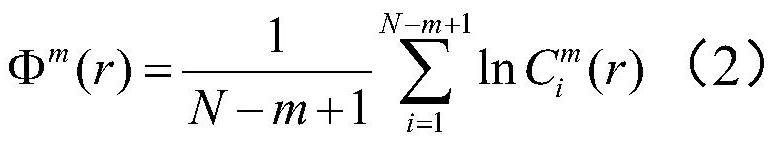Patents
Literature
64 results about "Marker analysis" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
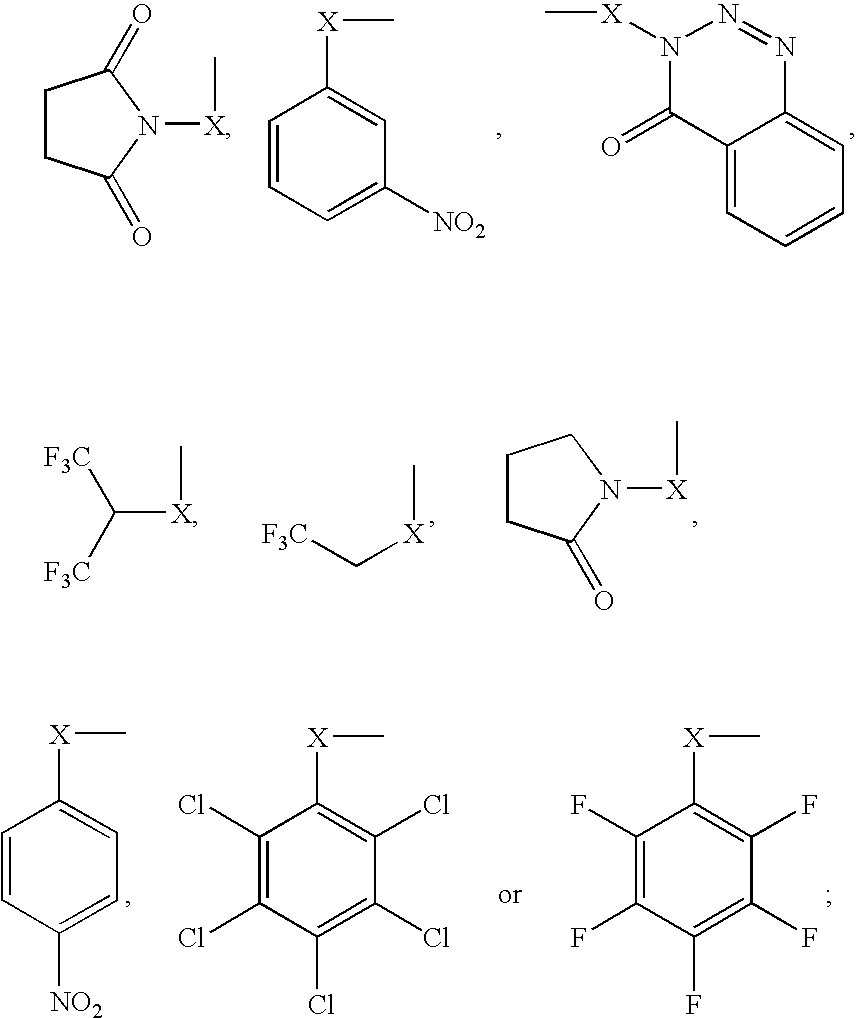
Methods and mixtures pertaining to analyte determination using electrophilic labeling reagents
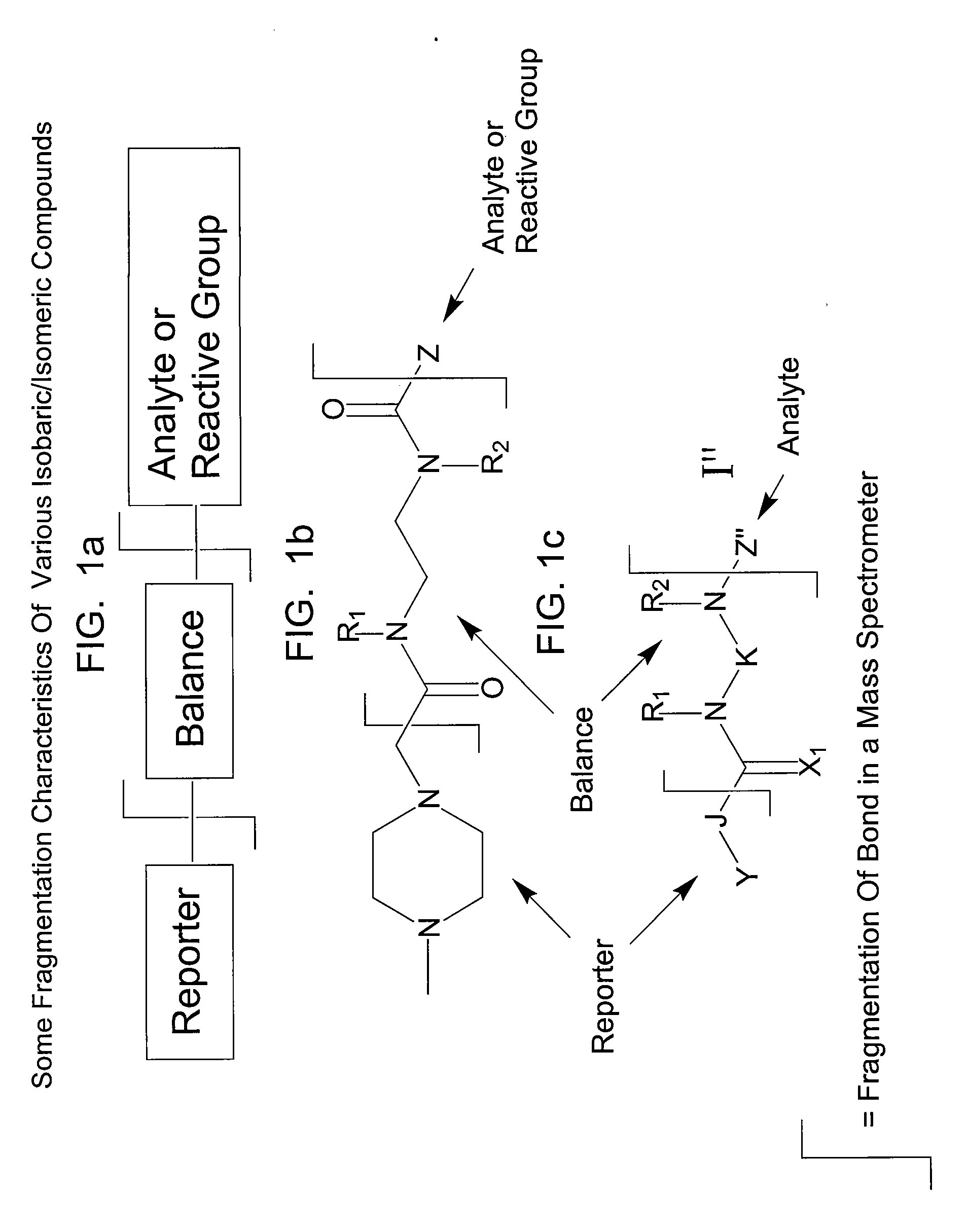
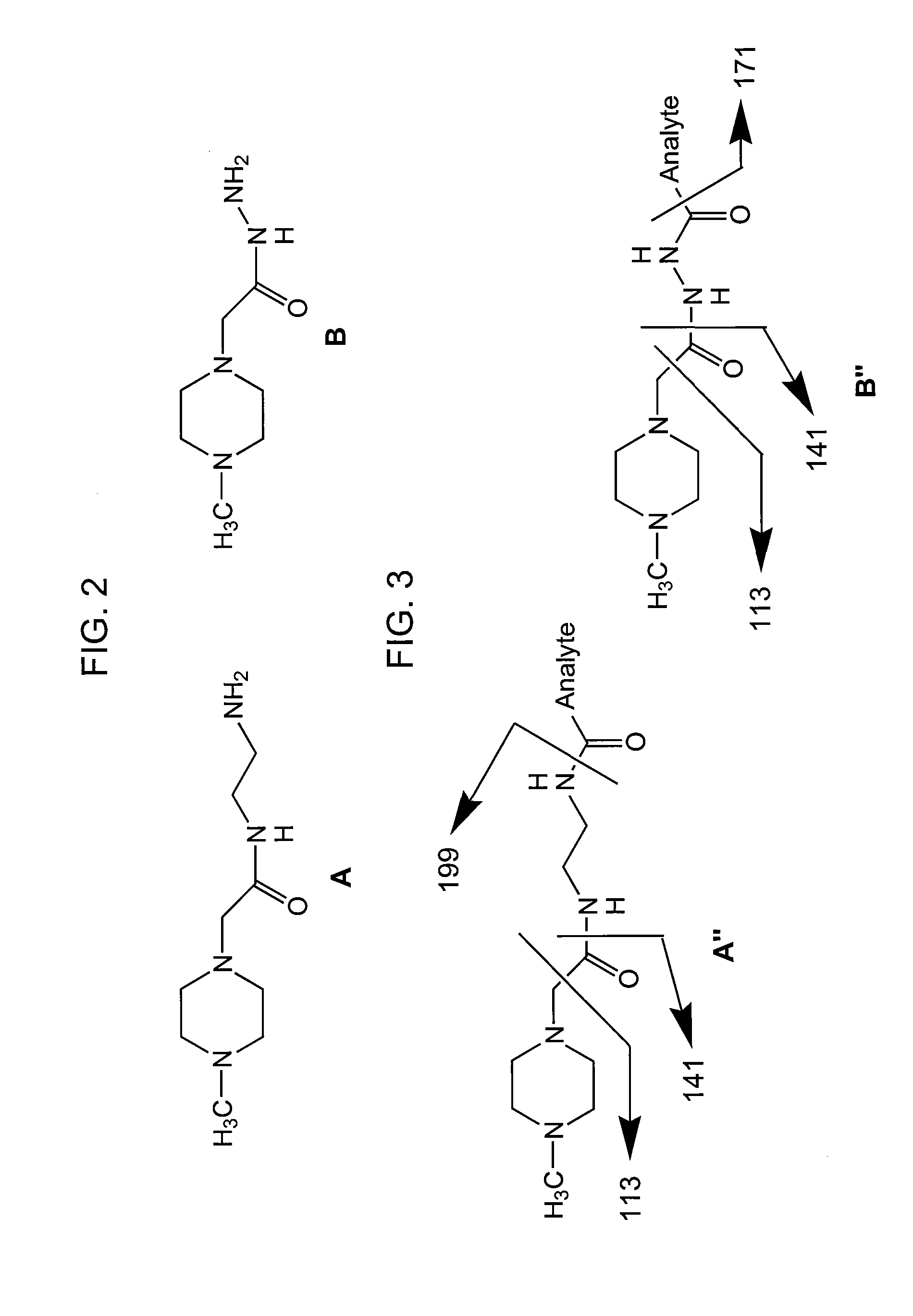
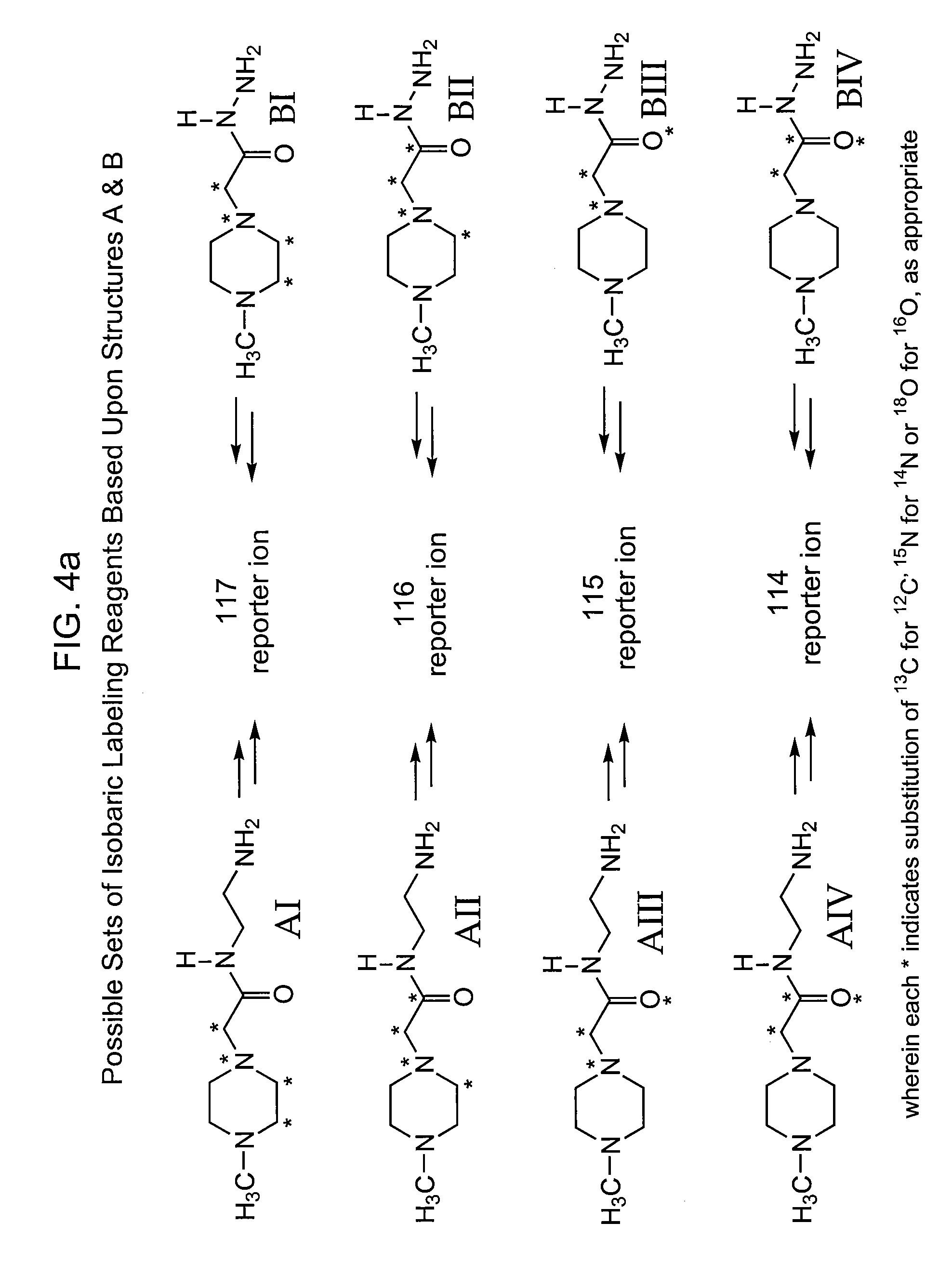
This invention pertains to methods, mixtures, kits and / or compositions for the determination of analytes by mass analysis using unique labeling reagents or sets of unique labeling reagents. The labeling reagents can be isomeric or isobaric and can be used to produce mixtures suitable for multiplex analysis of the labeled analytes.
Owner:DH TECH DEVMENT PTE
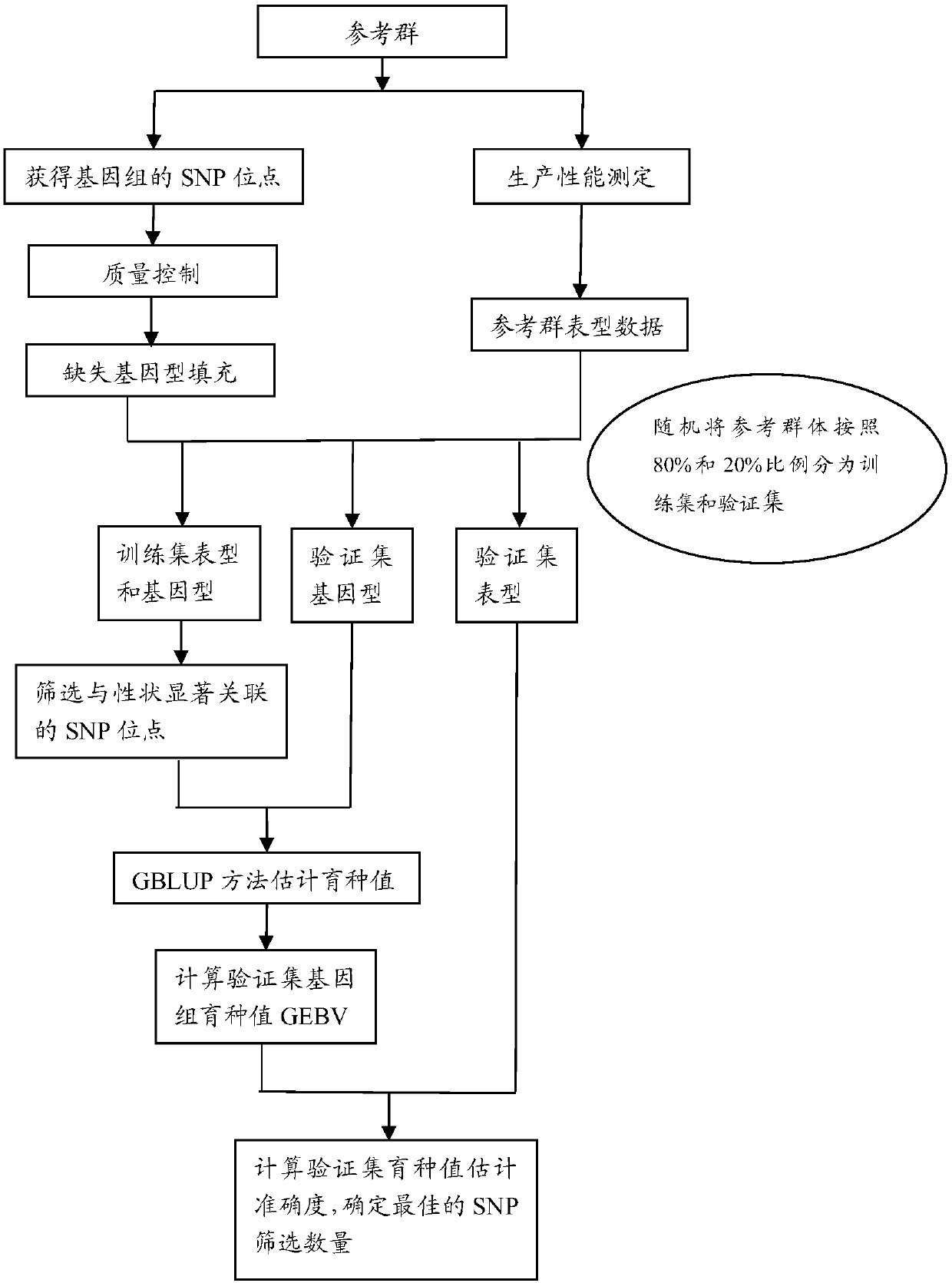
Method for determining optimal SNP quantity as well as performing genome selective breeding on production performance of large yellow croakers through selection markers
ActiveCN107338321AReduce breeding costsLow costMicrobiological testing/measurementProteomicsGenomic sequencingAgricultural science
The invention discloses a method for determining optimal SNP quantity as well as performing genome selective breeding on production performance of large yellow croakers through selection markers. The method comprises the following steps: performing phenotype determination and genomic sequencing on the production performance of individuals of a reference group to obtain SNP locus; screening out the qualified SNP locus and supplementing the deleted genotype; dividing the reference group into a training set and a validation set to perform hybridization validation; screening the SNP locus most remarkably associated with the character through single marker analysis, and then calculating GEBV of individuals of the validation set only by using the locus and through a GBLUP method; further obtaining breeding value estimation accuracy under each screening SNP quantity; finally determining the optimal quantity of SNP screening; and according to the optimal quantity, calculating the GEBV by the GBLUP method, further obtaining the breeding value estimation accuracy and performing genome selective breeding according to the value size. By the method, the genomic selection cost on the production performance of the large yellow croakers can be reduced remarkably.
Owner:JIMEI UNIV
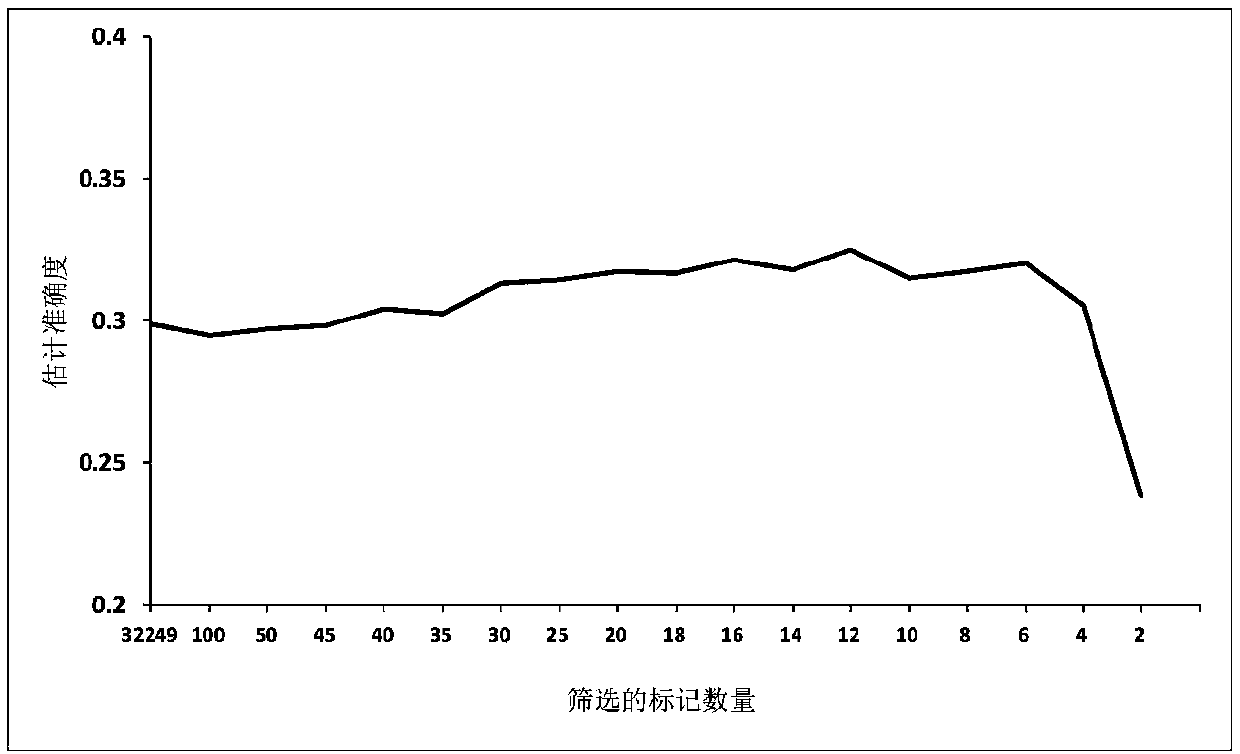
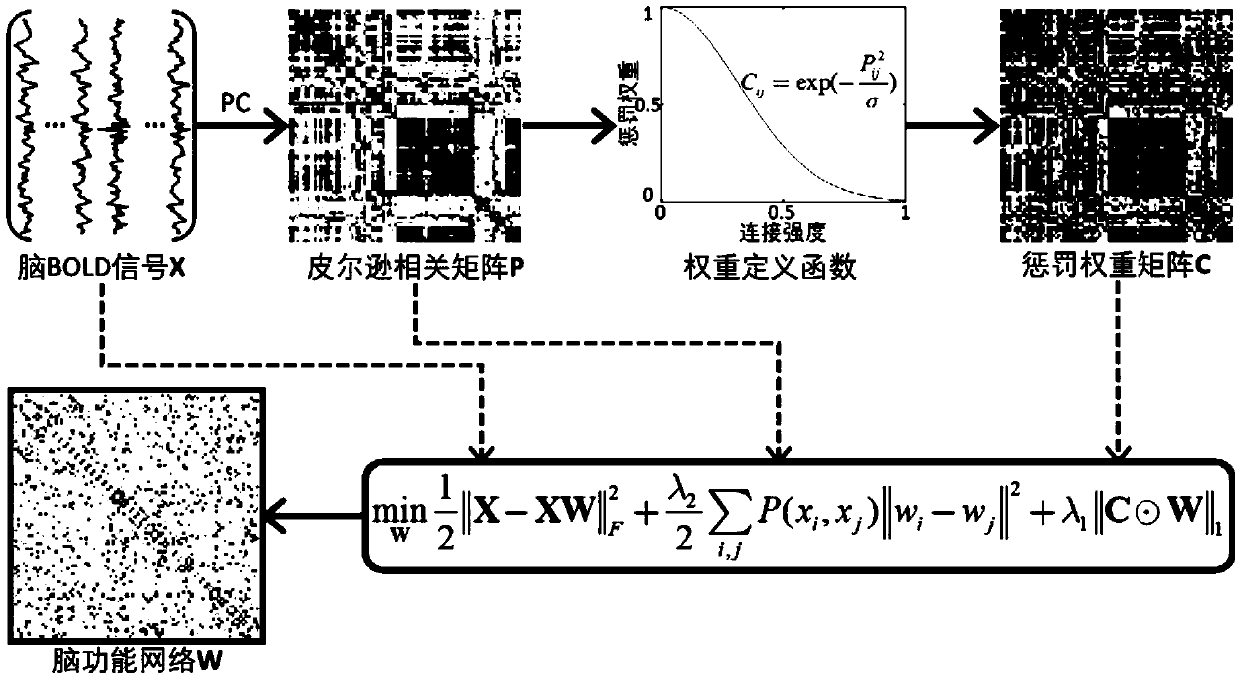
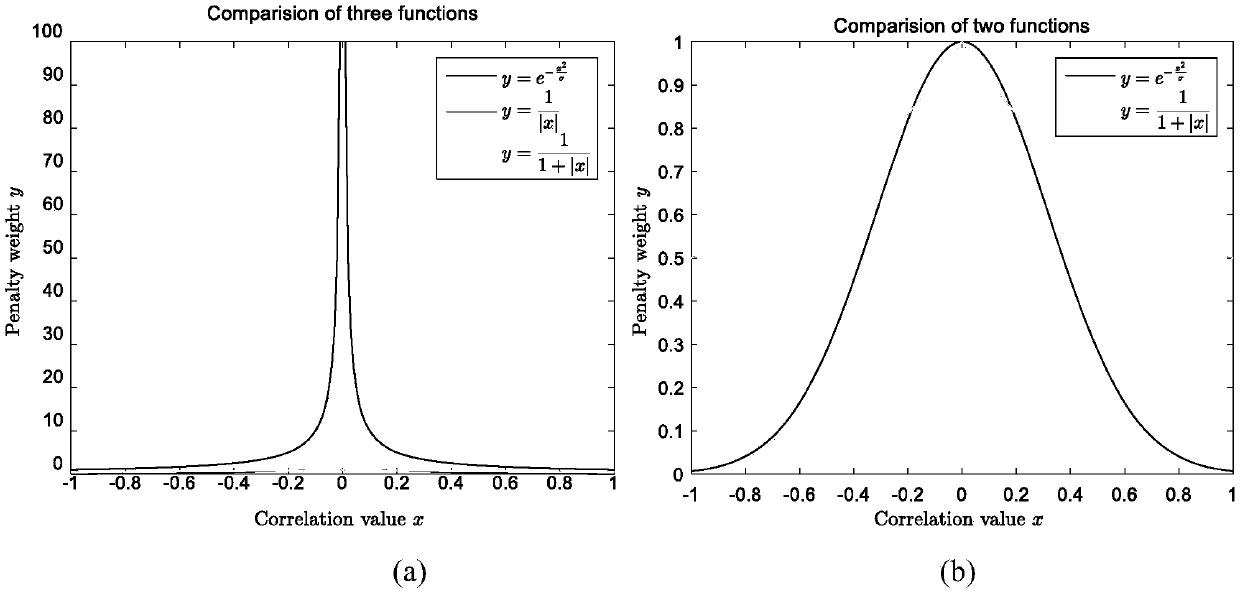
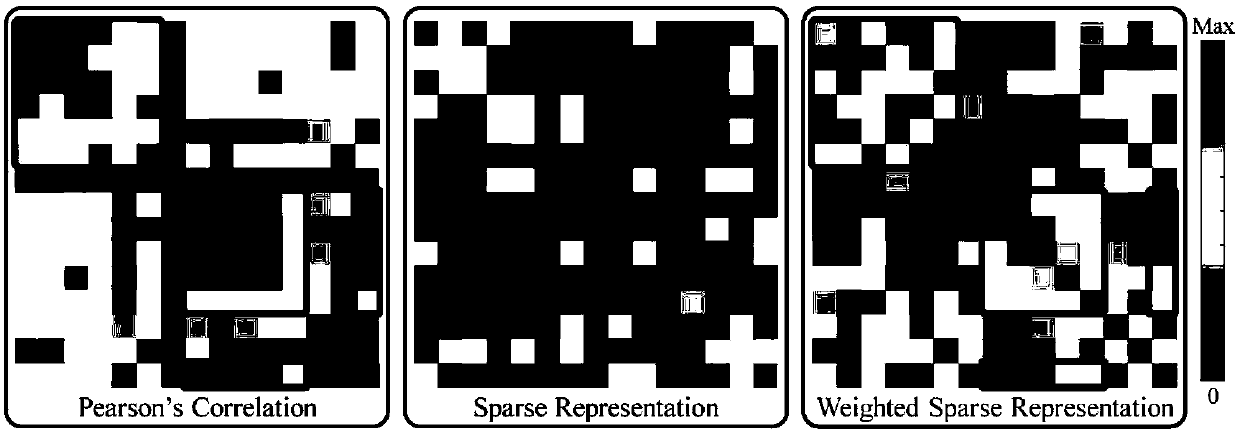
Weighted graph regularization sparse brain network construction method
PendingCN109065128ASolve the problem that the solution does not convergeImprove classification accuracyImage enhancementImage analysisDiseaseFunctional connectivity
The invention discloses a weighted graph regularization sparse brain network construction method. According to the method, on the basis of analysis of keeping similar local manifold characteristics ofsimilar data in original data space after projection, constrained modeling is carried out on association between brain connection by using a graph regularization item; with consideration of correlation analysis, the similarity degree of essences in a time sequence signal sequence of a brain area can be measured, sparse modeling is constrained by using measured functional connection strength priorinformation, and a weighted sparse regularization item is established; and then combined modeling is carried out on the whole brain function network and a brain network with the biological significance is constructed. According to the invention, on the basis of integration of correlation analysis, sparsity and graph regularization constraint into the unified modeling frame, a brain function network is constructed effectively by utilizing the similarity and locality of functional magnetic resonance brain image data fully; and the method is also used for neurological disease identification andbiological marker analysis.
Owner:ZHENGZHOU UNIV
Method for breeding raphanus sativus L. CMS (genic male sterility) lines by using marker assisted selection
InactiveCN101956007AImproved breeding efficiencyImprove breeding efficiencyMicrobiological testing/measurementPlant genotype modificationRaphanusMarker analysis
The invention relates to a method for breeding raphanus sativus L. CMS (genic male sterility) lines by using marker assisted selection, which belongs to the technical field of crop breeding and is used for efficient breeding of excellent male sterility lines in raphanus sativus L. hybrid advantage utilization. The invention adopts the technical scheme that: the method comprises the following steps of: screening inbred lines with excellent traits and stable fertility; selecting specific primers NAUmsF4 and NAUmsR4 to perform PCR amplification on the screened inbred lines, and determining that the inbred lines capable of amplifying 1069bp specific strips cannot be used as candidate maintenance lines; performing restored gene functional marker analysis on the inbred lines incapable of amplifying the specific strips, and screening the inbred lines capable of serving as the candidate maintenance lines; and performing cross and back cross of 4 to 5 generations on the original sterile lines and the identified candidate maintenance lines to obtain new sterile lines and corresponding maintenance lines with excellent traits. The method for quickly screening the candidate maintenance lines by using molecular makers is not affected by a growth period and an environment, can remarkably improve the breeding efficiency of the raphanus sativus L. CMS lines, and accelerates the breeding progress of excellent F1 hybrids.
Owner:NANJING AGRICULTURAL UNIVERSITY
Assay method and device
ActiveUS20110117674A1Simple and easy to performImprove accuracyComponent separationLaboratory glasswaresAnalysis methodAnalyte
A method for the analysis of at least two analytes in a liquid sample, in which a substrate is provided wherein at least two different types of capturing molecules are immobilized on the substrate and wherein each type of capturing molecule has specific affinity for an analyte. The sample is contacted with capturing molecules, wherein for at least one analyte to be analyzed contact is induced between the capturing molecules and a labelled detection molecule with specific affinity for the analyte, and for at least one another analyte to be contact is induced between the capturing molecules and a labelled version of the analyte. A detectable signal is measured from the labelled detection molecule and the labelled analyte on the substrate, wherein the concentration of the labelled analyte is adapted to the concentration of the analyte in the sample.
Owner:CRIMSON INT ASSETS LLC
Porphyra haitanensis germ plasma identification fingerprint construction method and its uses
InactiveCN101126746AImprove automationEasy extractionComponent separationMicrobiological testing/measurementMarker analysisReconstruction method
The utility model relates to a reconstruction method and application for identifying fingerprint chromatogram of porphyra haitaensis, belonging to fingerprint chromatogram of porphyra haitaensis, in particular to reconstruction method and application for identifying fingerprint chromatogram of porphyra haitaensis. The utility model provides a rapid accurate reconstruction method for identifying fingerprint chromatogram of porphyra haitaensis and the application for realizing the porphyra haitaensis identification automation, which is characterized in that porphyra haitaensis specimens are collected; genome DNA of various specimens is refined and purified; the PCR is enlarged, the refined genome DNA is utilized as a template, SRAP molecule marking analysis is performed by the optimized experimental system; the representative SRAP multi-state bars are selected for reconstructing the DNA fingerprint chromatogram of various specimens; the reconstructed DNA fingerprint chromatogram is converted to the identifiable digital fingerprint by computer, and then is inputted to the fingerprint chromatogram identification software, thereby the digital fingerprint database of porphyra haitaensis is established. The utility model is used for follow-up porphyra haitaensis automation identification.
Owner:JIMEI UNIV
Larimichthys crocea genome-wide SNP and InDel molecular marker method based on double enzyme digestion
ActiveCN105349675AReduce the scope of sequencingLow costMicrobiological testing/measurementDNA/RNA fragmentationSequence designAgricultural science
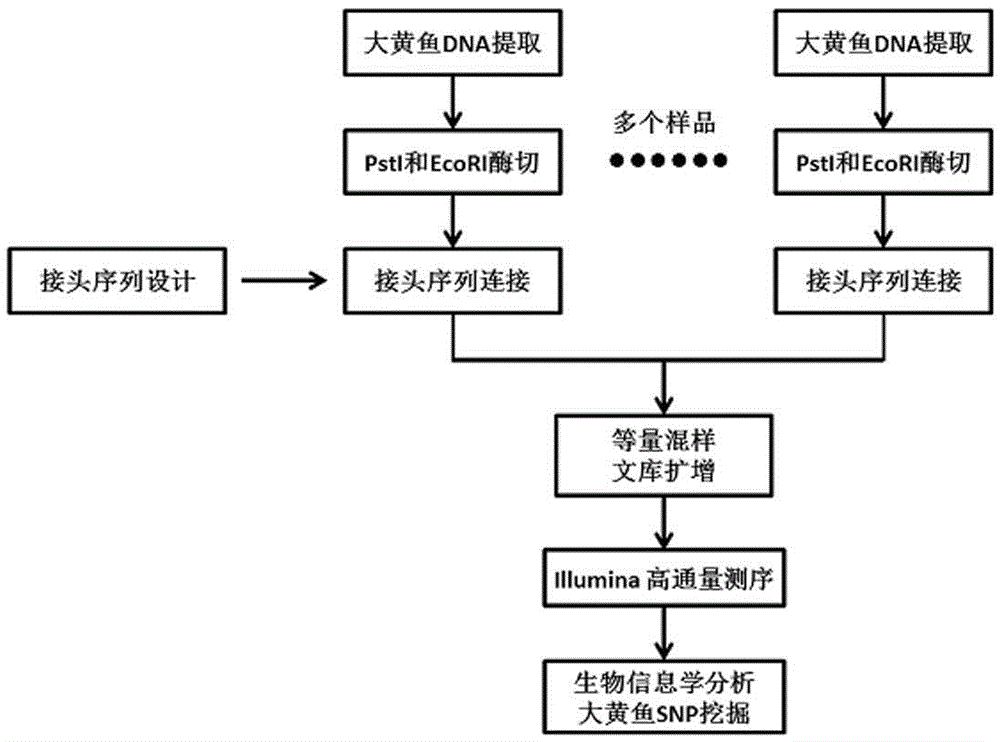
The invention discloses a larimichthys crocea genome-wide SNP and InDel molecular marker method based on double enzyme digestion. The method comprises the following steps: 1) joint sequence design; 2) genome DNA enzyme digestion; 3) joint sequence connection; 4) sample mixing and PCR amplification; 5) high-throughput sequencing; 6) SNP locus excavation and analysis through sequencing data. According to the method, the modern molecular biology and the advanced high-throughput sequencing technology are combined, the method of combination of EcoRII and NlaIII double enzyme digestion is used, the genome-wide DNA molecules are subjected to enzyme digestion, DNA segments with certain lengths are obtained for carrying out library establishment sequencing and SNP molecular marker excavation, and the uniformly distributed genome-wide SNP locus information is obtained; the workload and cost of genome-wide marker analysis are greatly reduced, and the method is also applicable to the genome-wide marker analysis of other species; the SNP marker excavated through the method is applicable to the study fields such as animal and plant species identification, species genetic genealogy analysis, germplasm resource genetic diversity analysis and genetic breeding.
Owner:JIMEI UNIV
SRAP molecular marker closely linked with male sterility genes of tomatoes and preparation method thereof
ActiveCN104120126AGuaranteed purityImprove protectionMicrobiological testing/measurementDNA/RNA fragmentationMarker analysisGenotype
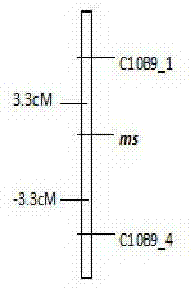
The invention discloses an SRAP molecular marker closely linked with male sterility genes of tomatoes and a preparation method thereof. The method comprises the following steps: taking purple-stem fertile tomatoes 87-5 as male parents and green-stem male sterile tomatoes at the seedling stage as female parents, hybridizing to generate F1 generation, and selfing to build an F2 segregation population; performing SRAP marker analysis on male sterility genes ms by adopting a bulked segregant analysis method, randomly combining primers before and after SRAP, selecting 544 pairs of primer combinations, and screening between male sterile and fertile pools to obtain two polymorphic markers C10B9_1 and C10B9_4 between two DNA pools; performing SRAP marker verification on the F2 segregation population by virtue of the two markers, wherein linkage analysis discovers that the linkage distance between the marker C10B9_1 and the sterility genes ms and the linkage distance between the marker C10B9_4 and the sterility genes ms are 3.3cM and -3.3cM respectively. By virtue of the molecular marker, assisted selection of male sterile tomatoes can be performed, the transfer cycle is shortened, the transfer efficiency is improved, a tedious program of identifying the sterility of each generation by selfing in a transfer process is omitted, conventional phenotype selection is transformed into genotype selection, and the accuracy and scientificity of selection are improved.
Owner:HORTICULTURE INST OF XINJIANG ACAD OF AGRI SCI
Rice seedling-stage salt-tolerant gene qST11 and molecular marker method thereof
ActiveCN104762298AIncreased salt tolerance at seedling stageImprove salt toleranceMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceMarker analysis
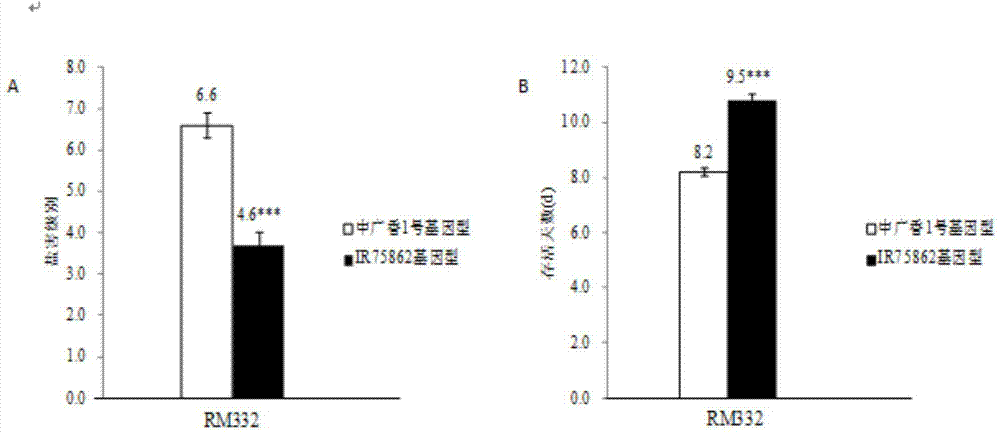
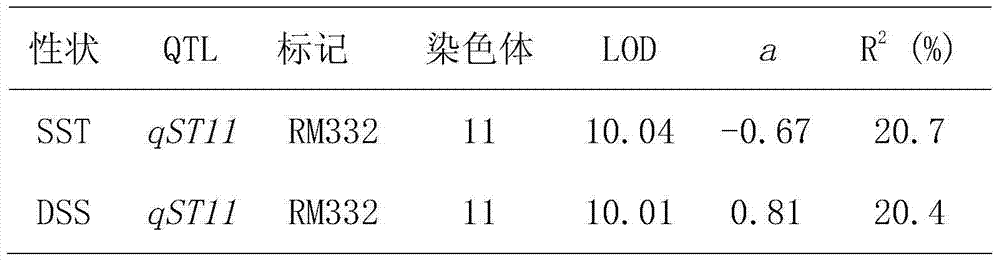
The invention relates to a rice seedling-stage salt-tolerant gene qST11 and a molecular marker method thereof and belongs to the fields of rice stress-resistance breeding and molecular genetics. The rice seedling-stage salt-tolerant gene qST11 is characterized in that the BC1F10 introgression line group constructed by the Zhongguangxiang No. 1 and the IR75862 as well as the F2 group constructed by cross derivation of the introgression lines are utilized and the single-marker analysis is used, so that the gene site qST11 related to the rice seedling-stage salt tolerance at the eleventh chromosome of the rice genome is positioned and a practical and economical marker RM332 which is closely linked to the gene site qST11 and is on the basis of the PCR amplification is obtained. The rice seedling-stage salt-tolerant gene qST11 is used in assisted selection breeding and polymerization breeding of rice seedling-stage salt tolerance, so that the defect of lack of salt-tolerant gene in prior art is effectively made up and the genotype selection to the low-generation breeding population in the seedling stage is realized; thus salt-tolerant individuals are obtained and timely hybrid trans-breeding is facilitated, so that the errors of phenotypic identifications during the salt tolerance identification process are reduced, the breeding efficiency is improved and the breeding process is fastened.
Owner:YANGTZE UNIVERSITY +1
Methods, Mixtures, Kits and Compositions Pertaining to Analyte Determination
ActiveUS20120196378A1Organic chemistryParticle separator tubesAnalyteMass Spectrometry-Mass Spectrometry
This invention pertains to methods, mixtures, kits and compositions pertaining to analyte determination by mass spectrometry using labeling reagents that comprise a nucleophilic reactive group that reacts with a functional group of an analyte to produce a labeled analyte. The labeling reagents can be used as isobaric sets, mass differential labeling sets or in a combination of isobaric and mass differential labeling sets.
Owner:DH TECH DEVMENT PTE
Electronic document processing device, electronic document display device, electronic document processing method, electronic document processing program, and storage medium
InactiveCN102549541ARead efficientlyEliminate repeated jumpsDigital data information retrievalText processingElectronic documentMarker analysis
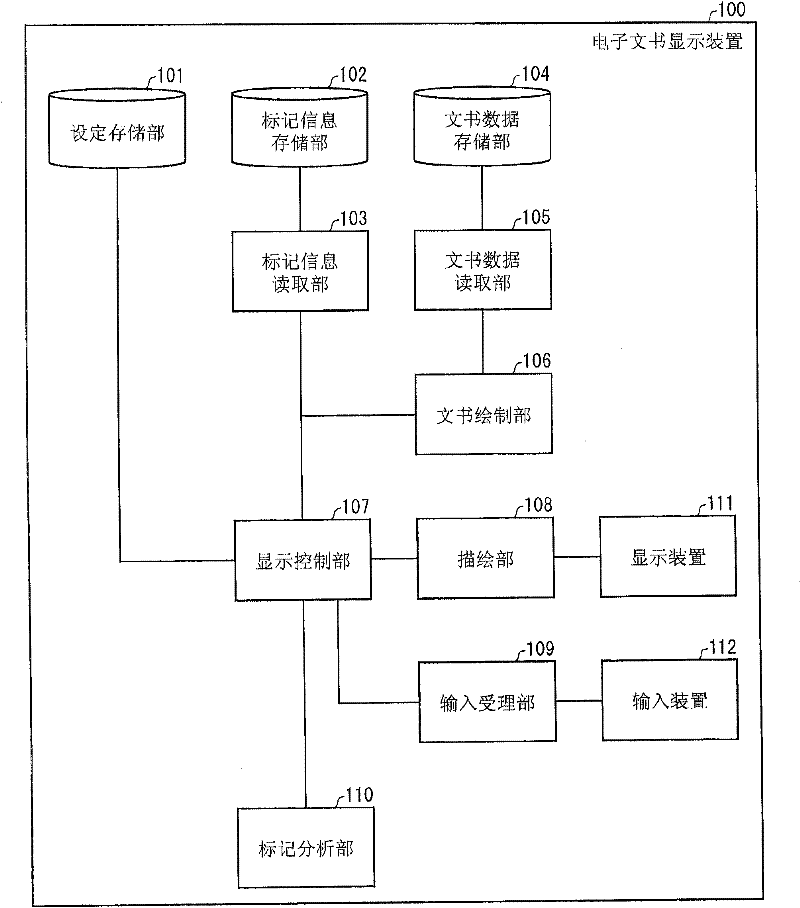
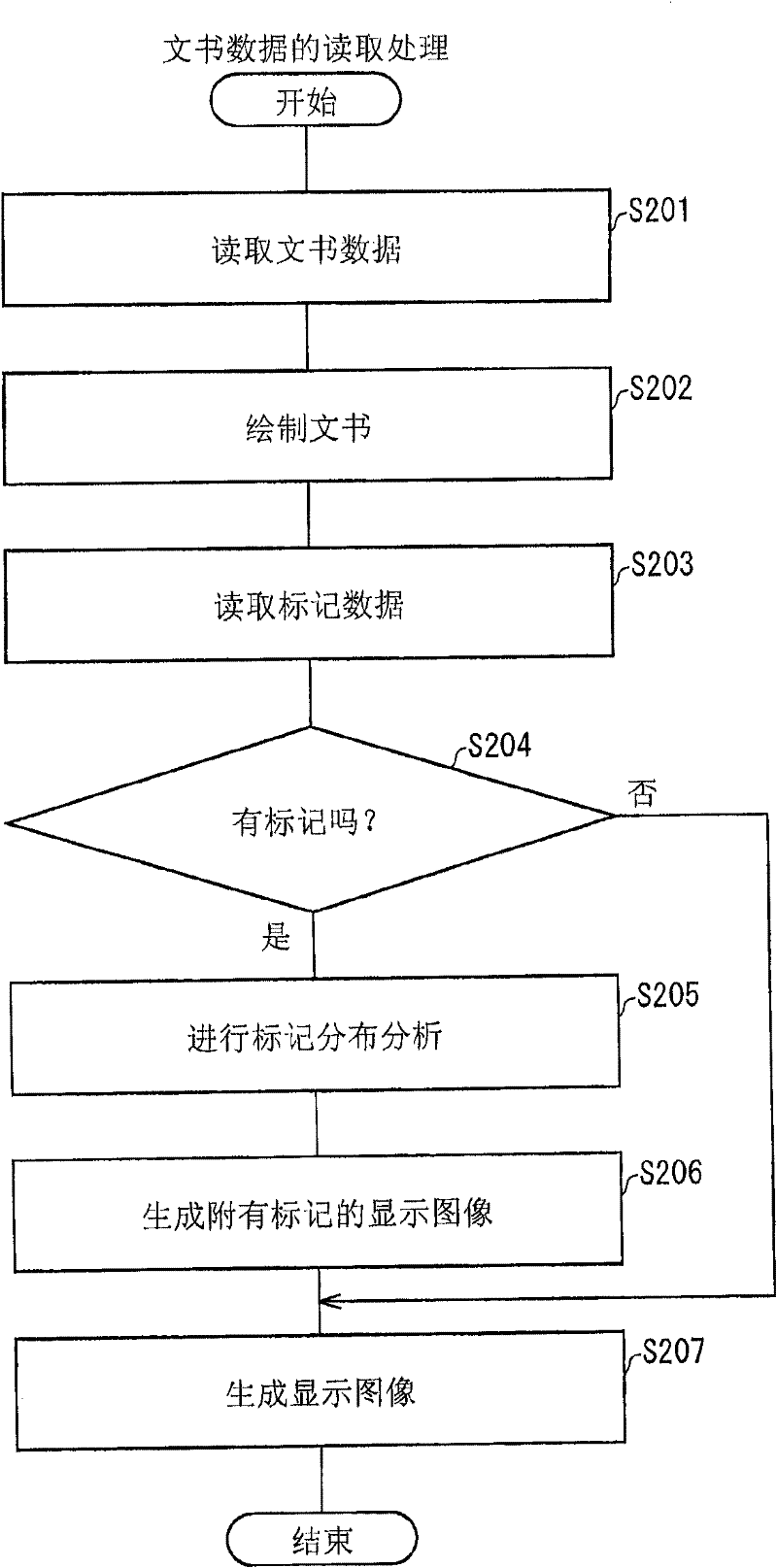
Disclosed is an electronic document display device (100) provided with a marker analysis unit (110) which analyzes distribution of markers in an electronic document; an input receiving unit (109) which receives designation of a range in the electronic document to be displayed on a display unit (111); and a display control unit (107) which, on the basis of the distribution of markers that has been analyzed by the marker analysis unit (110), displays on the display unit (111) a document-range that is different from the range that has been received by the input receiving unit (109), so as to increase the possibility that marker locations will be displayed on the display unit (111).
Owner:SHARP KK
Indel molecular marker of asparagus bean commonly used in different legume crops and development method and application thereof
ActiveCN110004245AImprove accuracyImprove selection efficiencyMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGenetic diversity
The invention discloses an Indel molecular marker of an asparagus bean commonly used in different legume crops and a development method and application thereof. The InDel molecular marker is preparedby using a legume DNA as a template and utilizing the following one or several sets of primer pairs as shown in SEQ ID NO.1-24 for PCR amplification. The method develops the InDel marker by performingtranscriptome sequencing on Suzi 41 and Susie 1419, performing BLAST alignment on transcriptome assembly sequences and screening for transcripts with insertion / deletion. After analyzing the feasibility of the developed InDel label, the universality of the different legume materials are analyzed by using the effective amplification marker obtained by screening. The InDel molecular marker has versatility on different legume materials, and can be used for genetic map construction, QTL localization, genetic diversity analysis and molecular marker assisted selection of the legume materials in thefuture.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
Method for detecting electrochemical luminescence
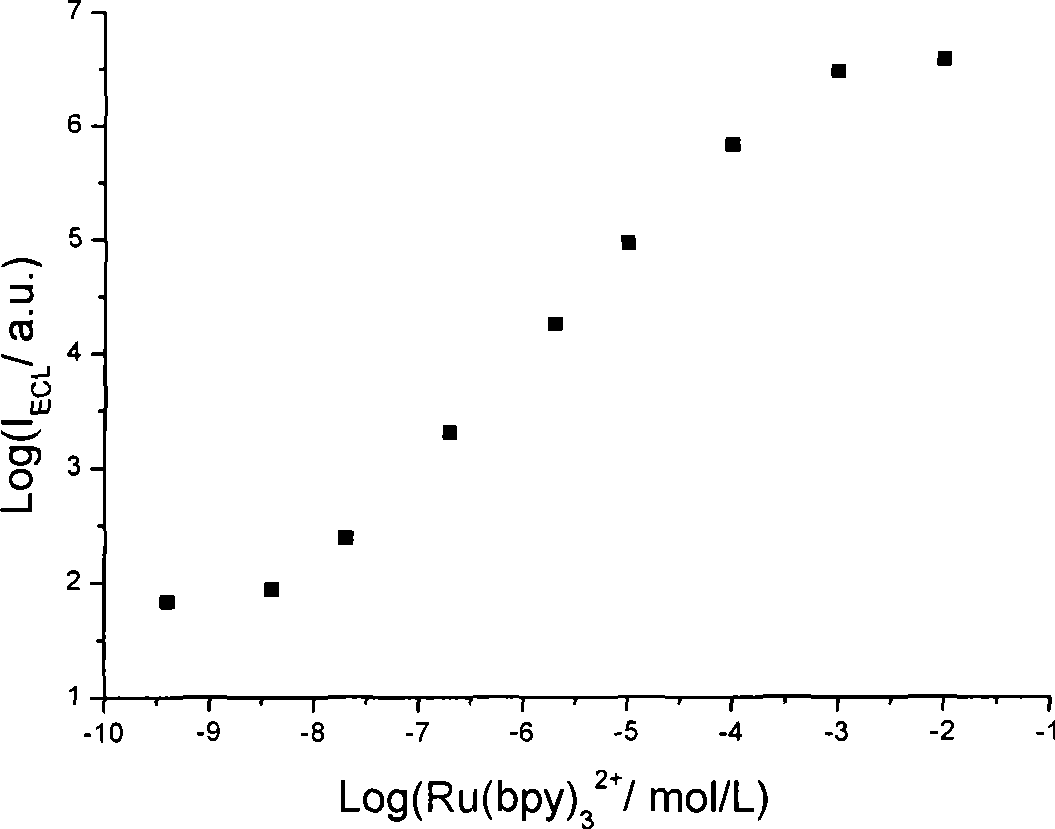
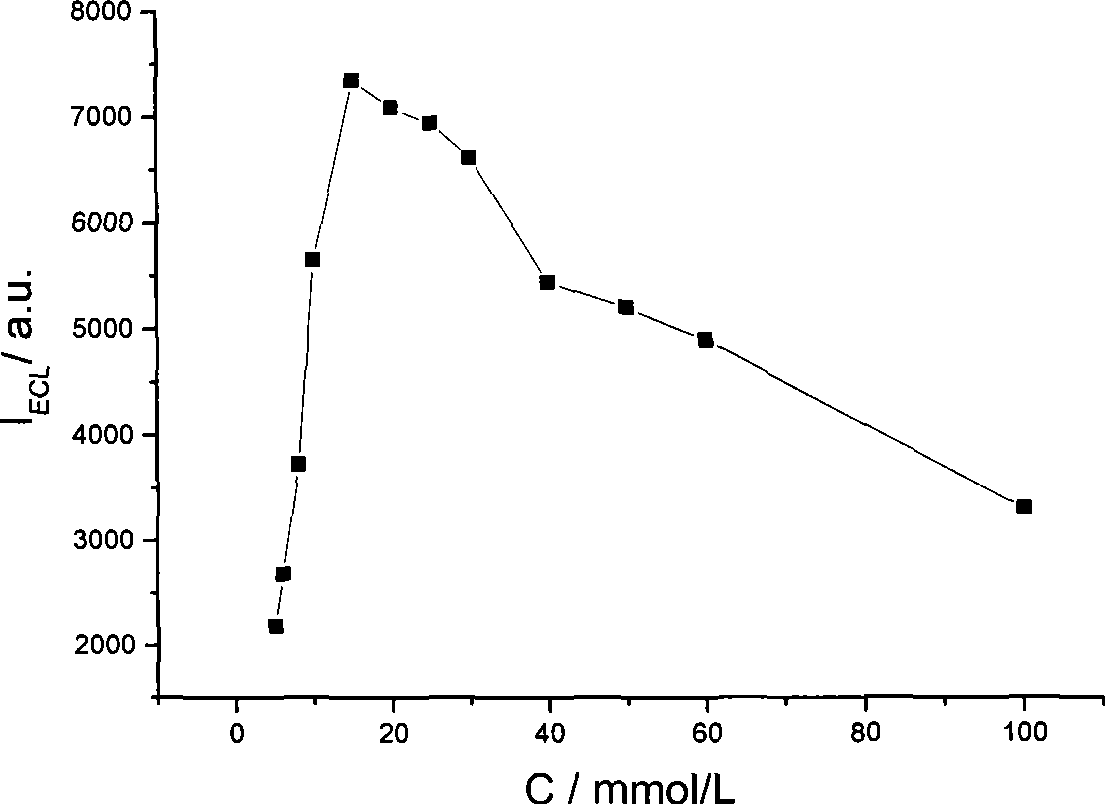
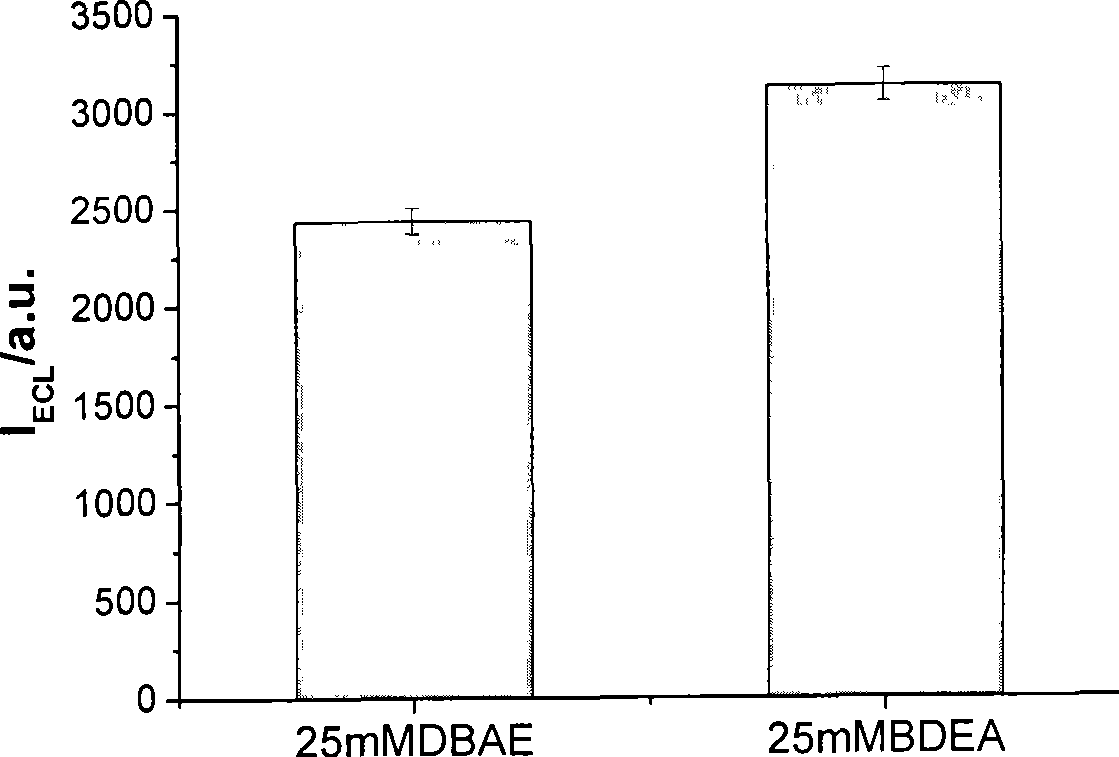
InactiveCN101532961AHigh ECL efficiencyReduce concentrationOrganic chemistryChemiluminescene/bioluminescenceElectrochemiluminescenceMarker analysis
The invention relates to a method for detecting electrochemical luminescence. A luminescence system of electrochemical luminescence detection is a pyridyl ruthenium / butyl diethanolamine luminescence system. Pyridyl ruthenium and butyl diethanolamine are mixed in phosphate buffer solution with pH between 7 and 9 to be simulated to be luminescent by an electrochemical method and analyzed. The concentration range of the pyridyl ruthenium is 1*10< [-10] >to 1mmol / L. The concentration range of the butyl diethanolamine is 0.05-50mmol / L. The electrochemical luminous efficiency of the pyridyl ruthenium / butyl diethanolamine luminescence system is high, and the concentration of co-reactant butyl diethanolamine can be very low. When the concentration of the pyridyl ruthenium is 1 mumol / L, and the concentration of the butyl diethanolamine is 25mmol / L, under the same condition, the electrochemical luminous intensity of the pyridyl ruthenium / butyl diethanolamine luminescence system at a gold electrode is approximately 1.3 times stronger than the electrochemical luminous intensity of the pyridyl ruthenium / butyl diethanolamine luminescence system. The invention can be widely applied to marker analysis.
Owner:CHANGZHOU INST OF ENERGY STORAGE MATERIALS &DEVICES
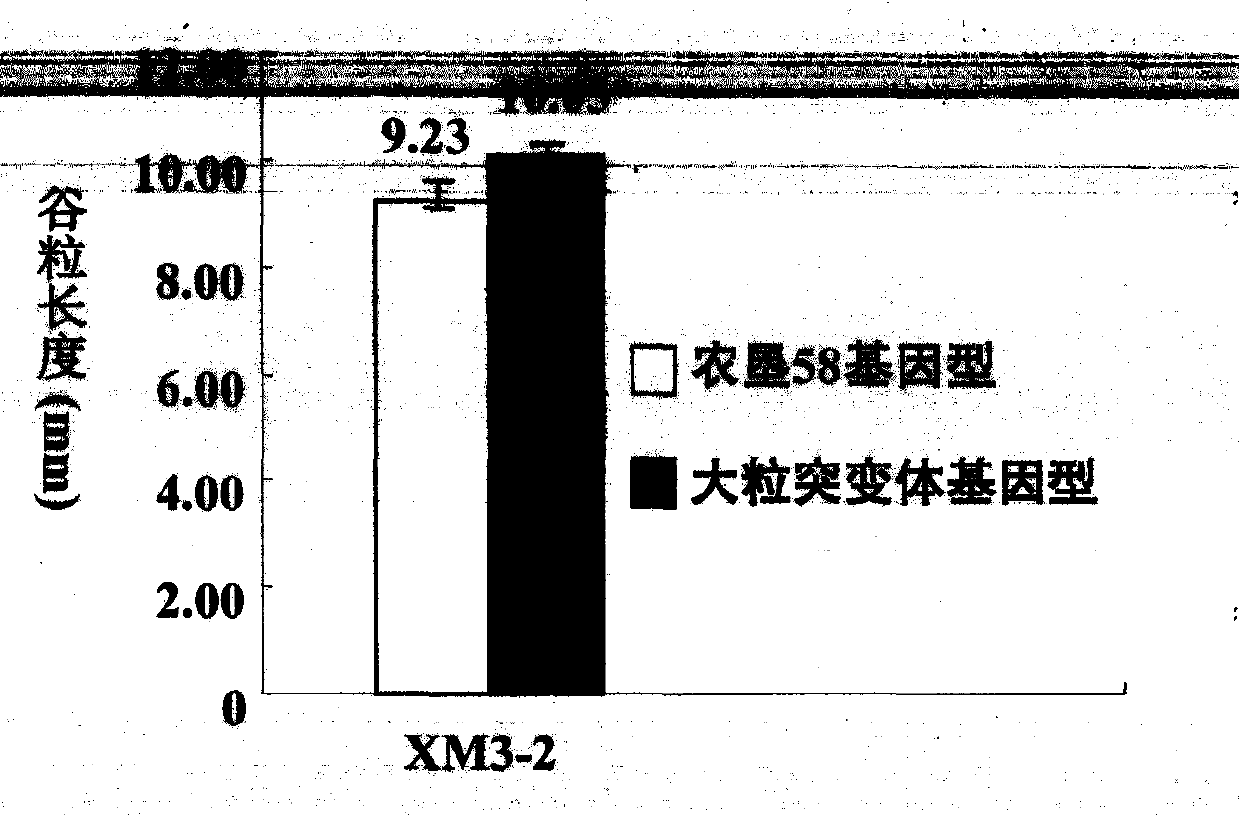
New space-mutated rice grain length gene qGS3a and molecular marker method thereof
InactiveCN103468715AGood appearanceGood yieldMicrobiological testing/measurementGenetic engineeringAgricultural scienceMarker analysis
The invention relates to a new gene qGS3a for simultaneously increasing rice grain length and grain weight and a molecular marker method thereof, belonging to the fields of rice high-yield good-quality breeding and molecular genetics. The invention is characterized in that single marker analysis is performed according to the linkage segregation law by using F2 population of large-grain mutant and original parent Nongken 58 and derivative F3 family to locate the new grain length gene qGS3a on the 3rd chromosome, thereby obtaining PCR (polymerase chain reaction) amplification-based practical and economical marker XM3-2 which is closely linked with the new qGS3a. The new gene qGS3a is used for auxiliary selective breeding and polymerization breeding of appearance quality and yield of rice, can effectively overcome the defects in spontaneous mutant genes, can perform genotype selection on the low-generation breeding population in the seedling stage to obtain the breeding material with better appearance quality and yield components, omits the grain identification process in the adult-plant stage, enhances the breeding efficiency and accelerates the breeding process.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
InDel molecular markers for assisted selection of early heading genes of rice and application of InDel molecule marker
ActiveCN111206113AAccelerate cultivationMicrobiological testing/measurementDNA/RNA fragmentationMarker analysisGenome resequencing
The invention discloses InDel molecular markers for assisted selection of early heading genes of rice and application of the InDel molecule markers, and belongs to the technical field of molecular markers. A whole-genome resequencing BSA technology is utilized to initially identify a candidate region related to the heading stage of rice, and then the InDel markers which are developed through parental whole-genome resequencing are utilized to carry out marking analysis on 500 early-heading single plants in an F2 segregation population which is constructed through hybridization of 9311 / CL33 in the candidate region; and it is revealed that a PSM8-8 marker has coseparation with the genes, and finally the gene qEH8-1 of the early heading stage is finely positioned within a range of about 270 kbbetween InDel-markers PSM8-6 and PSM8-9. Amplification is performed on a rice plant genome through the designed Indel molecular marker PSM8-8, and whether plants have the characteristics of early heading or not is judged and detected through denaturing polyacrylamide gel electrophoresis.
Owner:GUANGXI ZHUANG AUTONOMOUS REGION ACAD OF AGRI SCI
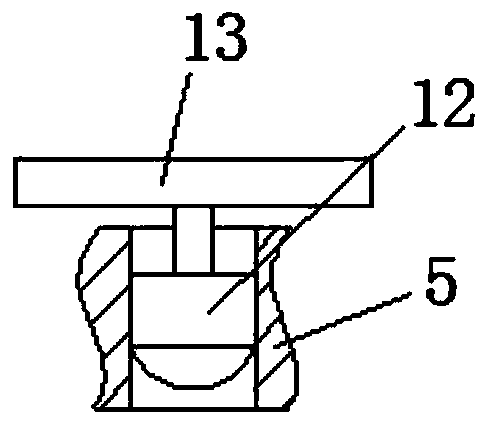
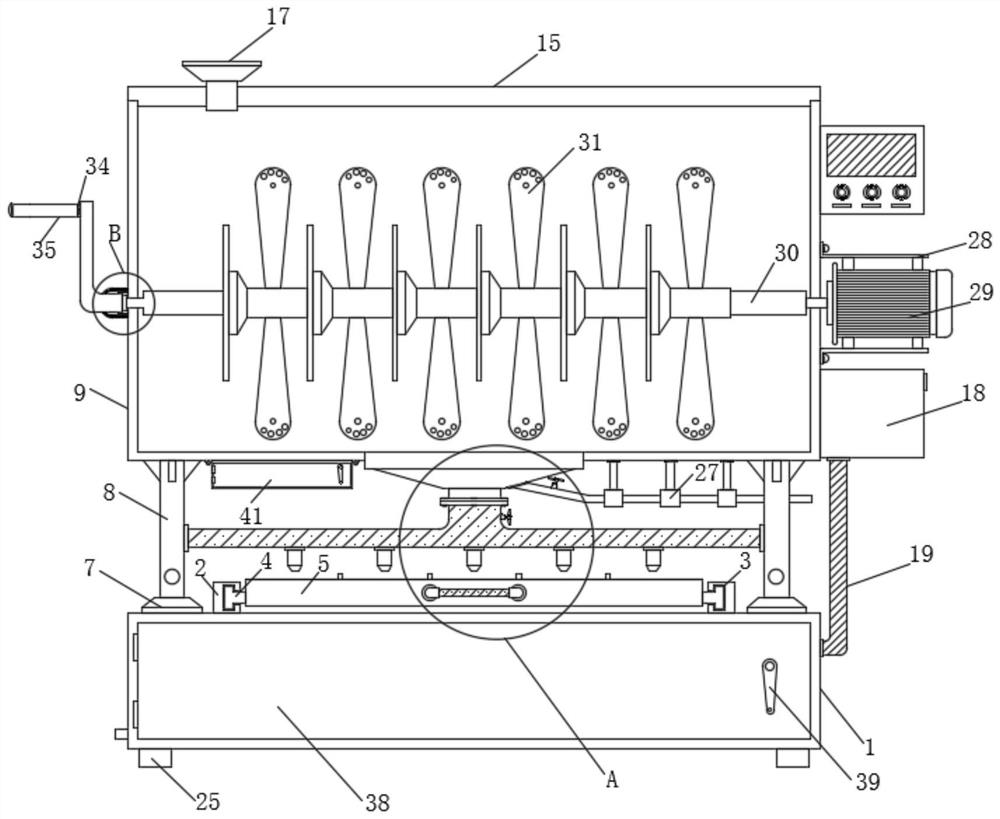
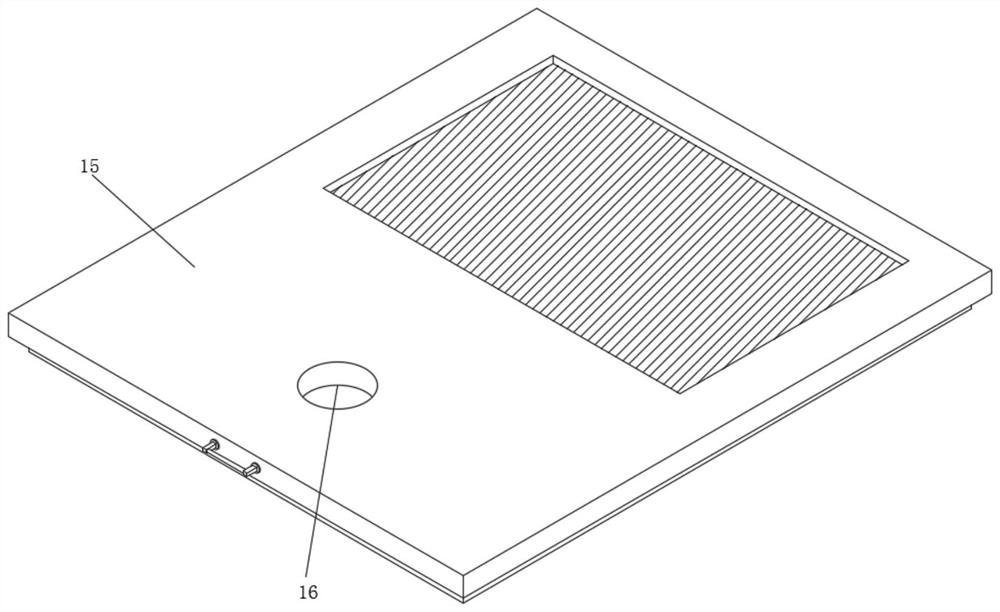
Blood tumor marker analysis device
InactiveCN111175115AWell mixedEasy to detectPreparing sample for investigationHematological TumorMarker analysis
The invention discloses a blood tumor marker analysis device. The device comprises an analysis box, wherein a cover plate is hinged to a front wall of the analysis box, supporting rods which are symmetrically distributed are fixedly mounted at the top of the analysis box, a first fixing plate and a second fixing plate are fixedly mounted on the supporting rods, the first fixing plate is positionedbelow the second fixing plate, the supporting rods which are symmetrically distributed are fixedly mounted at the top of the analysis box, the first fixing plate and the second fixing plate can be conveniently limited and fixed, a liquid inlet pipe is fixedly mounted at the top of a stirring roller, the liquid inlet pipe can conveniently drive the stirring roller to rotate through a first bearing, a driven gear disc is fixedly mounted on a liquid inlet pipe, so s motor drives s driving gear disc to rotate conveniently, the driving gear disc drives s driven gear disc to rotate, the stirring roller can be made to rotate, the stirring roller can better stir blood tumor markers, and the blood tumor markers can be mixed more uniformly.
Owner:新疆医科大学第三附属医院
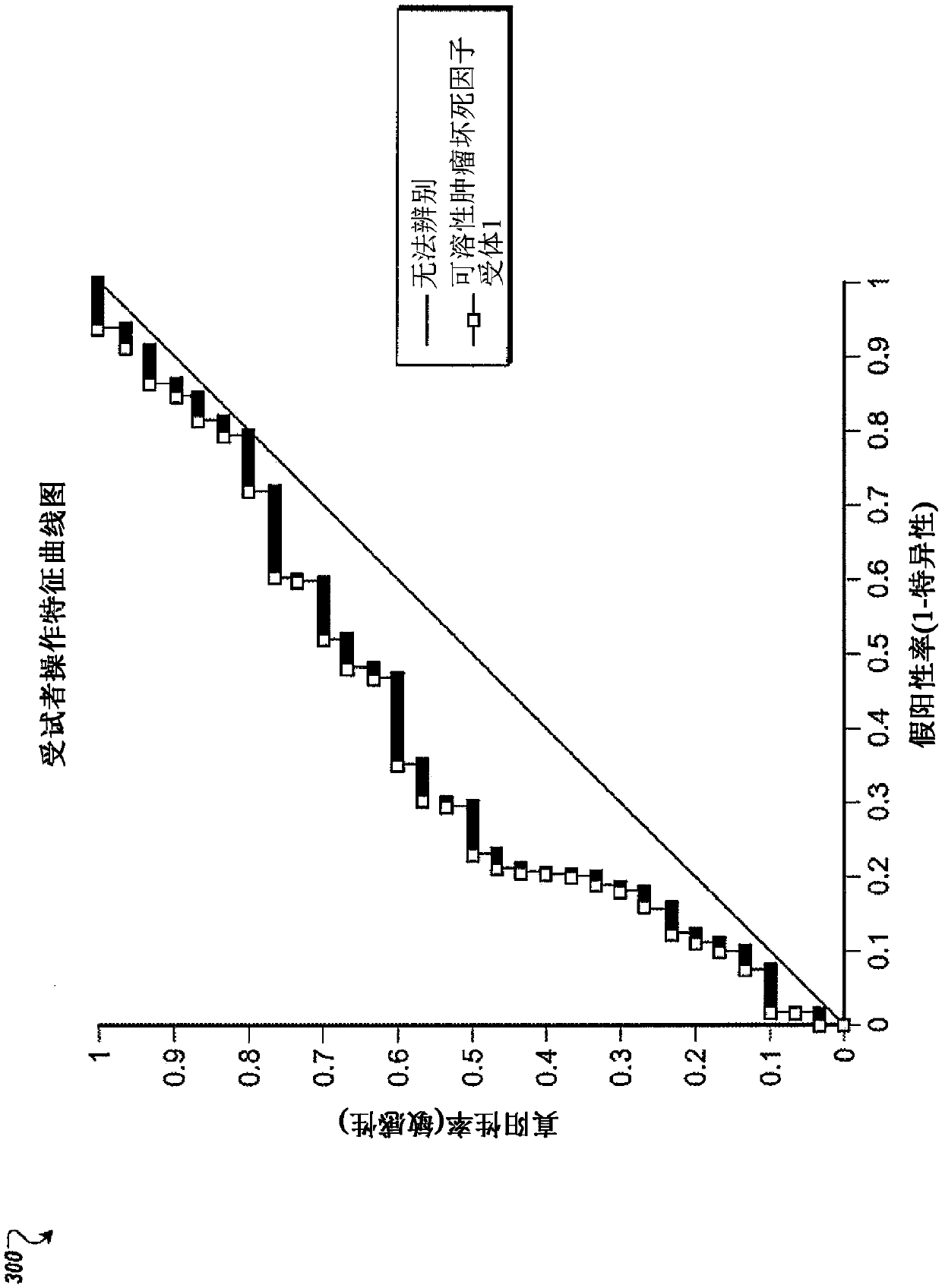
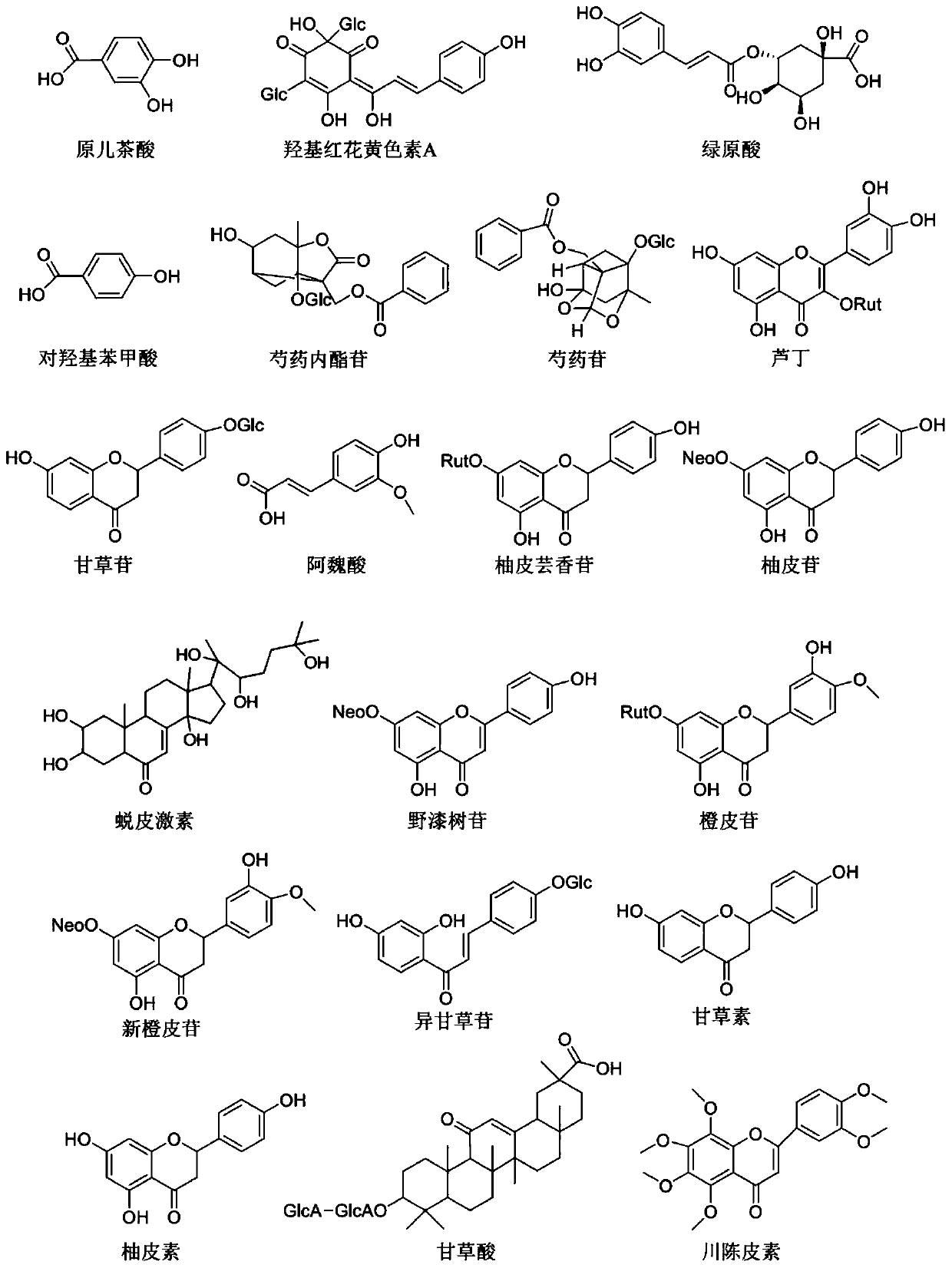
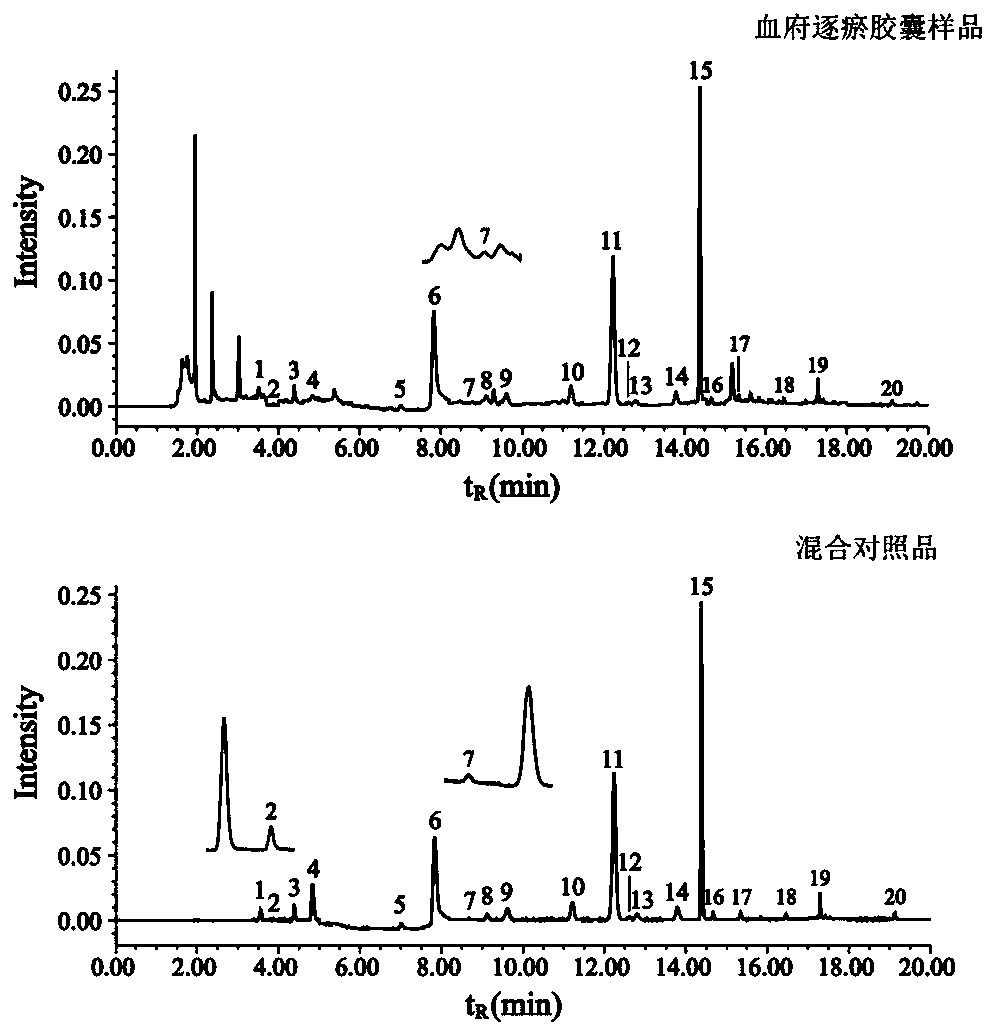
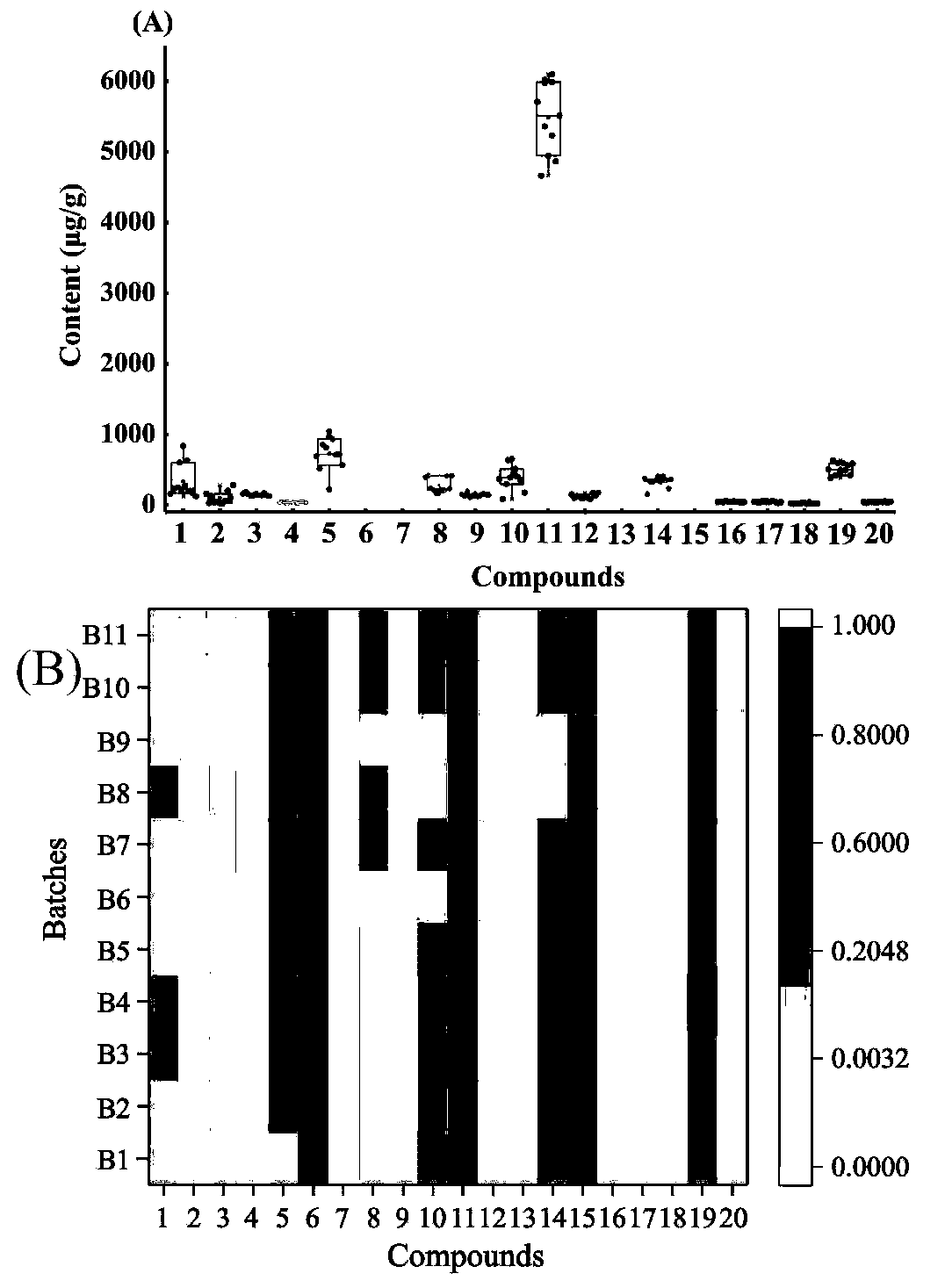
Xuefuzhuyu capsule quality control marker discovery method based on content-stability-activity multidimensional network mode
ActiveCN110441456AEnsure safetyGuaranteed validityComponent separationChemical compositionMarker Discovery
The invention relates to a Xuefuzhuyu capsule quality control marker discovery method based on a content-stability-activity multidimensional network mode. The method includes the following steps: stepone, obtaining a content dimension value of a Xuefuzhuyu capsule; step two, obtaining a stability value of the Xuefuzhuyu capsule; step three, obtaining an activity value of the Xuefuzhuyu capsule; and step four, constructing a content-stability-activity three-dimensional network for analysis, analyzing chemical components of the Xuefuzhuyu capsule by using regression areas of the three-dimensional network, and sorting the candidate components according to the regression areas to determine quality control markers of the Xuefuzhuyu capsule. The method selects the content, the stability and theactivity three dimensions of 20 candidate components of the Xuefuzhuyu capsule, constructs quality control marker analysis technology based on the content-stability-activity multidimensional networkmode, and evaluates comprehensive contribution of the candidate components in the compound by calculating the regression areas of the components, which provides a basis for optimizing quality controlindexes.
Owner:TIANJIN HONGRENTANG PHARM CO LTD
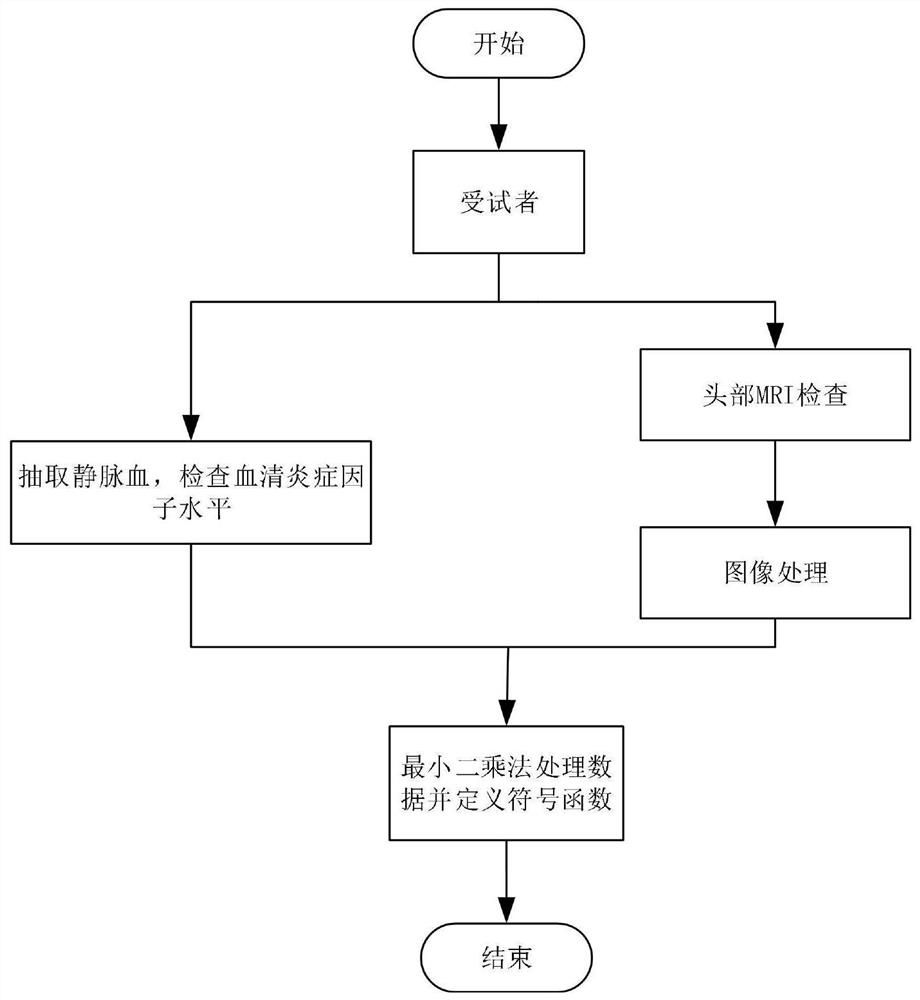
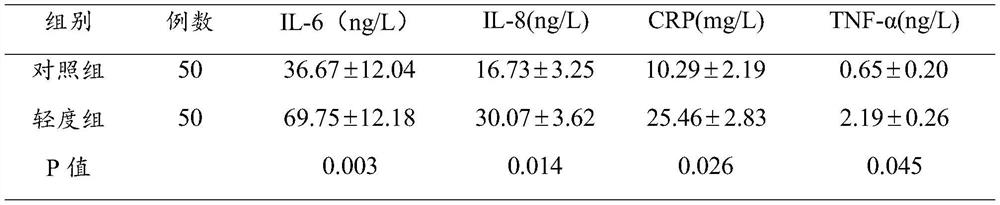
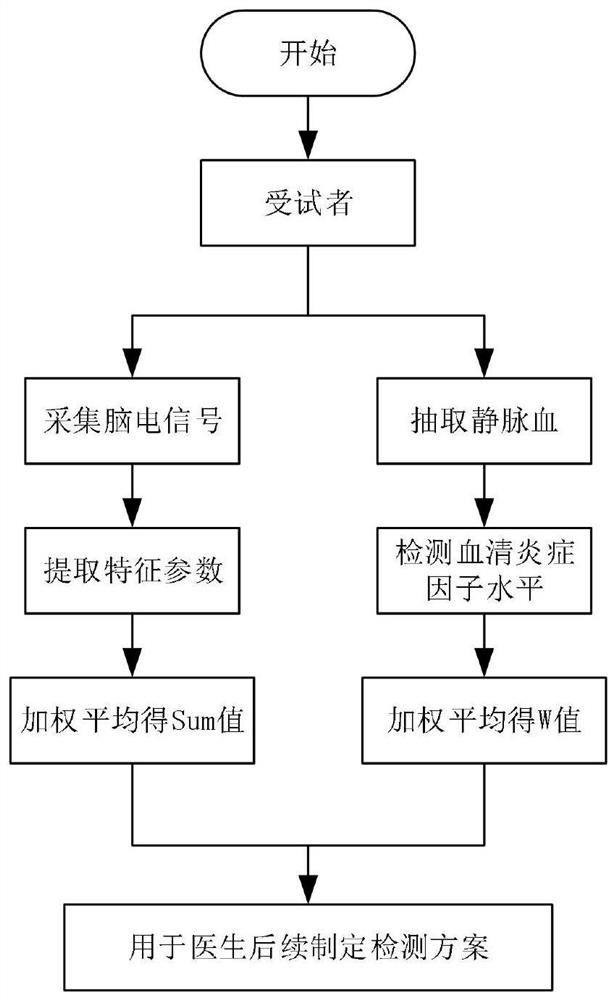
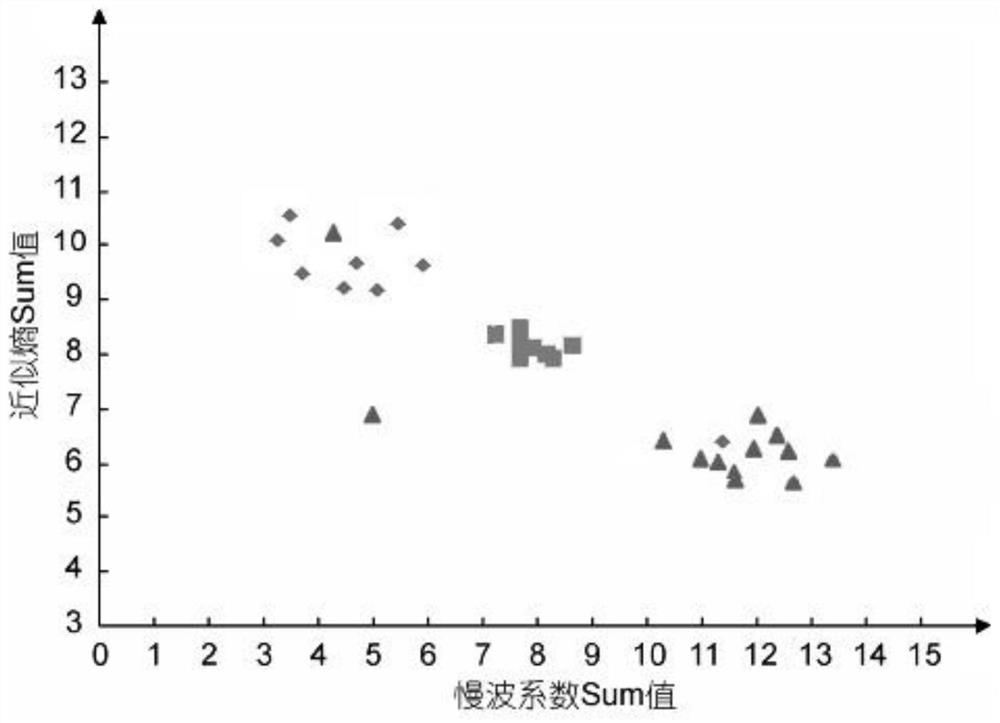
Brain injury marker analysis system based on DTI and serum factors
The invention relates to a brain injury marker analysis system based on diffusion tensor imaging (DTI) and a serum factor, the system comprises a DTI scanning module, a serum detection module and a data processing module, and the system is controlled according to the following modes: firstly, detecting the serum inflammatory factor level of venous blood extracted from an empty stomach by adopting an enzyme-linked immunosorbent assay; then, carrying out DTI scanning, carrying out image processing and data analysis on the collected images, and obtaining change data of the FA and the ADC; normalizing a serum inflammatory factor level parameter and an FA value and an ADC value obtained by magnetic resonance diffusion imaging by using a least square method, then defining a sign function, and taking a result of the sign function as a reference for a doctor to analyze and formulate a next detection scheme.
Owner:YANSHAN UNIV
Resource system evaluation and screening method of pholiota nameko flowers
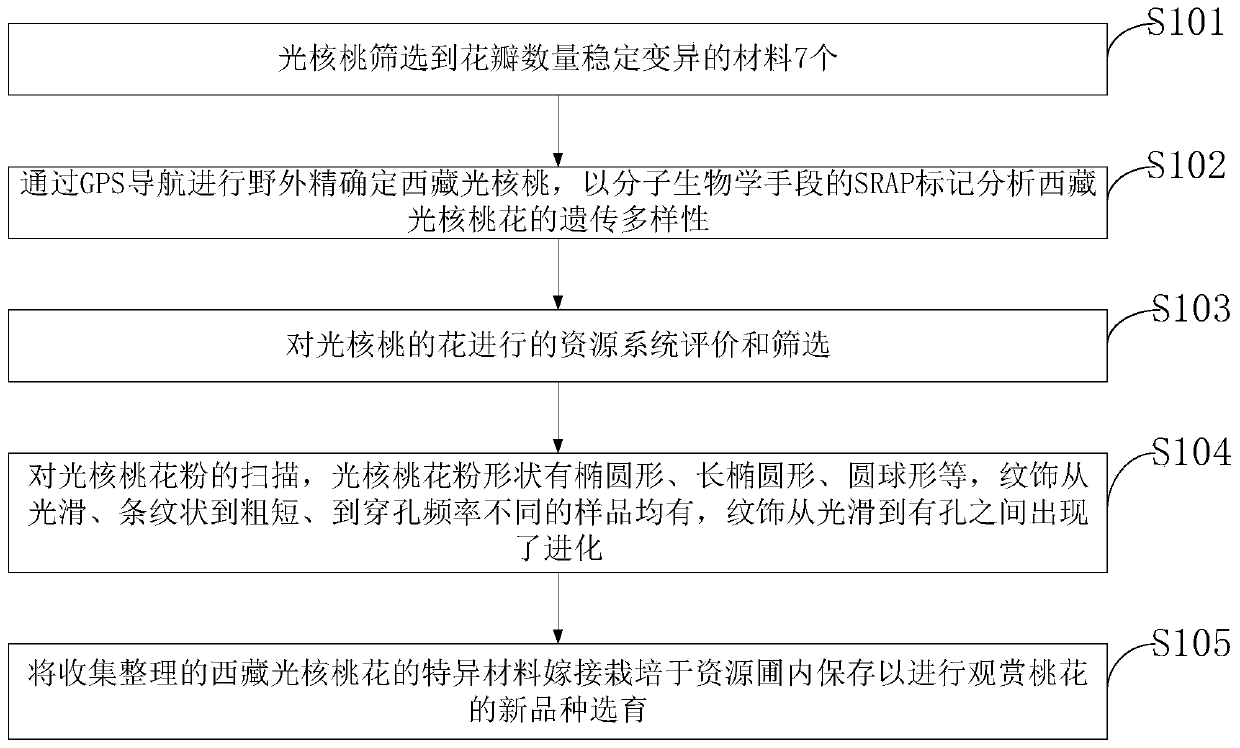
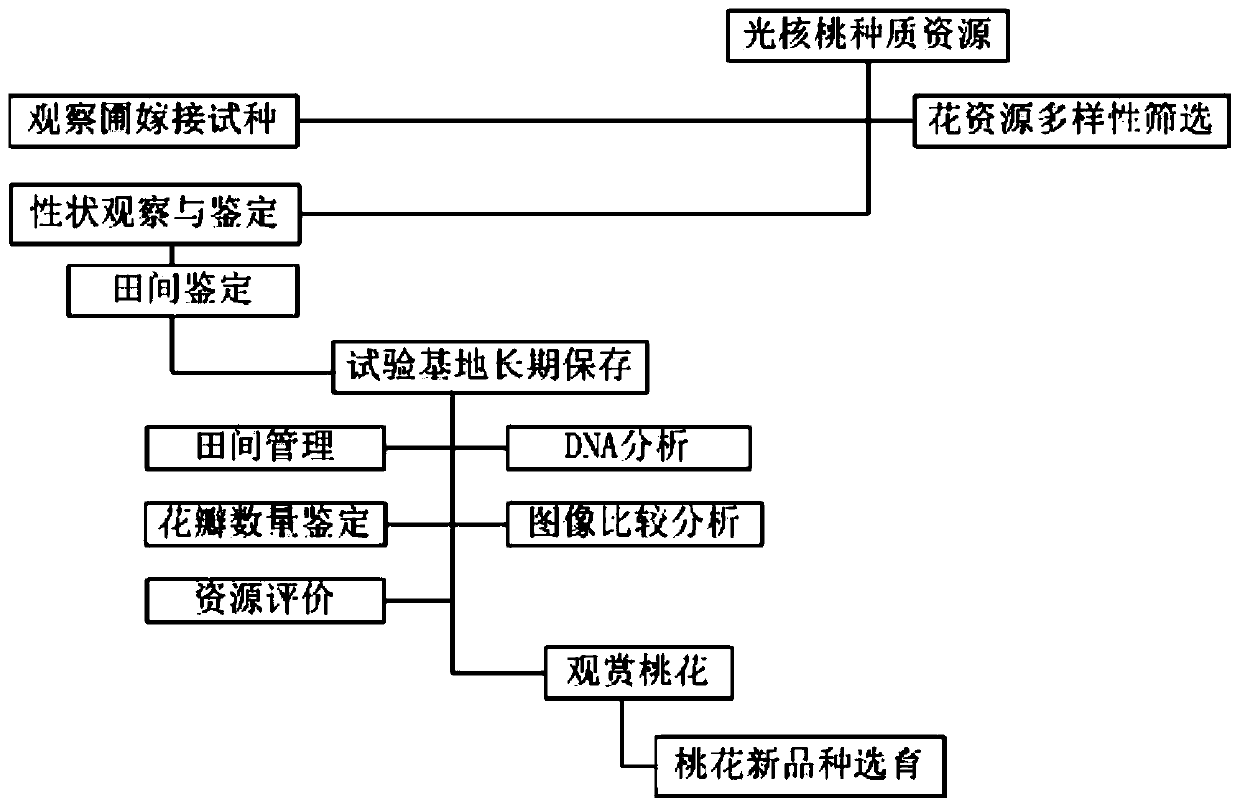
PendingCN111024897ARaise the level of developmentUnique ornamental charmMaterial analysis using wave/particle radiationPreparing sample for investigationPrunus miraMarker analysis
The invention belongs to the technical field of light walnut flowers and discloses a resource system evaluation and screening method of Tibetan light walnut flowers. The method comprises steps that characteristics of Tibetan wild light walnut flowers are observed and screened to obtain seven materials with stable and variable petal number; the Tibetan light walnuts are accurately positioned in thefield through GPS navigation, and genetic diversity of the Tibetan light walnut flowers is analyzed through SRAP markers of molecular biological means; resource system evaluation and screening of theflowers of the light walnuts are performed; when the walnut pollen is scanned, the ornamentation is evolved from smooth to porous; the collected and sorted specific resources of the Tibetan light walnut flowers are grafted and cultivated in a resource garden. The method is advantaged in that screening and evaluation of Tibetan light walnut flower specific resources and related reports of the system in domestic and overseas literatures and time limits are not found, and the method is provided for breeding of new varieties of Tibetan light walnut ornamental peach flowers.
Owner:VEGETABLE RES INST OF TIBET ACADEMY OF AGRI & ANIMAL HUSBANDRY SCI
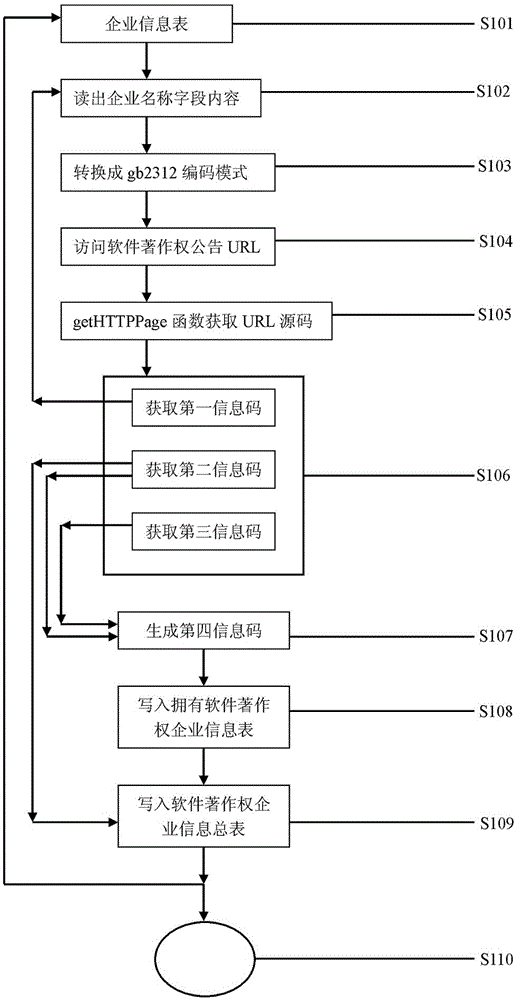
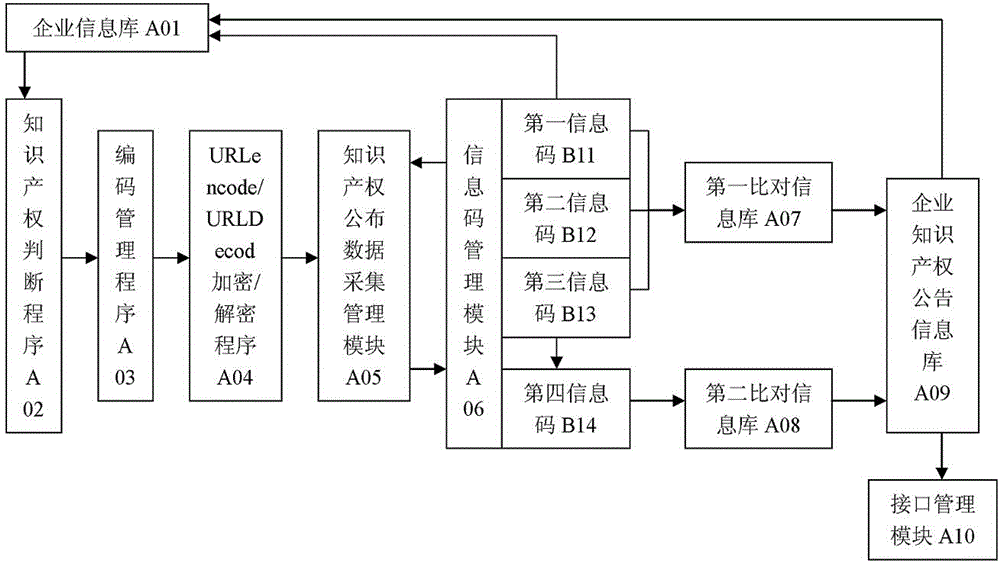
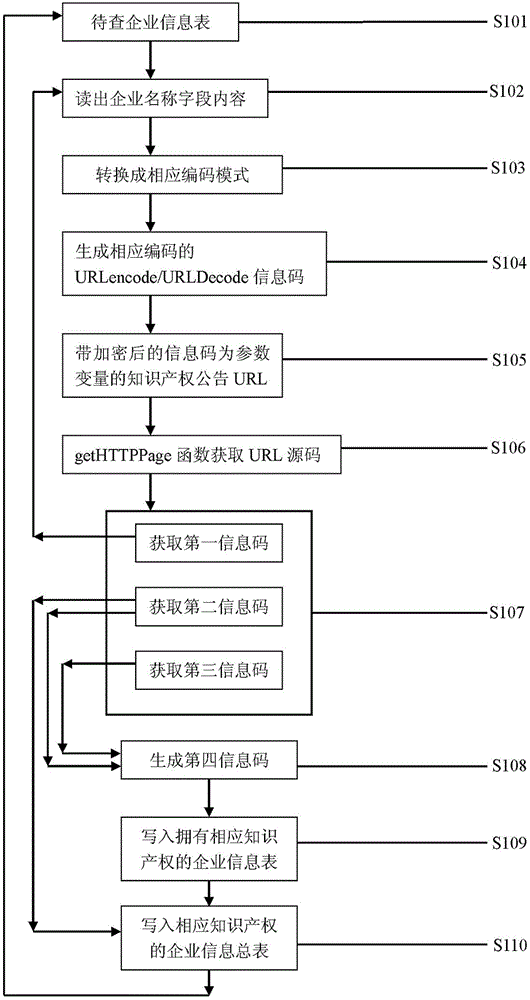
Enterprise software copyright announcement information grasping and managing system
InactiveCN105160472AResourcesSpecial data processing applicationsInformation repositoryIntellectual property
The invention discloses an enterprise software copyright announcement information grasping and managing system. The invention discloses a method, wherein a getHTTPPage method is used in a page-level technology for grasping public registration or change announcement data of software copyright; the grasping technology is combined with a marker analysis method to obtain a first information code, a second information code, and a third information code; by comparing the information codes, a fourth information code is generated by using a corresponding program; and according to a corresponding method, the fourth information code is written into a first intellectual property information base and a second intellectual property information base so as to be used in different scenarios.
Owner:FOSHAN HENGNAN MICRO TECH
Pre-clinical rapid experiment method of drug
InactiveCN106388966ASpeed up the development processReduce dosageDiagnostic recording/measuringSensorsDiseaseMarker analysis
The invention relates to a pre-clinical rapid experiment method of a drug. The method comprises the following steps: constructing an experiment animal disease model; feeding the drug to an experiment animal for the first time; after pre-set time, starting to carry out trace blood drawing and carrying out drug-time curve analysis on a blood sample; feeding the drug to the experiment animal for a plurality of times; observing a treatment effect on diseases and drug toxicity; after the drug effect is finished, feeding the drug to the experiment animal for the last time and carrying out the trace blood drawing again; analyzing an accumulation condition of the drug; dissecting the animal and carrying out mass spectrum scanning analysis on organ slices to obtain distribution data of the drug in each organ; proving pharmacology and toxicology according to the distribution data; and taking relative biological matrixes and carrying out bio-marker analysis. According to the pre-clinical rapid experiment method of the drug, provided by the invention, the pre-clinical experiment time can be greatly shortened and a drug development progress is accelerated; pharmacokinetic-pharmacological function-toxicology correlation analysis is relatively accurate; and injuries to the animal are reduced and the pre-clinical experiment cost is reduced.
Owner:GUANGDONG RANGER BIOSCI CO LTD
Brain injury marker analysis system based on EEG and serum inflammatory factor analysis
Owner:YANSHAN UNIV
Enterprise intellectual property information capture and management system
The invention discloses an enterprise intellectual property information capture and management system. According to the system, based on a technology for capturing the page level of registration or announcement change data, open to the public, of three common intellectual properties including patents, trademarks and software copyrights, a first information code, a second information code and a third information code are obtained in combination with a marker analysis method and are compared; a fourth information code is generated through a corresponding program; and with corresponding methods, the information codes are written in a first intellectual property information base and a second intellectual property information base for later use in different occasions.
Owner:FOSHAN HENGNAN MICRO TECH
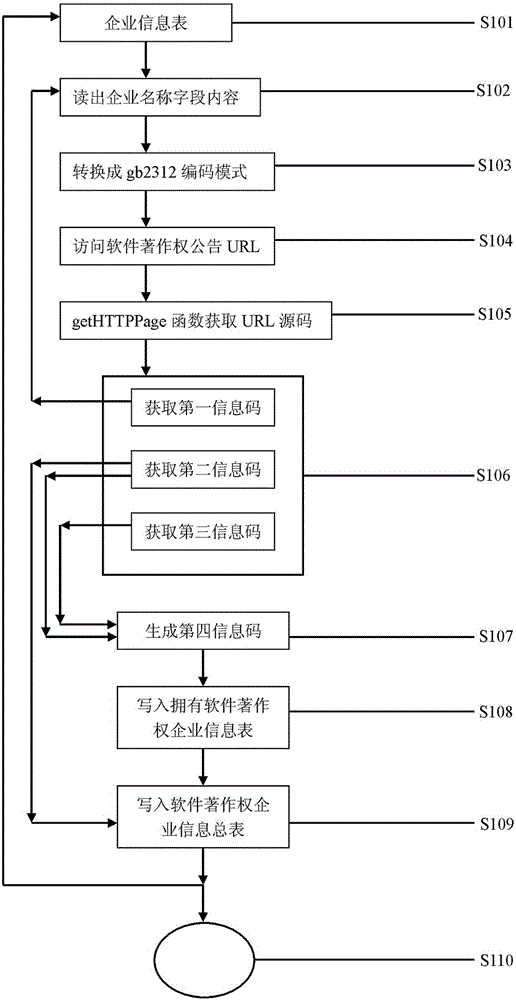
Enterprise software copyright announcement information capture and management method
InactiveCN105139309AData processing applicationsSpecial data processing applicationsInformation repositoryMarker analysis
The invention discloses an enterprise intellectual property information capture and management method which captures information based on the page level of public registration or announcement data change of software copyright through a getHTTPPage method, obtains a first information code, a second information code and a third information code based on a marker analysis method, compares the information codes, generates a fourth information code under a corresponding program, and writes the information codes into a first intellectual property information database and a second intellectual property information database according to corresponding methods in order to be used on different occasions.
Owner:FOSHAN HENGNAN MICRO TECH
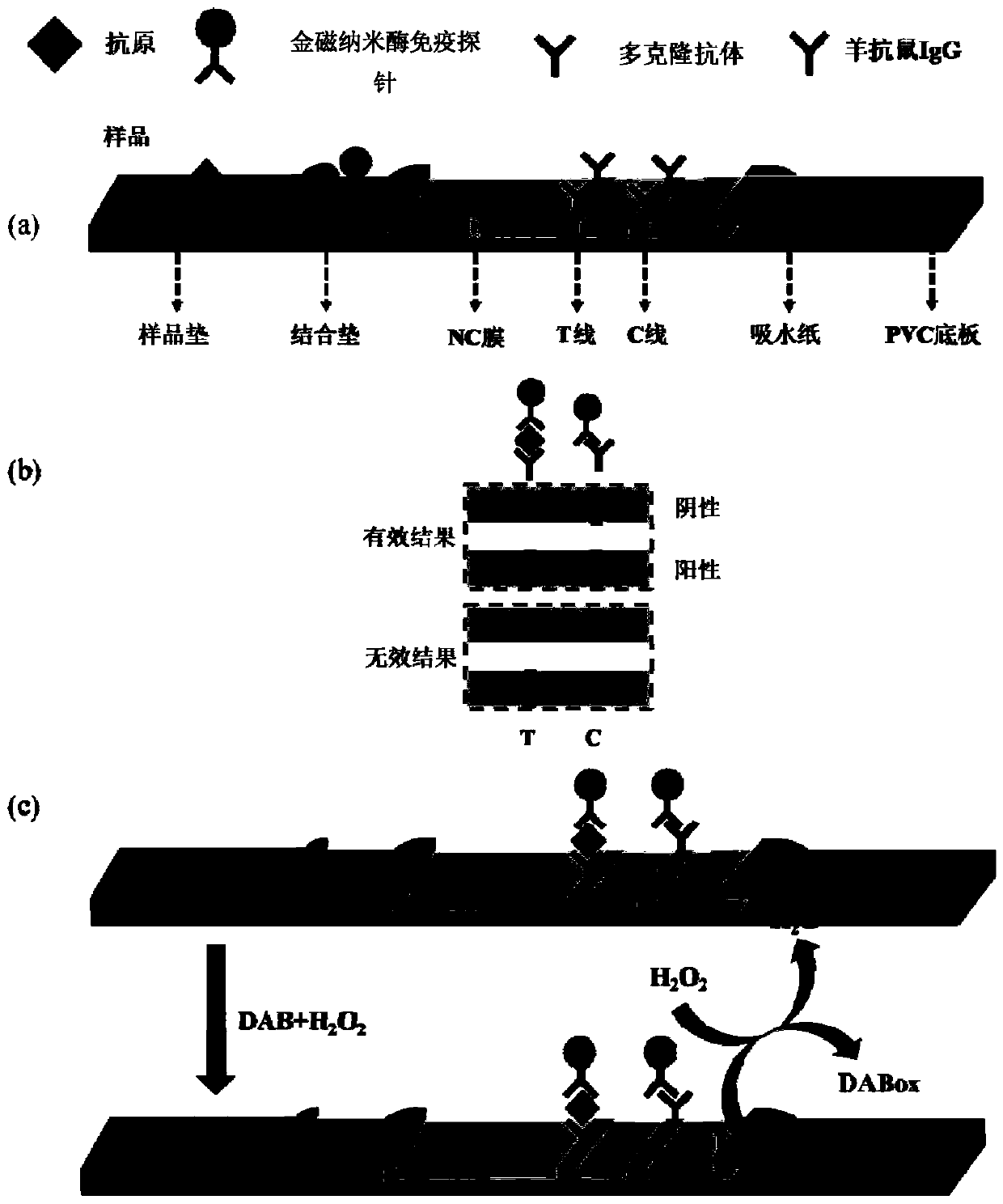
Lateral flow immunochromatographic assay test paper based on gold magnetic nano-enzyme immune probe as well as preparation method and application of lateral flow immunochromatographic assay test paper
ActiveCN111596065AImprove the rate of enzyme-catalyzed color reactionSimple processBiological testingMarker analysisEnzyme catalysis
The invention discloses lateral flow immunochromatographic assay test paper based on a gold magnetic nano-enzyme immune probe, as well as a preparation method and an application of the lateral flow immunochromatographic assay test paper, and belongs to the technical field of biological analysis. The preparation method comprises the following steps of: preparing Fe3O4 / Au composite nanoparticles byadopting an in-situ reduction method, then preparing the gold magnetic nano-enzyme immune probe by adopting a physical or chemical method, and finally finishing membrane material treatment and assembly of lateral flow immunochromatographic test paper to obtain the lateral flow immunochromatographic assay test paper based on the gold magnetic nano-enzyme immune probe. In the preparation method, more enzyme catalytic active sites are provided by utilizing a Fe3O4 / Au composite nanoparticle phase, an enzyme catalytic chromogenic reaction rate is effectively improved, and the preparation method hasthe advantages of simple process, low cost and easiness in large-scale production; and compared with the existing lateral flow immunochromatographic test paper, the lateral flow immunochromatographicassay test paper based on the gold magnetic nano-enzyme immune probe has the advantages that the detection sensitivity can be improved by 2-3 orders of magnitudes, the specificity of a detection system is high, and an efficient detection technology is provided for marker analysis with low detection limit.
Owner:SHAANXI UNIV OF SCI & TECH
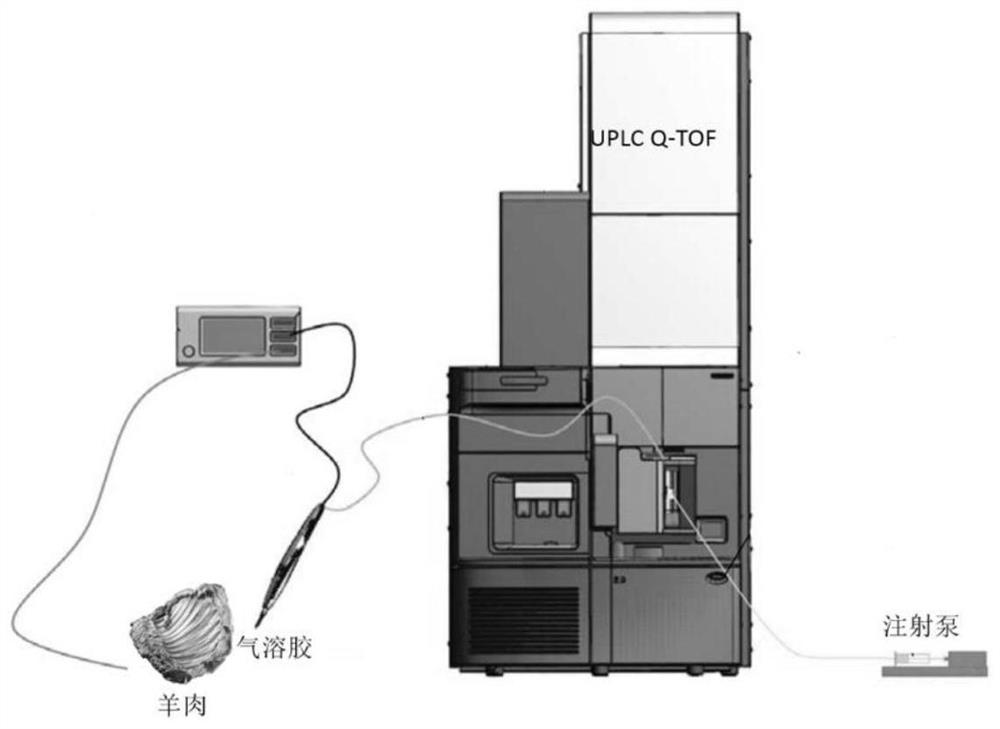
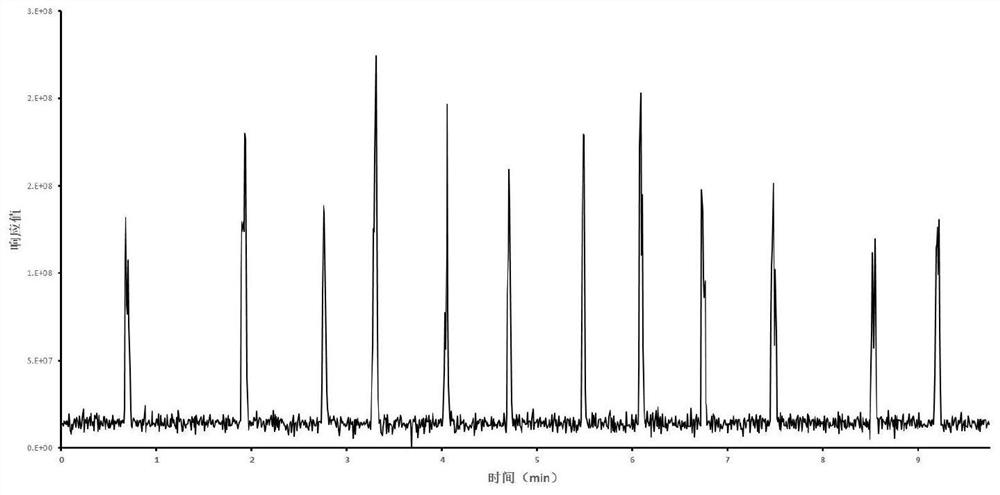
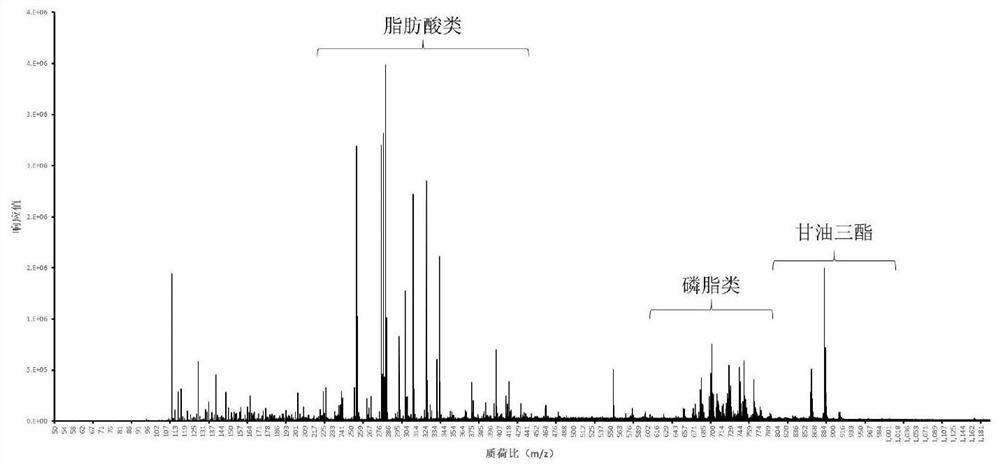
Detection method applied to origin tracing based on IKnife-REIMS technology
PendingCN114062477AShort analysis timeThe result is stablePreparing sample for investigationMaterial analysis by electric/magnetic meansBiotechnologyResolution (mass spectrometry)
The invention discloses a method for rapidly identifying Shuanghuama sheep by adopting an REIMS rapid evaporation ionization mass spectrometry technology and a high-resolution mass spectrometry. The real-time source-tracing detection method for the Shuanghuama sheep is obtained by on-site collection of mutton samples from different producing areas, instrument acquisition parameter optimization and model establishment. Recognition accuracy of the method is 97.81%; differential marker analysis is conducted on the data by using principal component analysis and a partial least square discriminant model, and a series of characteristic compounds with large differences are identified. The method does not need pretreatment, and is short in detection time, high in recognition rate and stable in result.
Owner:ZUNYI INST OF PROD QUALITY INSPECTION & TESTING
Reproduction updating method for soybean heredity integrity analysis
ActiveCN106386116AImprove reliabilityImprove accuracyPlant cultivationCultivating equipmentsMarker analysisMorphological trait
The invention provides a reproduction updating method for soybean heredity integrity analysis. The reproduction updating method includes: performing artificial aging treatment on parent seeds, selecting parent treatments at different germination rate gradients, raising the treated seeds, and transplanting the seedlings to a field; harvesting single plants; and performing seedling raising, transplanting, morphological trait investigation, and single plant harvesting on each descendant treatment according to the step of parent treatments. One of keys of the reproduction updating method is the fact that descendant single plants are corresponding to the parent single plants thereof in a one-to-one manner, i.e., the number of each single plant of each descendant treatment is corresponding to that of each parent single plant in a one-to-one manner; morphological marker analysis and SSR molecular marker analysis is performed during reproduction and updating of a parent generation and a descendant generation so as to detect the soybean heredity integrity. The reproduction updating method is complete and feasible, is normative in system, can reproduce and update stored high-viability soybean so as to maintain the heredity integrity of the soybean.
Owner:SHANDONG CROP GERMPLASM CENT
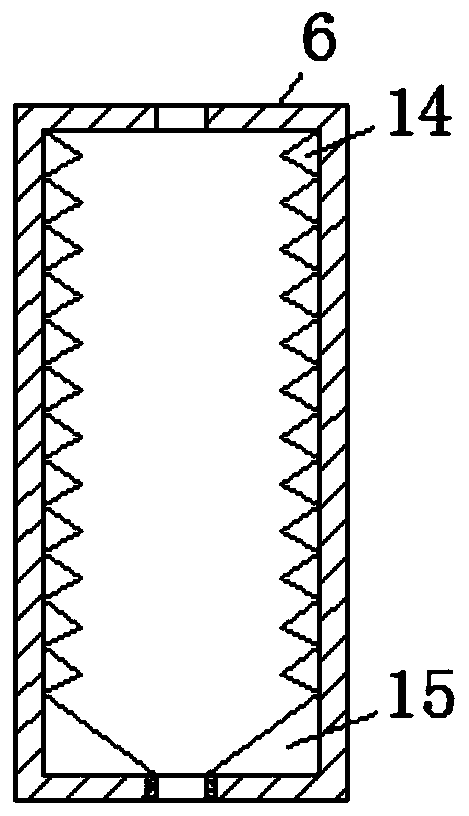
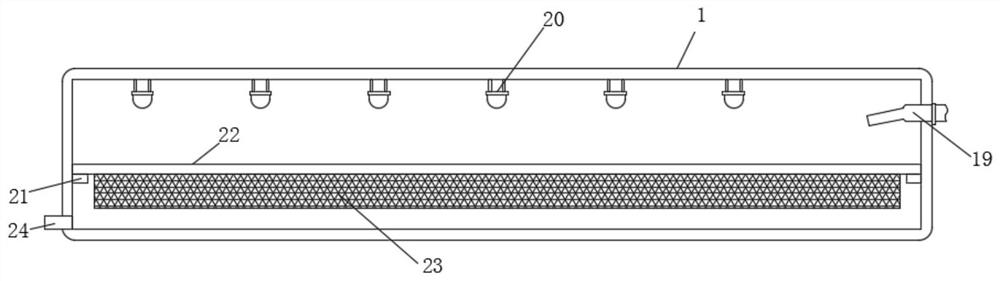
Hematologic tumor marker analysis device
InactiveCN114814187AEasy to useImprove practicalityPreparing sample for investigationHollow article cleaningMarker analysisUltraviolet lights
The invention discloses a blood tumor marker analysis device, and belongs to the technical field of blood analysis devices. The blood tumor marker analysis device comprises a rectangular box, a fixed block is fixedly connected to the upper surface of the rectangular box, a sliding groove is formed in one side of the fixed block, a sliding block is slidably connected to the interior of the sliding groove, an experiment box is fixedly connected to one side of the sliding block, a partition plate is fixedly connected to the interior of the experiment box, and the partition plate is fixedly connected to the interior of the rectangular box. Supporting seats are fixedly connected to the two sides of the upper surface of the rectangular box; after cleaning, a worker pulls a push-pull handle, so that an experiment box can be pulled out of a sliding groove, then the cleaned experiment box is placed in a steel wire net in a rectangular box, a drying machine is started, and water on the surface of the experiment box can drip to the inner bottom wall of the rectangular box and then is discharged through a water discharging pipe due to the arrangement of the steel wire net; the drying machine and the ultraviolet lamp can dry and disinfect the cleaned experiment box, so that the medical staff can use the experiment box conveniently.
Owner:化静
A Reproductive Renewal Method for Analysis of Wild Soybean Genetic Integrity
ActiveCN106358729BImprove reliabilityImprove accuracyFabaceae cultivationCultivating equipmentsBiotechnologyMarker analysis
The invention provides a reproduction and regeneration method applied to genetic integrity analysis of Glycine soja Sieb.et Zucc. The method comprises the following steps: performing artificial aging treatment on parental seeds, and selecting parent processes of different germination rate gradients; breaking hard seeds, growing seedlings, and transplanting the seedlings into a field; harvesting individual plants; treating filial generations, and performing seedling growing, transplanting, morphological character investigation and individual plant harvesting according to the same steps of the parent processes. Filial generation individual plants correspond to parental generations one by one, namely, the numbers of individual plants in processes of the filial generations correspond to parental individual plants one by one. In the reproduction and regeneration period of the parental generations and the filial generations, morphological marker analysis and SSR (Simple Sequence Repeat) molecular marker analysis are performed to detect the genetic integrity. A method for breaking hard seeds of the Glycine soja Sieb.et Zucc.. is innovated. The method is complete, feasible, systematic and standardized; the method can be applied to reproduction and regeneration of stocked Glycine soja Sieb.et Zucc.. with high activity in order to better maintain the genetic integrity.
Owner:SHANDONG CROP GERMPLASM CENT +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com