Indel molecular marker of asparagus bean commonly used in different legume crops and development method and application thereof
A molecular marker, bean technology, applied in biochemical equipment and methods, microbial determination/inspection, recombinant DNA technology, etc., to save costs, improve accuracy and selection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1 Extraction of Plant Material Leaf Genomic DNA
[0053] The seeds of bean materials were sterilized with 1% hypochlorous acid solution for 10 minutes, and then rinsed with distilled water for 5 times. Sow Su Zi 41, Su Cow 1419, 10 cowpea resources and 34 different bean crops in plastic cups filled with vermiculite (the lower diameter is 7 cm, the upper diameter is 9 cm, and the height is 15 cm), and then covered with 2 cm of vermiculite ;
[0054] DNA was extracted from Suzi 41 and Su cowpea 1419, 10 cowpea resources and 34 different bean plant leaves.
[0055] The total DNA of leaves was extracted by the CTAB method, and the specific steps were as follows:
[0056] (1) Take 0.1 gram of fresh leaf sample, add 650 μL of extract solution (1.4M NaCl, 100mM Tris, pH 8.0, 20mM EDTA, pH 8.0, 2% CTAB) to grind, then put it into a 1.5mL centrifuge tube and place it in a constant temperature water bath at 65°C 60 minutes, during which mixed 2-3 times;
[0057] (2) A...
Embodiment 2I
[0060] Example 2 Development of InDel molecular marker primers and screening of polymorphisms
[0061] (1) Use the lllumina Hiseq2000 platform to sequence the transcriptomes of cowpea varieties Suzi 41 and Su cowpea 1419, and perform BLAST comparison based on the transcriptome assembly sequences of Suzi 41 (salt-tolerant) and Su cowpea 1419 (salt-sensitive), and screen for Insertion / deletion (InDel) transcripts.
[0062] (2) Select sequences with relatively large differences in the sequence length of the insertion / deletion (InDel), and compare parameters: a. Alignment sequence ≥ 300bp, b. Insertion / deletion length ≥ 3bp, c. A pair of sequences of Suzi 41 and Suyu 1419 1. Alignment, d. The similarity of the two aligned sequences is greater than 90%. Then use Primer5.0 software to design InDel labeled primers, and the length of the amplified product is about 200bp.
[0063] (3) 845 pairs of InDel primers were synthesized by a biological company for polymorphism screening. The...
Embodiment 3
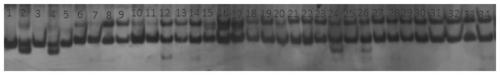
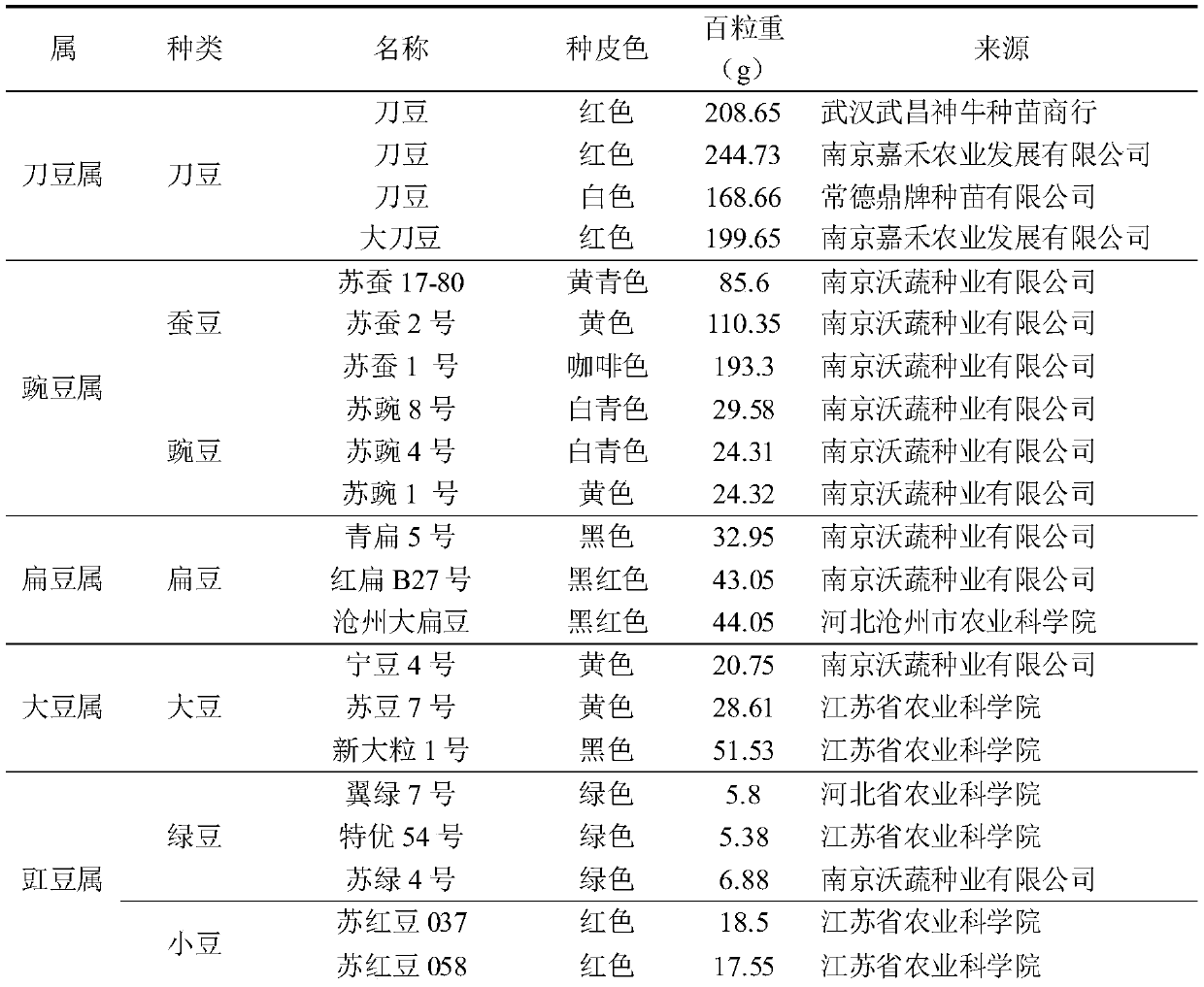
[0087] The feasibility of the developed InDel markers was analyzed using 10 cowpea resources (including long cowpea and rice cowpea). 107 pairs of marker primers were randomly selected from the InDel markers designed in Example 2, and PCR amplification and electrophoresis analysis were performed on 10 cowpea resources respectively, wherein the DNA extraction was the same as in Example 1, and the methods such as PCR amplification and electrophoresis were the same as in Example 2. The material names of 10 cowpea resources are shown in Table 2 (not limited to the materials in Table 2). After electrophoresis, the amplification rate was counted.
[0088] Table 2
[0089]
[0090]The results are shown in Table 3. There were 93 markers (87%) that could amplify bands in 10 cowpea materials, and 14 markers (13%) could only amplify bands in some cowpea materials. Among these markers, 98 markers (91.6%) had polymorphisms in two or more cowpea materials, and 9 markers (8.4%) had no p...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Lower mouth diameter | aaaaa | aaaaa |
| Upper mouth diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com