-glycoamidase gene from Pseudomonas meningitidis ATCC 33958 (purchased from ATCC), PCR amplification conditions: 50 μl reaction system, 10×PCR buffer 5 μl , MgCl 2 (25mmol / L) 3μl, dNTP (25mmol / L) 1μl, primers 1 and 2 (20pmol / L) 1μl each, genomic DNA 1μl, Taq DNA polymerase 0.5U, after pre-denaturation at 94°C for 5min, according to the following parameters: 94 Denaturation at ℃ for 45s, annealing at 56℃ for 45s, extension at 72℃ for 2min, 28 cycles, and the last cycle of extension at 72℃ for 10min. The XbaI and NotI sites inserted into the pBluescriptSK(-) vector af...
Embodiment 3
[0059] Production of non-N-glycosylated ribonuclease B in yeast strains
[0060] 1. Materials: ribonuclease B gene, pPIC9K vector (Invitrogen Ltd), Pichia GS115 strain containing PNGase F
[0061] 2. Method:
[0062] 1), the construction of surface expression N-glycoamidase Pichia strain:
[0063] Using 5-ATATGAATTCGCTCCGCCAGATAATACCGT-3; 5-CACGTCTAGAGTTTGTAACTACCGGAG-3 as primers to clone the N-glycoamidase gene from Pseudomonas meningitidis ATCC 33958 (purchased from ATCC), PCR amplification conditions: 50 μl reaction system, 10×PCR buffer 5 μl , MgCl 2 (25mmol / L) 3μl, d NTP (25mmol / L) 1μl, primers 1 and 2 (20pmol / L) 1μl each, genomic DNA 1μl, Taq DNA polymerase 0.5U, pre-denaturation at 94°C for 5min, according to the following parameters: Denaturation at 94°C for 45 s, annealing at 56°C for 45 s, extension at 72°C for 2 min, 28 cycles, and the last cycle of extension at 72°C for 10 min. The XbaI and NotI sites inserted into the pET22b vector after digestion with XbaI a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com