Larimichthys crocea genome-wide SNP and InDel molecular marker method based on double enzyme digestion
A genome-wide, molecular marker technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial assay/inspection, etc., to control the cost of marker development, low cost, and simplify genome building and sequencing.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
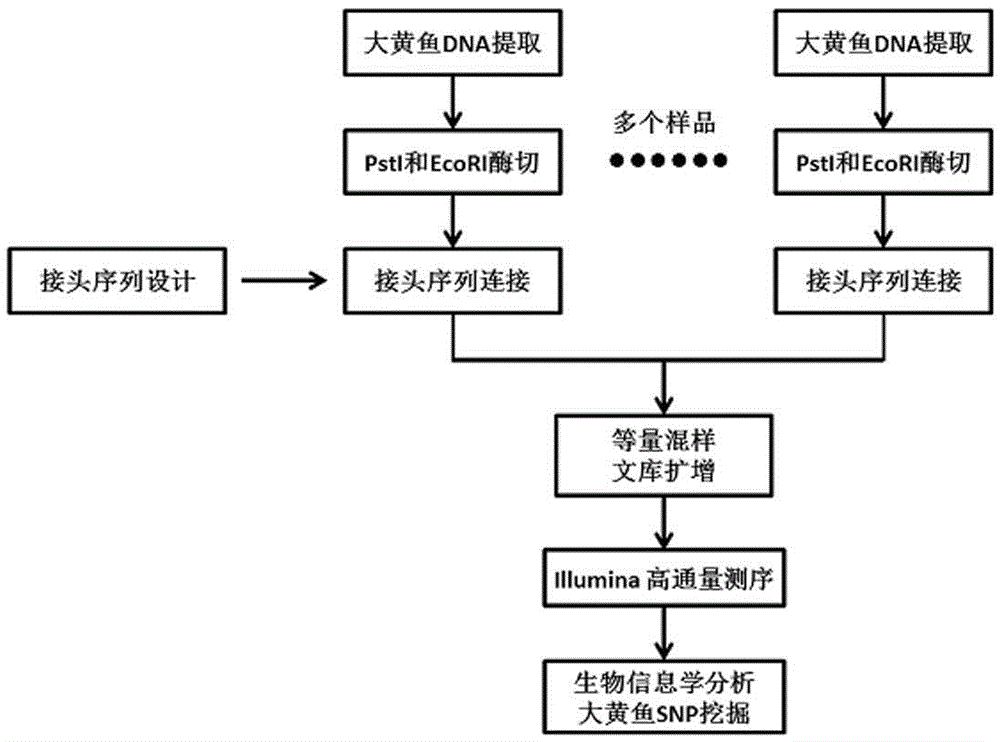
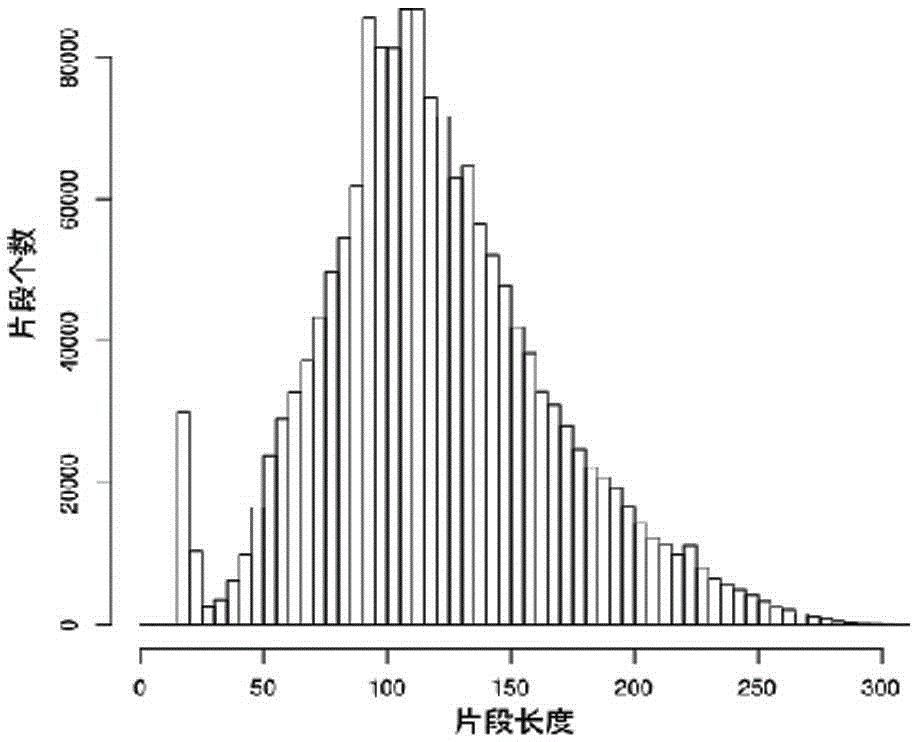
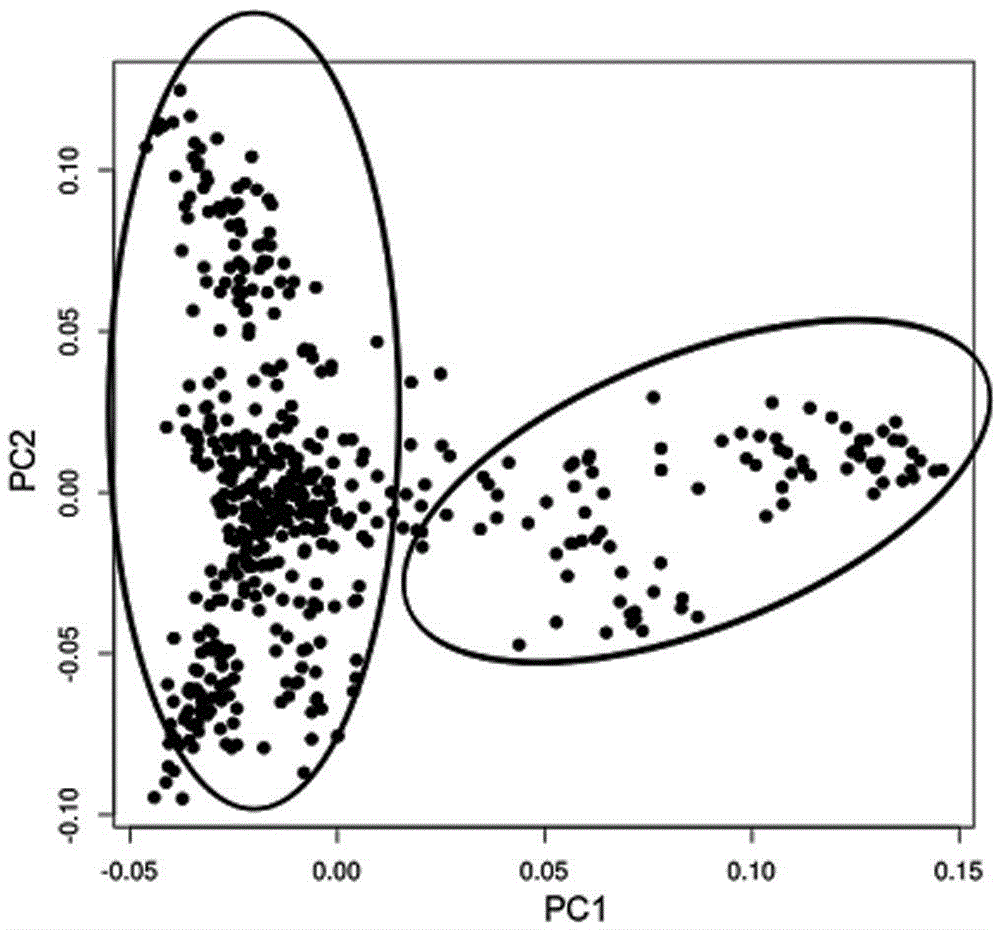
[0029] see Figure 1-3 , a method for whole-genome SNP and InDel molecular markers of large yellow croaker based on double enzyme digestion, comprising the following steps:
[0030] 1) Select experimental materials
[0031] In this experiment, a random group of 96 large yellow croakers at the age of 1 was taken in the same period, with similar body weight and body length ranges; after recording the basic traits of all large yellow croakers, the fin rays were kept, preserved with 70% alcohol by volume, and placed in Store at -20°C for later use.
[0032] 2) Genomic DNA extraction and preservation of large yellow croaker
[0033] a. Grind the frozen fish fins into powder with a mortar in liquid nitrogen, take a centrifuge tube with a specification of 1.5ml, add 1ml of digestion buffer, slowly add the powder into 1ml of digestion buffer, and make the powder in the digestion buffer Wet the surface of the solution evenly, then shake to submerge the sample, and digest at 37°C for...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com