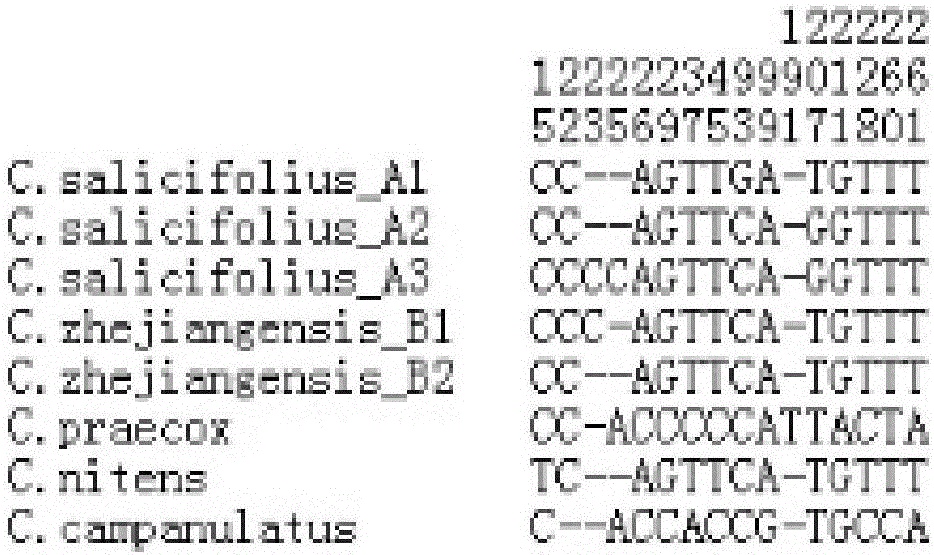
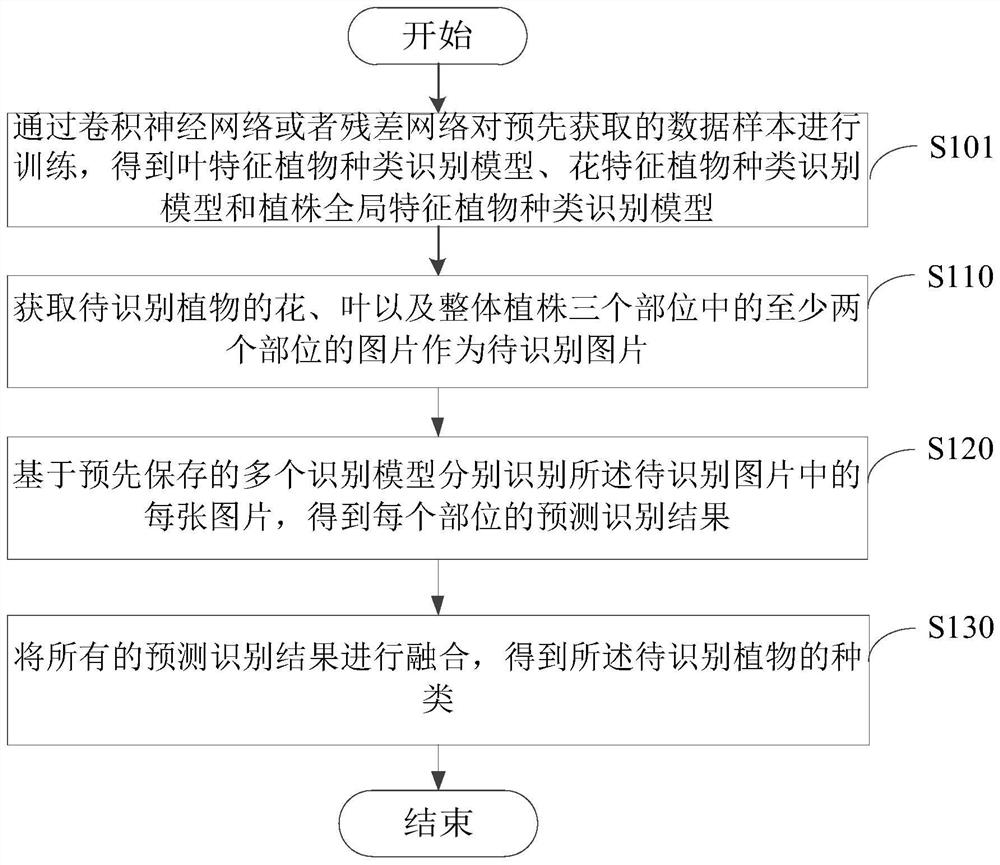
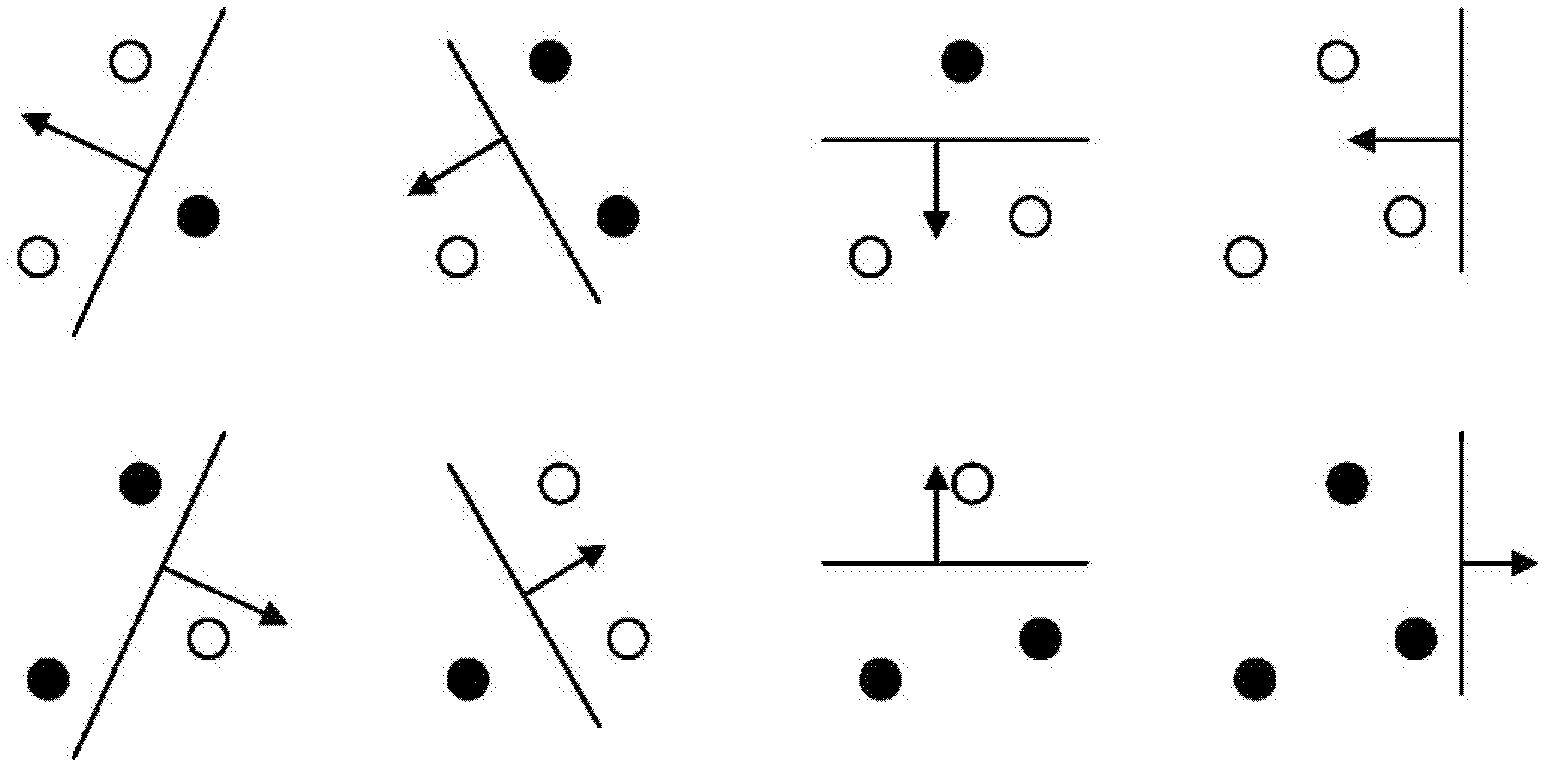Patents
Literature
30 results about "Plant species identification" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Forestry forest fire prevention safety intelligent monitoring and management system based on Internet of Things
InactiveCN111930049AAvoid damageRealize quantitative displayProgramme controlComputer controlForest industryEnvironmental resource management
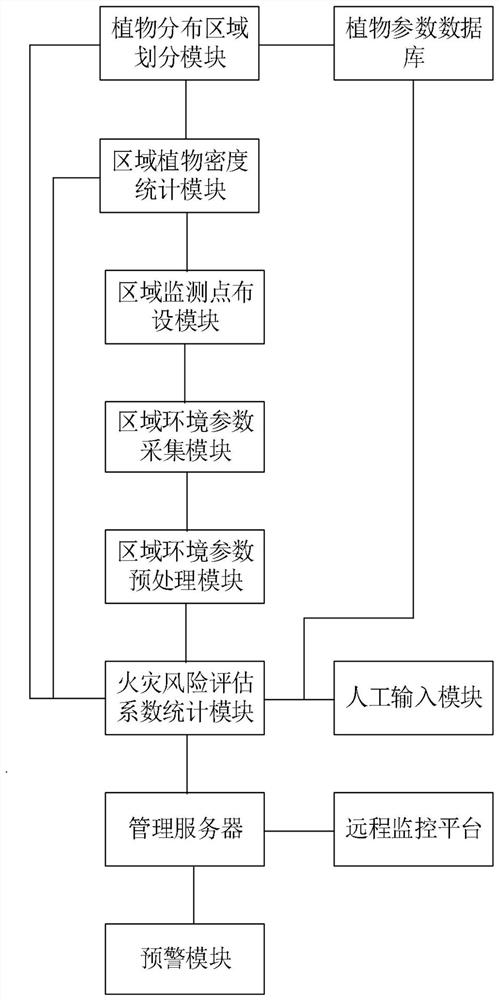
The invention discloses a forestry forest fire prevention safety intelligent monitoring and management system based on the Internet of Things. The system comprises a plant distribution area division module, a regional plant density statistics module, a regional monitoring point layout module, a regional environment parameter acquisition module, an environment parameter preprocessing module, a plant parameter database, a manual input module, a fire risk assessment coefficient statistics module, a management server, an early warning module and a remote monitoring platform. According to the invention, plant species identification and plant distribution region division are carried out on the whole forest region; meanwhile, a plurality of monitoring points are arranged in each sub-region; environmental parameter acquisition processing is carried out at each monitoring point; therefore, the fire risk assessment coefficient of each sub-region is counted, the possibility of a fire in each sub-region can be predicted in advance, the sub-regions of which the fire risk assessment coefficients are greater than the preset safety fire risk assessment coefficient are subjected to prevention processing, the occurrence of the fire is avoided to the maximum extent, and the damage to forest resources is reduced.
Owner:广州立信电子科技有限公司
Leaf image multi-feature integrated plant species identification method and device
InactiveCN106295661AImprove accuracyImprove efficiencyCharacter and pattern recognitionImaging processingPrincipal component analysis
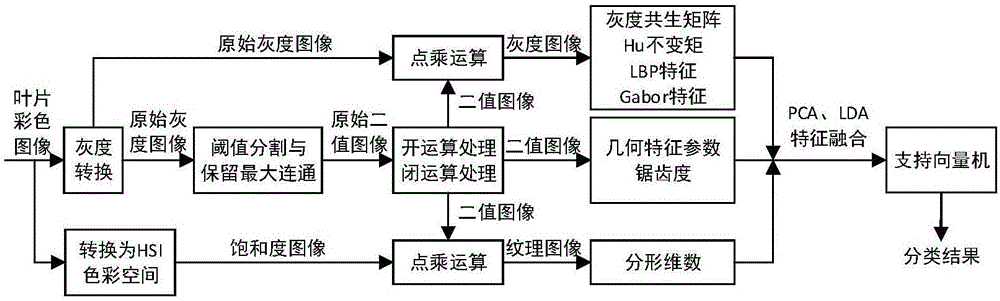
The invention relates to a leaf image multi-feature integrated plant species identification method and device. The method comprises the steps of obtaining plant leaf sample images of known species; obtaining binary images, leaf gray images and leaf texture images according to the leaf sample images; extracting leaf shape features and texture factures according to the leaf gray images, the binary images and the texture images; carrying out feature integration on the extracted leaf shape features and the texture factures through a main component analysis and linear judgment analysis combined method and carrying out dimension reduction on feature integration results, thereby obtaining training sample feature data; and training a support vector machine classifier according to the training sample feature data and corresponding plant species, thereby carrying out species identification on the leaf images of to-be-tested plants according to training results. According to the method and the device, image processing and machine learning principles are combined for plant species identification, and the plant classification and identification accuracy and efficiency are improved.
Owner:BEIJING FORESTRY UNIVERSITY
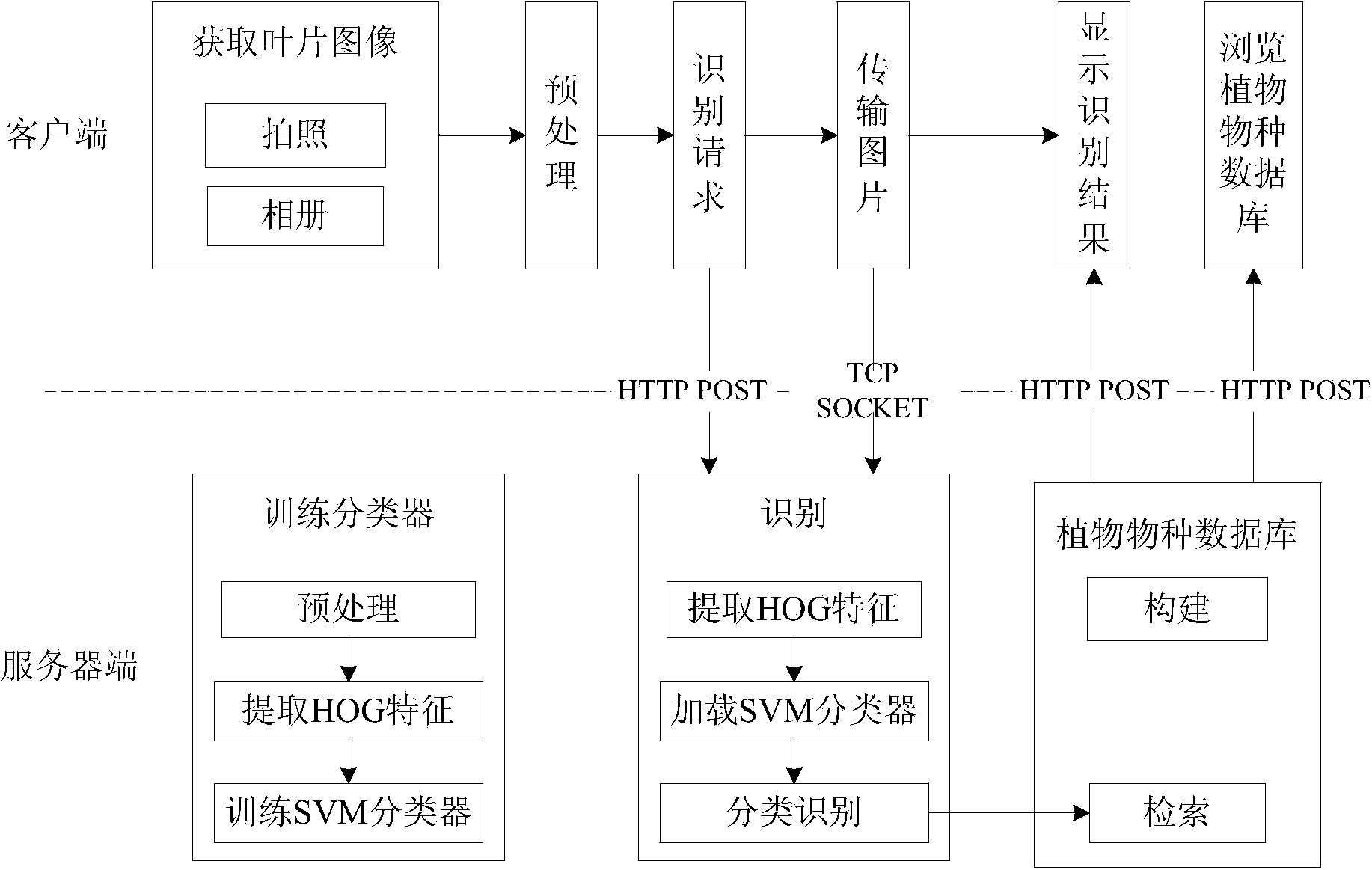
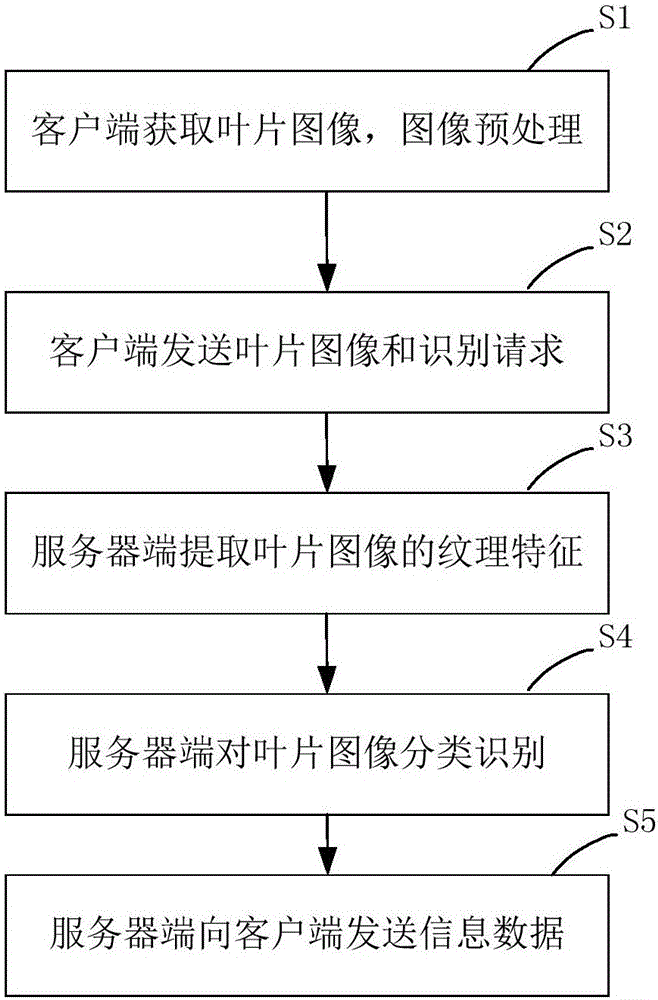
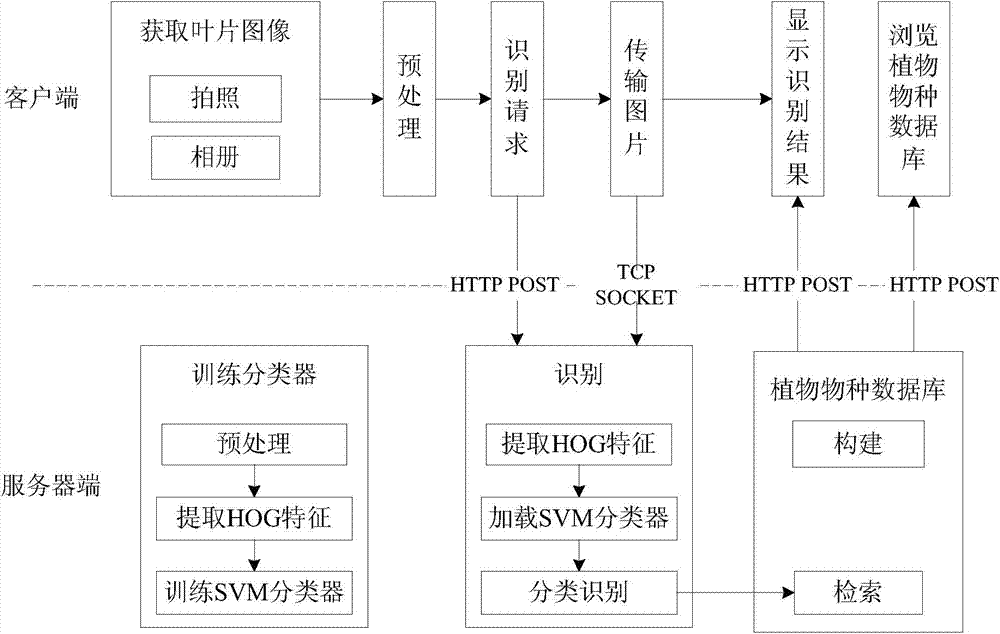
Plant species identification method based on leaf HOG features and intelligent terminal platform
InactiveCN104036235AEfficient identificationImprove accuracyCharacter and pattern recognitionHistogram of oriented gradientsSvm classifier
The invention relates to a plant species identification method based on leaf HOG (Histograms of Oriented Gradients) features and an intelligent terminal platform. The method comprises the following steps that: (1) a client obtains leaf images; (2) the obtained leaf images are preprocessed and are then transmitted to a server end, and meanwhile, an identification request is sent to the server end; (3) after the server end receives the identification request, the leaf images are subjected to HOG feature extraction; (4) the extracted HOG features are used as the input of a trained SVM (Support Vector Machine) classifier, and the leaf images are subjected to classification identification; (5) plant species information of categories corresponding to the current leaf images is found from a plant species database according to identification results, and is sent to the client; and (6) the client displays the identification results and the corresponding plant species information. Compared with the prior art, the plant species identification method has the advantages that the accuracy is high, the use is convenient, and the like.
Owner:TONGJI UNIV
Plant species identification method based on digital image
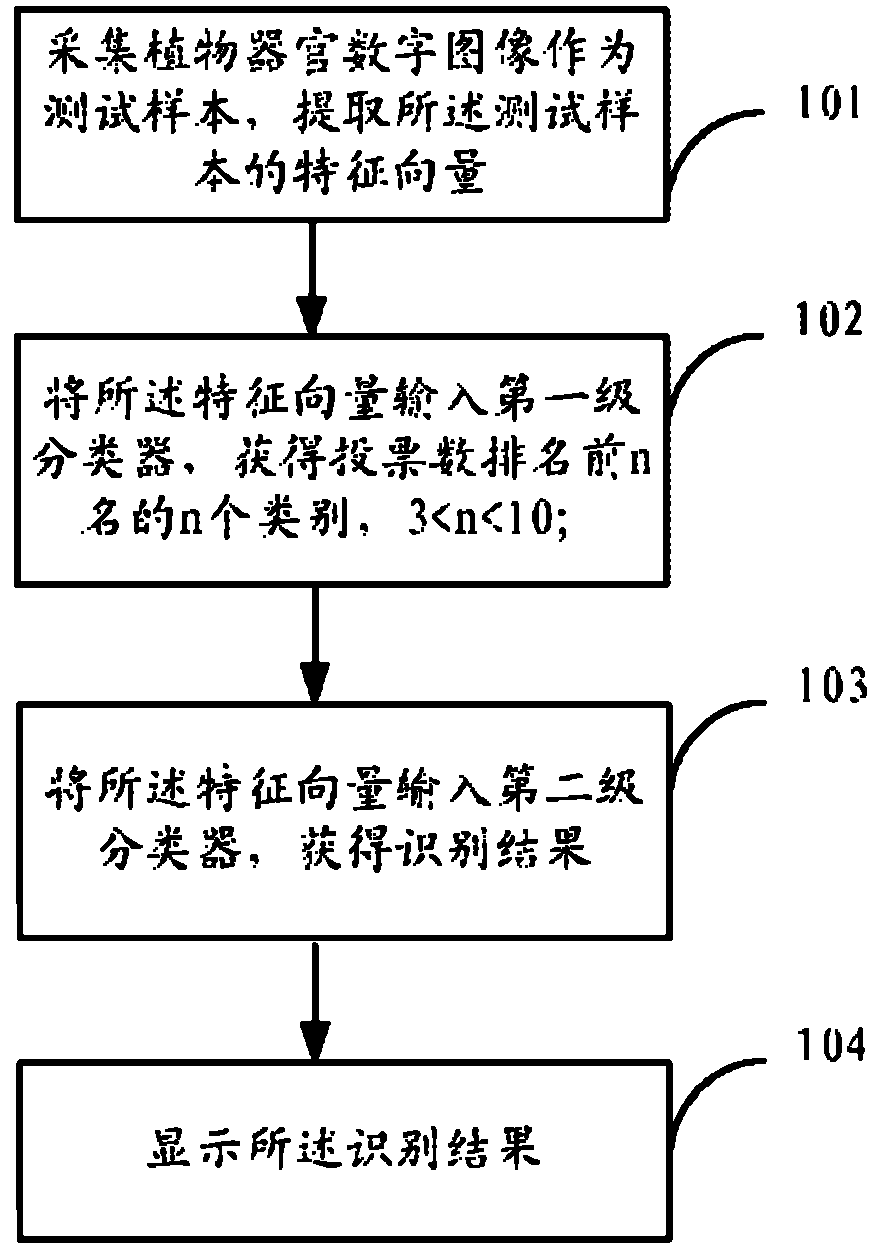
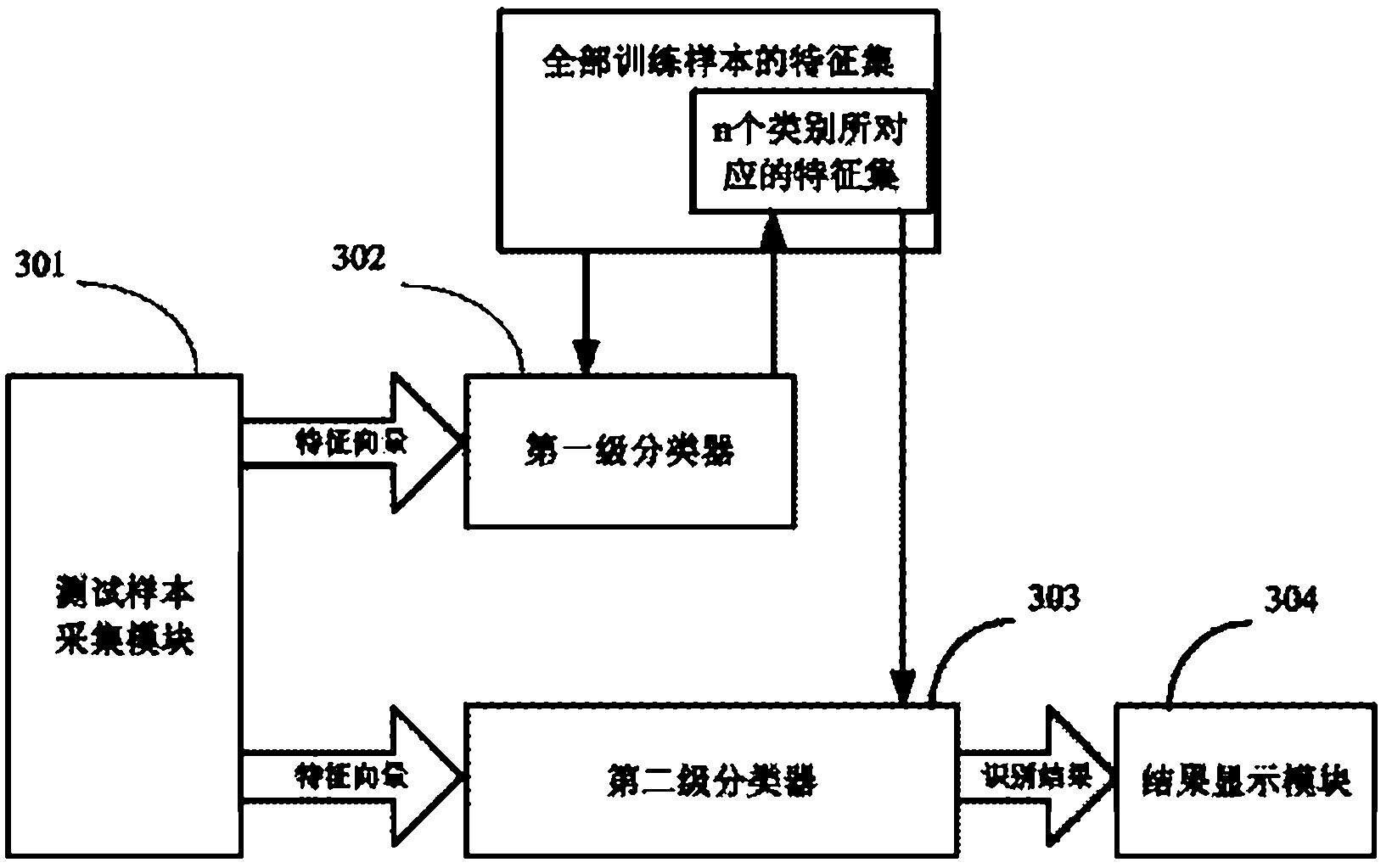
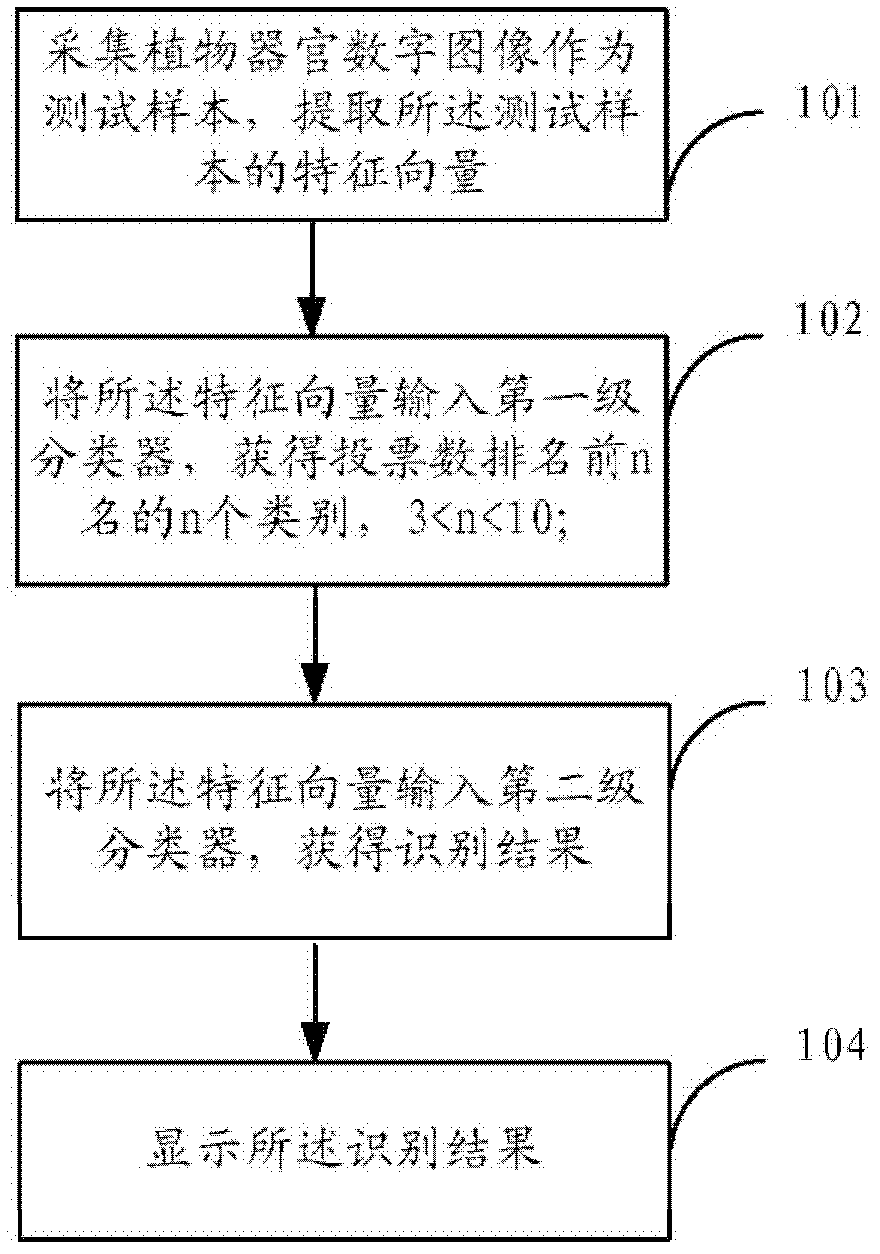
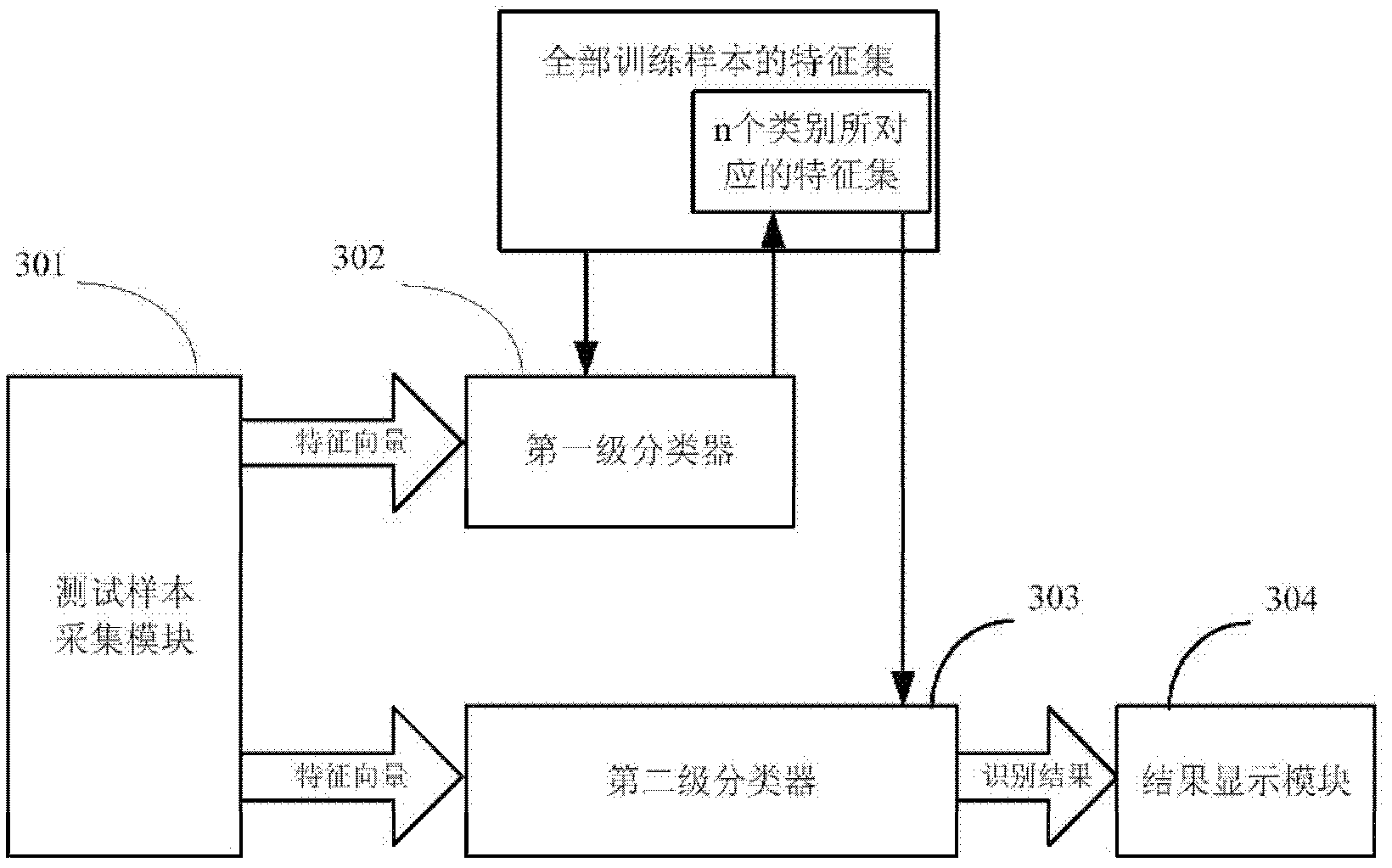
ActiveCN102324038AEliminate the impact of recognition accuracyKnow the kindCharacter and pattern recognitionFeature vectorTest sample
The invention provides a plant species identification method based on a digital image. The method comprises the following steps: collecting a plant organ digital image as a test sample, and extracting a characteristic vector; inputting the characteristic vector into a first stage classifier, and obtaining first n species in a vote number ranking, wherein n is larger than 3 and smaller than 10; the first stage classifier is obtained through the following modes: carrying out classifier training based on a characteristic set of all training samples; inputting the characteristic vector into a second stage classifier, and obtaining an identification result; the second stage classifier is obtained through the following modes: extracting a characteristic set corresponding to the n species from the characteristic set of all training samples to carry out classifier training. According to the invention, through a grading SVM classifier, sensitivity of the classifier to sample kind quantity is effectively reduced, influence of sample species increase on identification accuracy is eliminated, a problem of low accuracy of large sample size identification by the SVM classifier is overcome, and plant identification accuracy is raised.
Owner:BEIJING FORESTRY UNIVERSITY
Online plant species identification method based on geometric invariant shape features
InactiveCN104361342AEasy pretreatmentRemove complex operationsCharacter and pattern recognitionFeature vectorHypersphere
The invention relates to an online plant species identification method based on geometric invariant shape features. The method includes 1, allowing a browser to acquire leaf image and transmit to a server after pre-processing; 2, allowing the server to calculate the leaf shape features according to pre-processed leaf images, establishing feature vectors, storing a trained moving center hypersphere classifier in the server, allowing the moving center hypersphere classifier to perform species identification according to the feature vectors, outputting the plant species code, querying a leaf information database according to the plant species code, and feeding the final identification result back to the browser; 3, allowing the browser to display the final identification result including the leaf, flower, fruit and seed images of the identified species and the scientific instruction information of the species. Compared with the prior art, the method has the advantages of fine real-time performance, convenience and high accuracy.
Owner:TONGJI UNIV
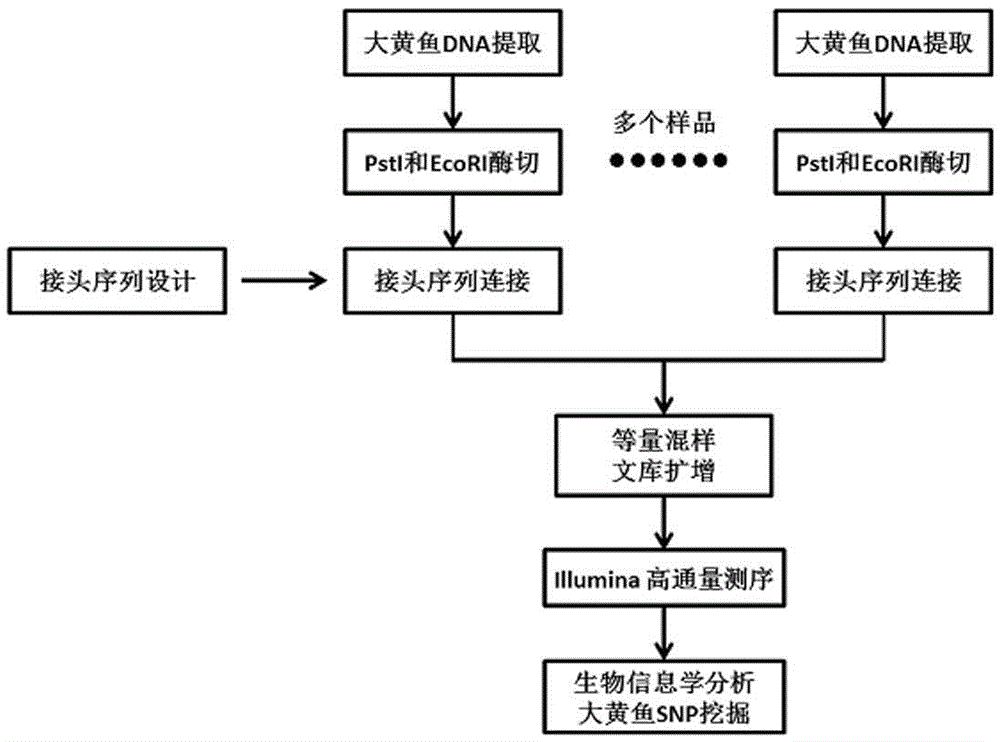
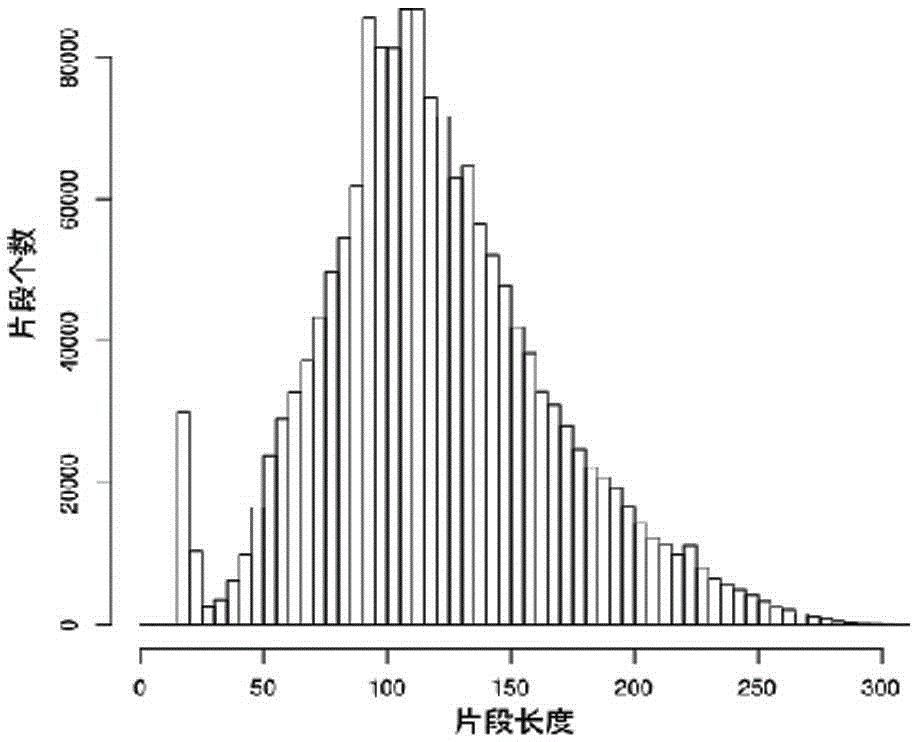
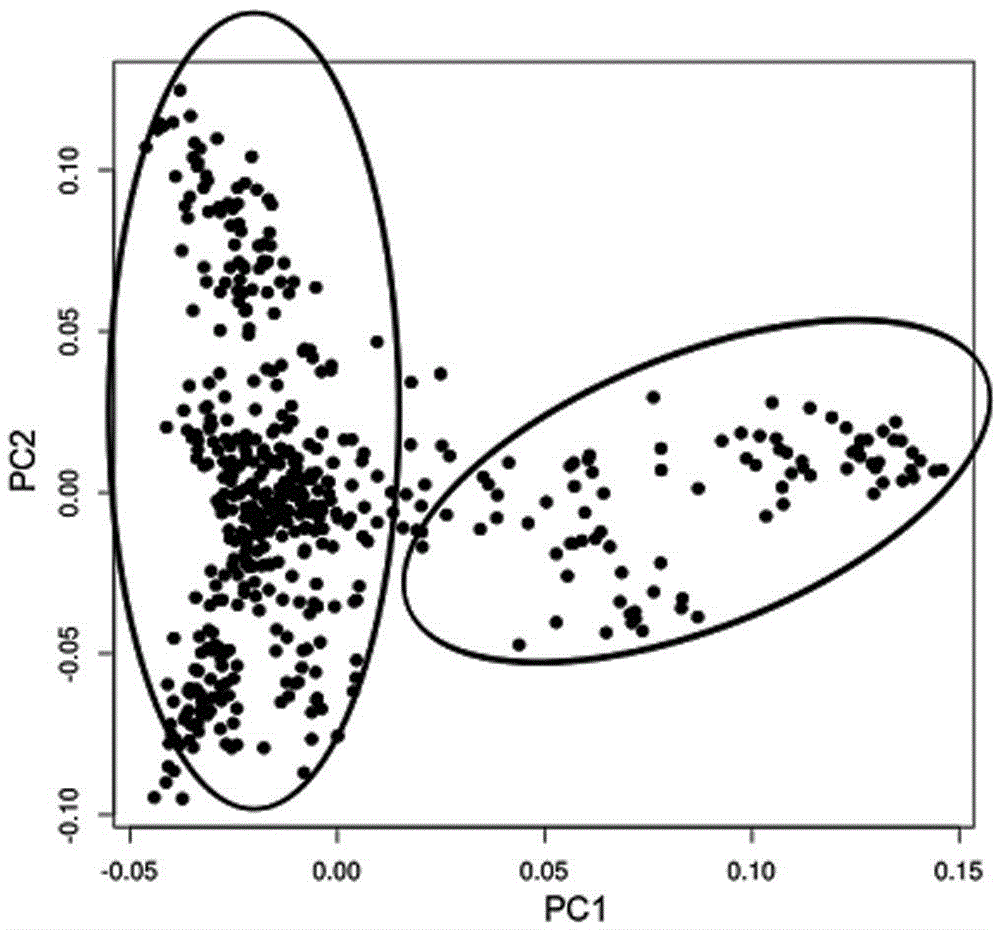
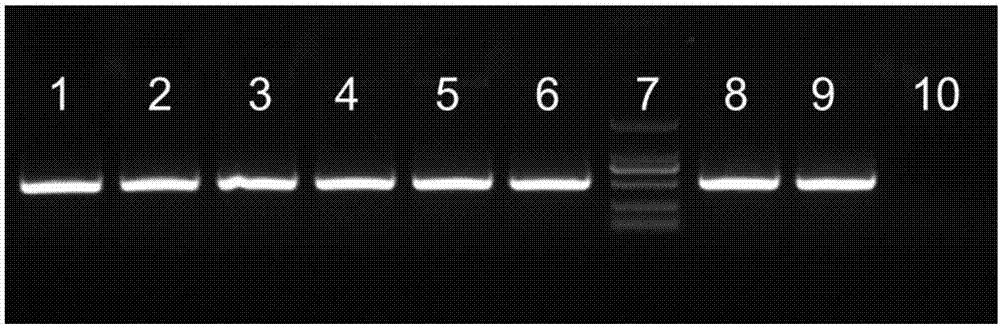
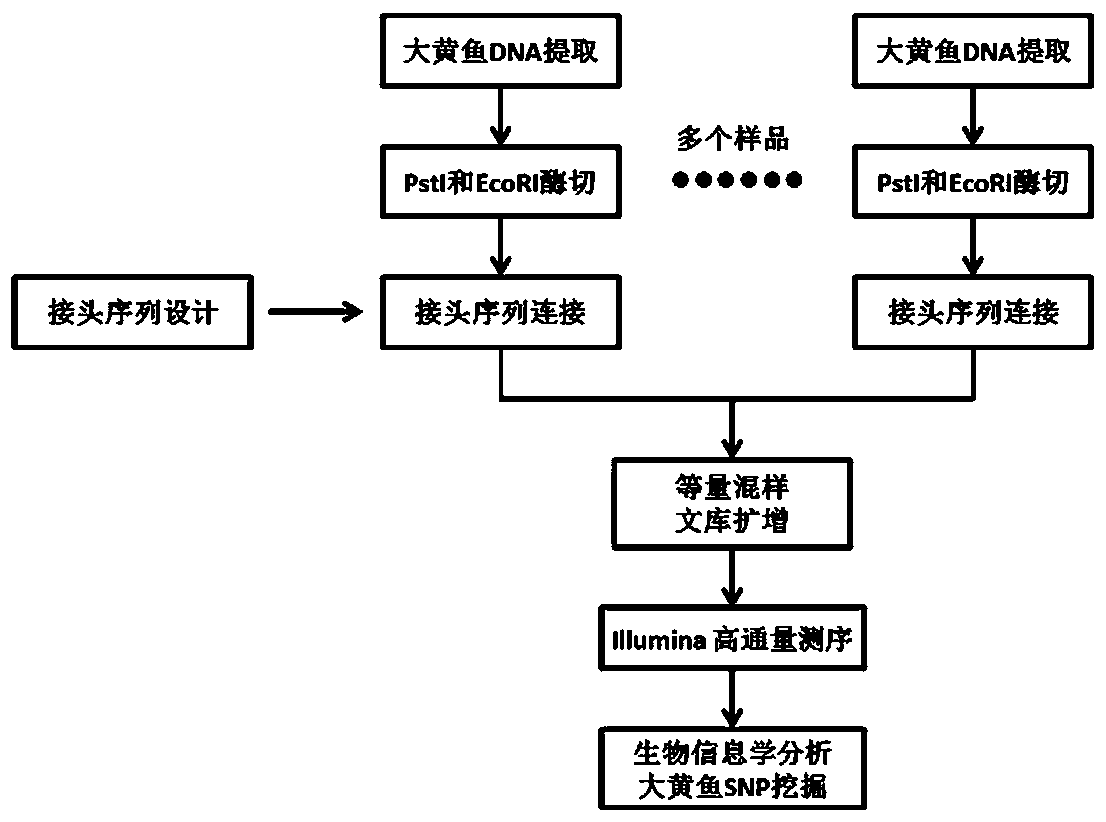
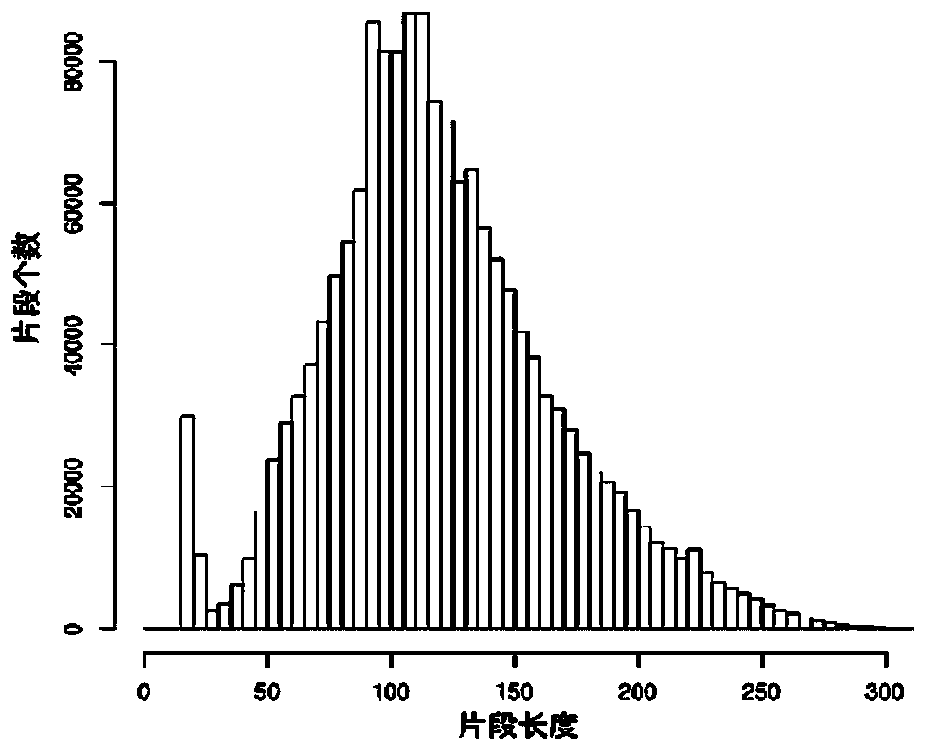
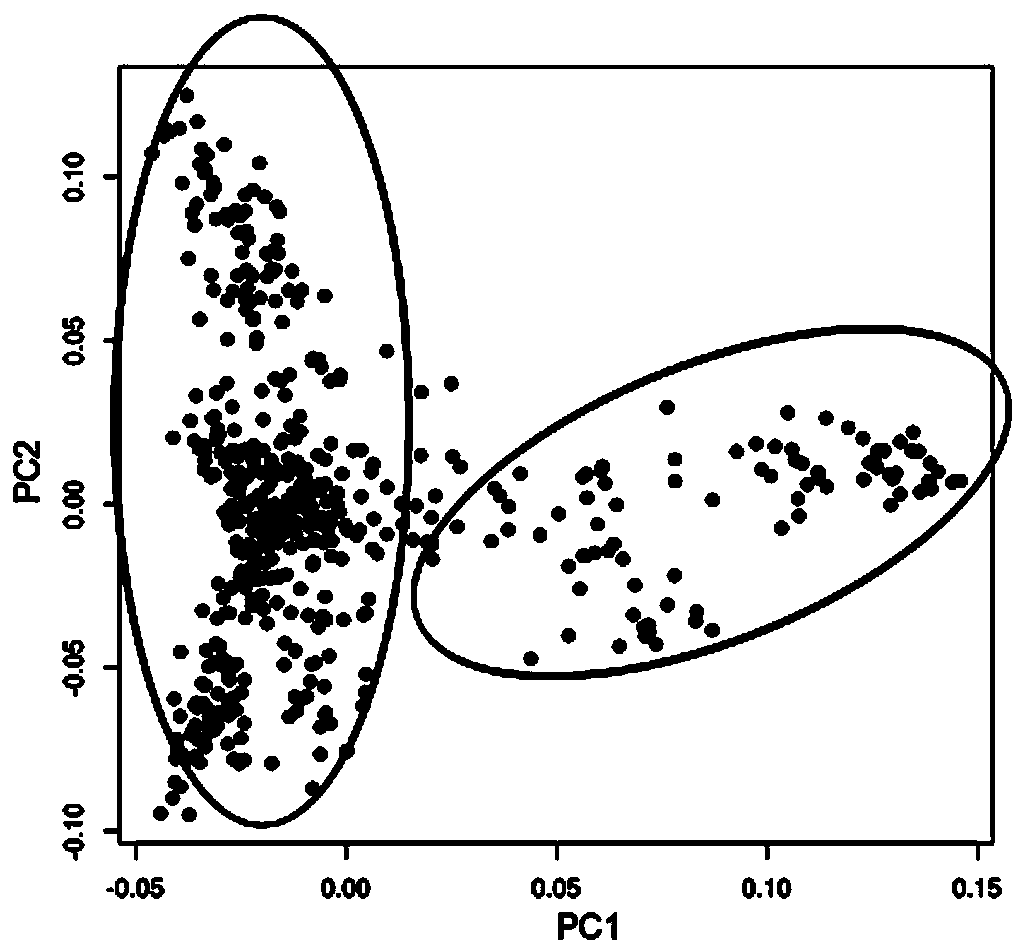
Larimichthys crocea genome-wide SNP and InDel molecular marker method based on double enzyme digestion
ActiveCN105349675AReduce the scope of sequencingLow costMicrobiological testing/measurementDNA/RNA fragmentationSequence designAgricultural science
The invention discloses a larimichthys crocea genome-wide SNP and InDel molecular marker method based on double enzyme digestion. The method comprises the following steps: 1) joint sequence design; 2) genome DNA enzyme digestion; 3) joint sequence connection; 4) sample mixing and PCR amplification; 5) high-throughput sequencing; 6) SNP locus excavation and analysis through sequencing data. According to the method, the modern molecular biology and the advanced high-throughput sequencing technology are combined, the method of combination of EcoRII and NlaIII double enzyme digestion is used, the genome-wide DNA molecules are subjected to enzyme digestion, DNA segments with certain lengths are obtained for carrying out library establishment sequencing and SNP molecular marker excavation, and the uniformly distributed genome-wide SNP locus information is obtained; the workload and cost of genome-wide marker analysis are greatly reduced, and the method is also applicable to the genome-wide marker analysis of other species; the SNP marker excavated through the method is applicable to the study fields such as animal and plant species identification, species genetic genealogy analysis, germplasm resource genetic diversity analysis and genetic breeding.
Owner:JIMEI UNIV
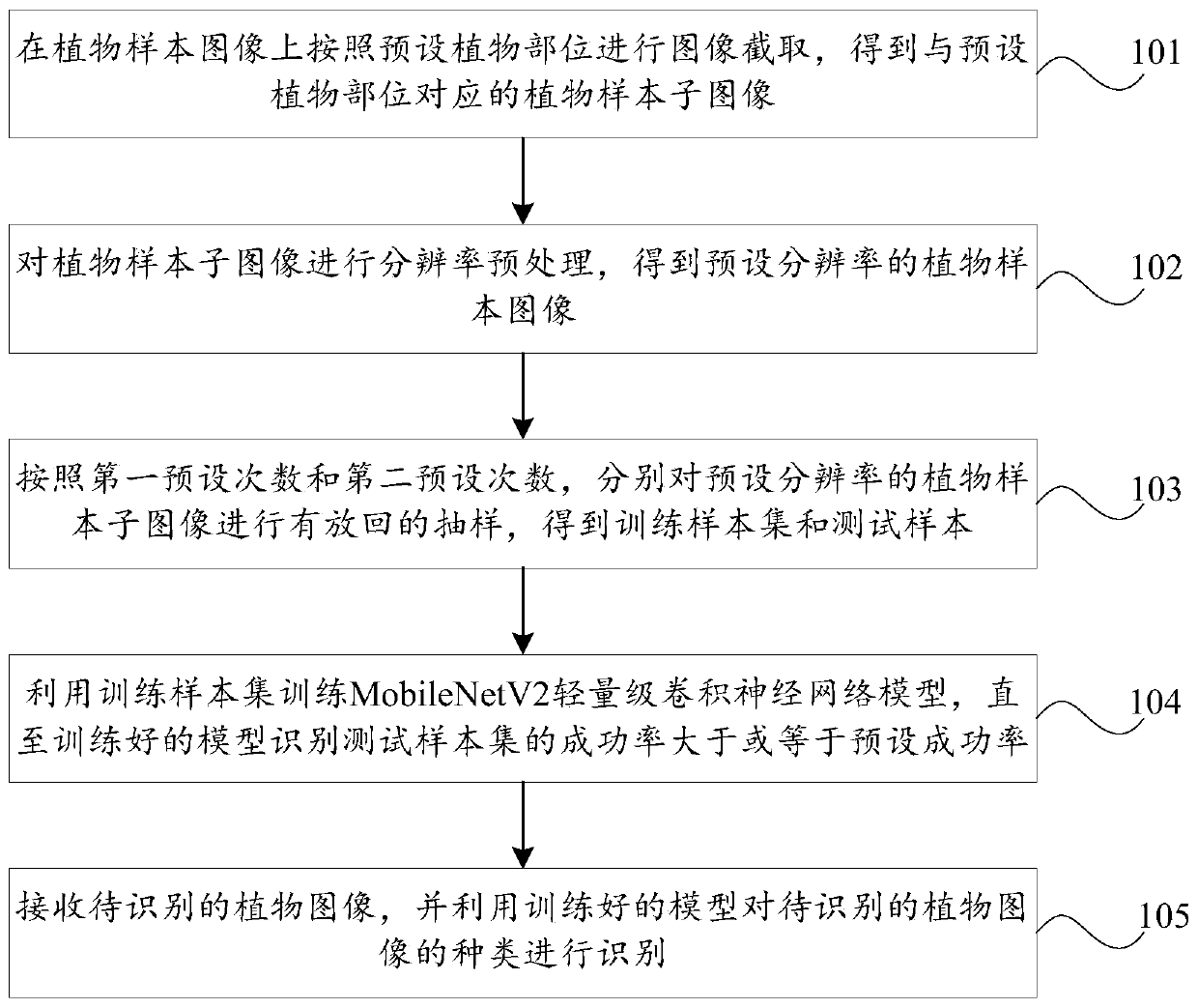
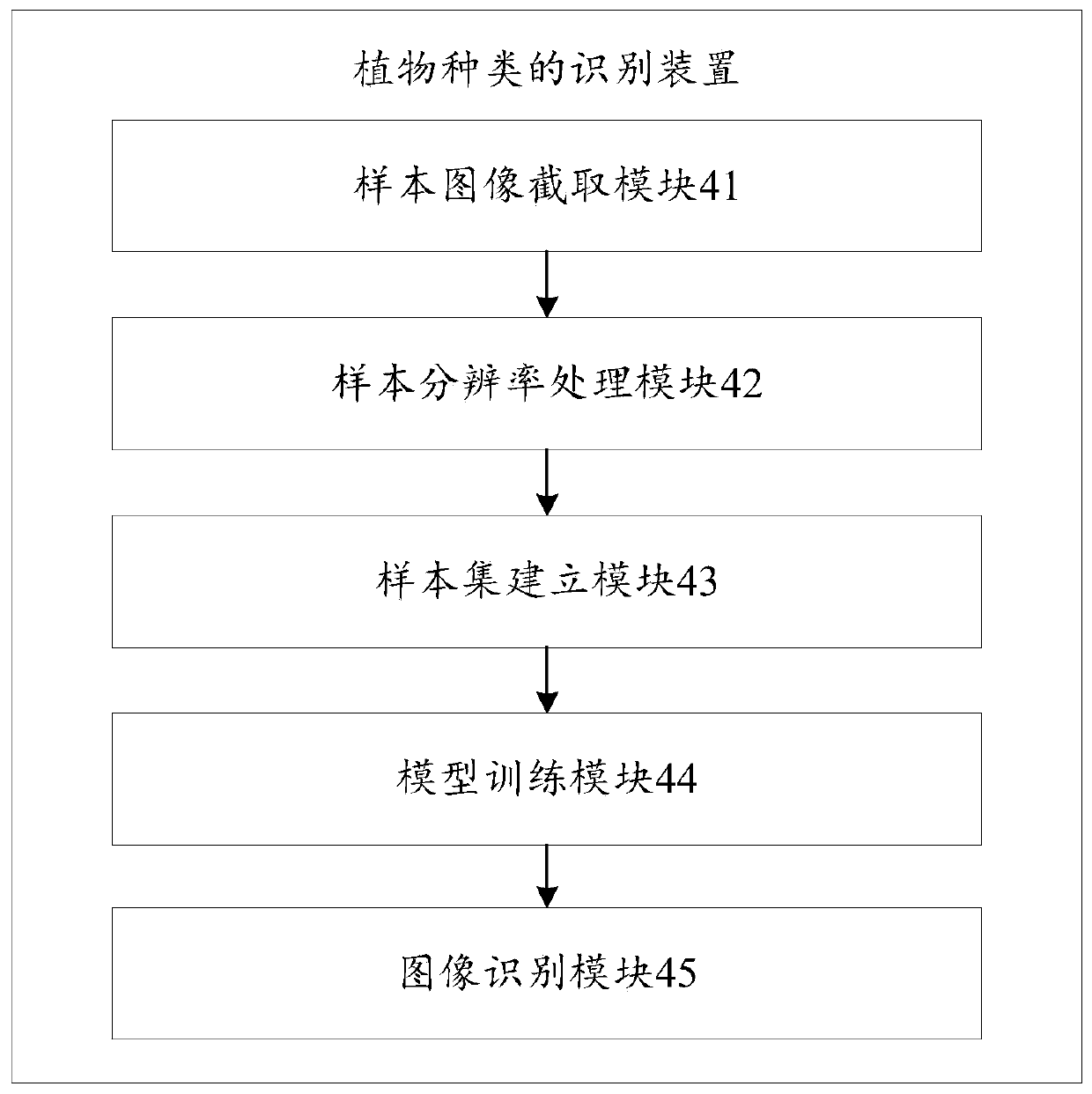
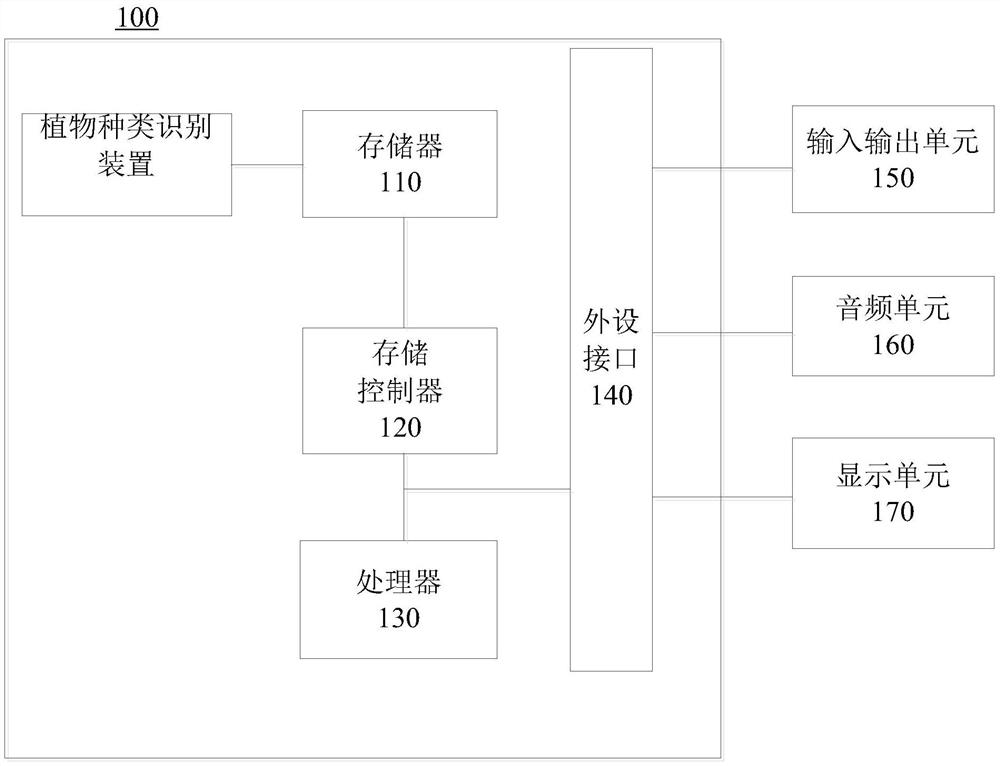
Identification method and device for plant species, storage medium and computer equipment
PendingCN110070101AImprove recognition efficiencyReduce labor costsCharacter and pattern recognitionTest sampleImage resolution
The invention discloses an identification method and device for plant species, a storage medium and computer equipment, and the method comprises the steps: carrying out the image interception on a plant sample image according to a preset plant part, and obtaining a plant sample sub-image corresponding to the preset plant part; carrying out resolution preprocessing on the plant sample sub-image toobtain a plant sample sub-image with a preset resolution; according to a first preset number of times and a second preset number of times, carrying out put-back sampling on the plant sample sub-imagewith the preset resolution to obtain a training sample set and a test sample; using the training sample set to train the MobileNetV2 lightweight convolutional neural network model until the success rate of the trained model for identifying the test sample set is greater than or equal to a preset success rate; and receiving a to-be-identified plant image, and identifying the type of the to-be-identified plant image by using the trained model. According to the invention, the plant species identification efficiency is improved, and the labor cost is saved.
Owner:PING AN TECH (SHENZHEN) CO LTD
Plant species identification method based on blade textural features and iOS platform
InactiveCN106803093AGood multiresolutionGood directionImage enhancementImage analysisState of artFeature vector
The present invention relates to a plant species identification method based on blade textural features and an iOS platform. The method employs a C / S construction, takes an iOS mobile phone platform as a client and takes a computer as a server, the client is configured to collect a blade image to send an identification request to the server and display a plant species information database, and the server is configured to extract the feature vectors of the blade images, perform classification identification of the blade image, construct the plant species information database and send the species information data to the client. Compared to the prior art, the identification efficiency is high and the accuracy is high.
Owner:TONGJI UNIV
A plant species identification method based on leaf and flower fusion for locally distinguishing CCA
PendingCN109948652AAccurate identificationImprove performanceCharacter and pattern recognitionHat matrixFeature set
The invention relates to a plant species identification method based on leaf and flower fusion for local discrimination of CCA, comprising the following steps: constructing two weighted adjacent graphs according to a double-view feature set comprising a leaf image and a flower image; Obtaining two dimensionality-reduced projection matrixes by enabling the adjacent samples in the class to be most correlated, enabling the adjacent samples between the classes to be most uncorrelated and enabling the correlation between leaves and flowers of the same species to be maximum; identifying plant species using a 1-NN classifier with a geodesic distance. According to the method, the idea of local discrimination embedding (LDE) is introduced into canonical correlation analysis (CCA); The method has the advantages that the identification is accurate, the performance is stable, the identification characteristics can be extracted from the two plant organs, the identification information and the datastructure can be well reserved, and the method is further popularized and applied on the basis.
Owner:TIANJIN UNIVERSITY OF SCIENCE AND TECHNOLOGY
DNA barcode identification method of traditional She medicinal Shiliang herb tea based original species and related confusable species
InactiveCN106834471AResolve Species IdentificationSolve resource problemsMicrobiological testing/measurementDNA barcodingGermplasm
The invention relates to the field of Chinese herbal medicine based original plant species identification technology, specifically to a method for identifying traditional She medicinal Shiliang herb tea based original species and related confusable species by the utilization of ITS2 fragments of ribosomal DNA and application thereof. A DNA barcode identification method of traditional She medicinal Shiliang herb tea based original species and related confusable species comprises the following steps: 1) DNA sample extraction; 2) PCR amplification of fragments containing ITS2 sequences; 3) splicing of PCR amplification products; 4) comparison of ITS2 sequences of multiple species; and 5) construction of phylogenetic tree based on the ITS2 sequences. The DNA extraction method of traditional She medicinal Shiliang herb tea based original species and related confusable species and the ITS2 preparation method can effectively solve the problems such as species identification of Shiliang herb tea and related species, development and utilization of germplasm resources, etc. The method of the invention has extensive applicability, is simple to operate, is easy to grasp and has high accuracy. By the method, rapid and accurate identification of the Shiliang herb tea based original species and related confusable species can be successfully realized.
Owner:LISHUI AGRI SCI +1
Method for identifying walnut species by analyzing leaves based on GC-IMS
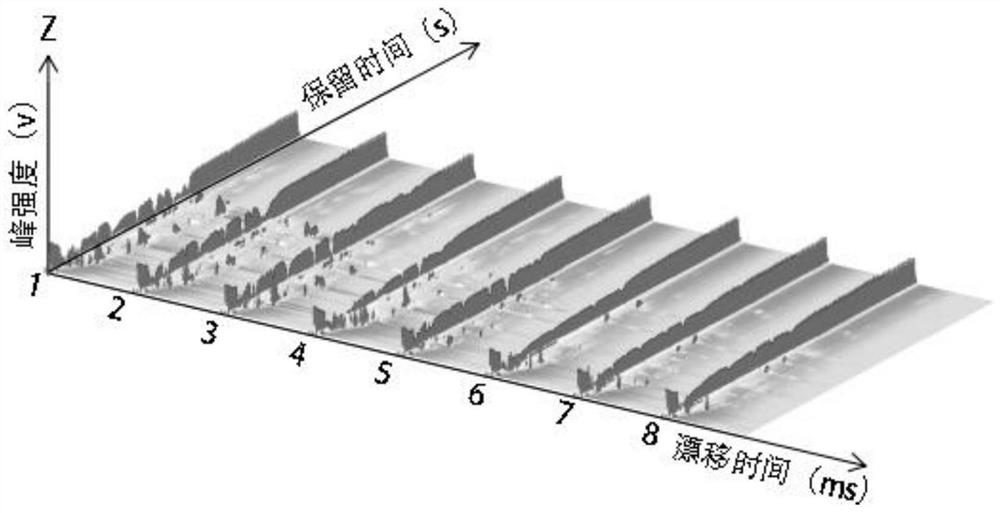
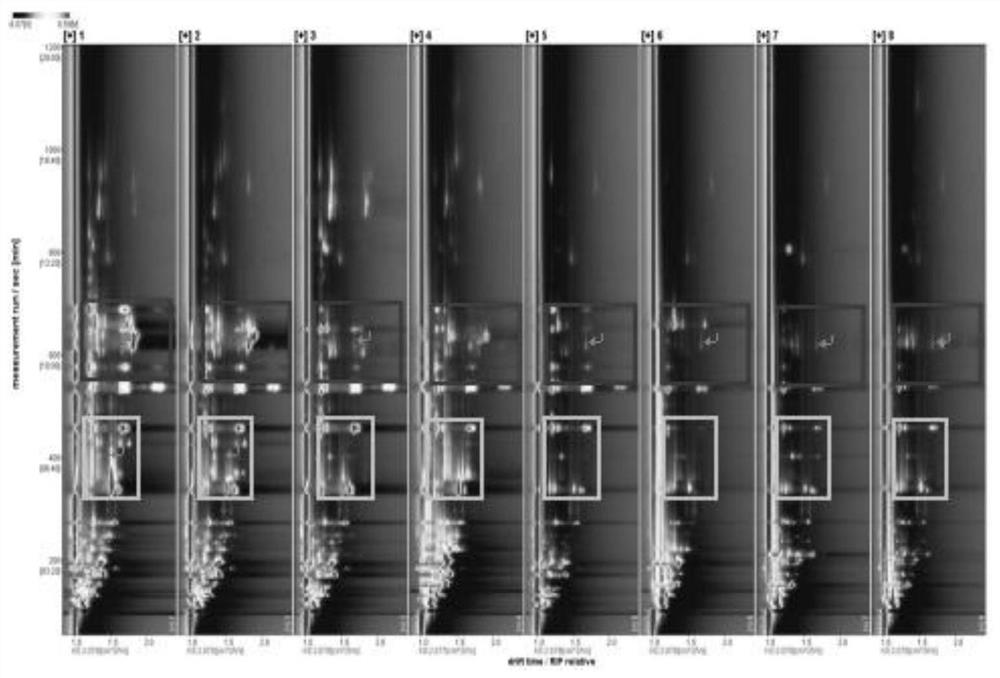
PendingCN113109486ADetermination of integrityRapid detection of volatile organic compoundsComponent separationGas liquid chromatographicKetone
The invention discloses a method for identifying walnut species by analyzing leaves based on gas chromatography-ion mobility spectrometry, and belongs to the technical field of plant species identification. The method comprises the following steps: obtaining a leaf sample of a walnut to be identified, performing liquid nitrogen treatment, and grinding the sample; analyzing volatile substances in the leaves by using gas chromatography-ion mobility spectrometry; and comparing the obtained volatile substance with a reference seed so as to obtain the walnut seed to be identified. According to the method, volatile substances such as terpenes, esters, aldehydes, alcohols and ketones in the walnut leaves are analyzed through gas chromatography-ion mobility spectrometry, a certain research basis is provided for development and utilization of walnut leaf resources, and technical basis and support are further provided for walnut seed identification.
Owner:XINJIANG ACADEMY OF FORESTRY SCI
Method for identifying traditional she medicine Rubi Radix et Rhizoma original plant and congeneric similar confusable species thereof on the basis of ITS2 sequence gene fragments
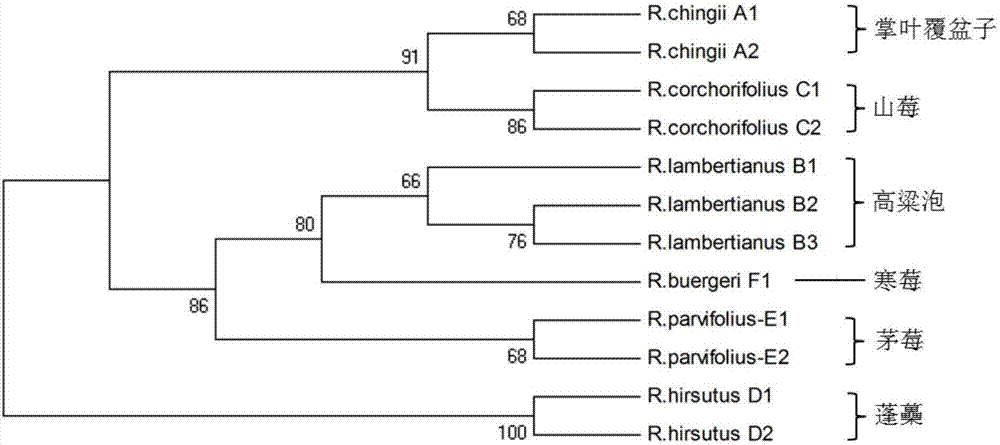
InactiveCN107541547AAccurate identificationRapid identificationMicrobiological testing/measurementDNA/RNA fragmentationRibosomal DNARubus
The invention relates to the field of traditional Chinese medicinal material original plant species identification technology, specifically to a method for identifying leaf, root and fruit of traditional she medicine Rubi Radix et Rhizoma original plant and congeneric similar confusable species thereof by the utilization of ITS2 fragments of ribosome DNA and application thereof. The method for identifying traditional she medicine Rubi Radix et Rhizoma original plant and congeneric similar confusable species thereof on the basis of ITS2 sequence gene fragment comprises the following steps: 1) extraction of multiple rubus plant leaf, root and fruit DNA samples; 2) PCR amplification of fragments containing ITS2 sequence; 3) assembling of PCR amplification fragments; 4) comparison of ITS2 sequences of multiple rubus species; and 5) construction of a phylogenetic tree on the basis of ITS2 sequence. The method of the invention has extensive applicability, is simple and convenient to operate,is easy to master, has high accuracy, and can successfully realize efficient and accurate identification of Rubi Radix et Rhizoma original plant and congeneric similar confusable species thereof.
Owner:LISHUI AGRI SCI
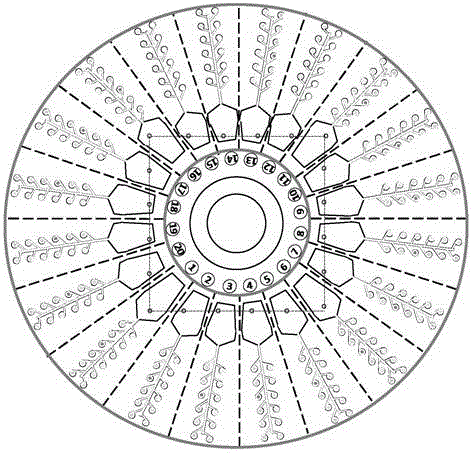
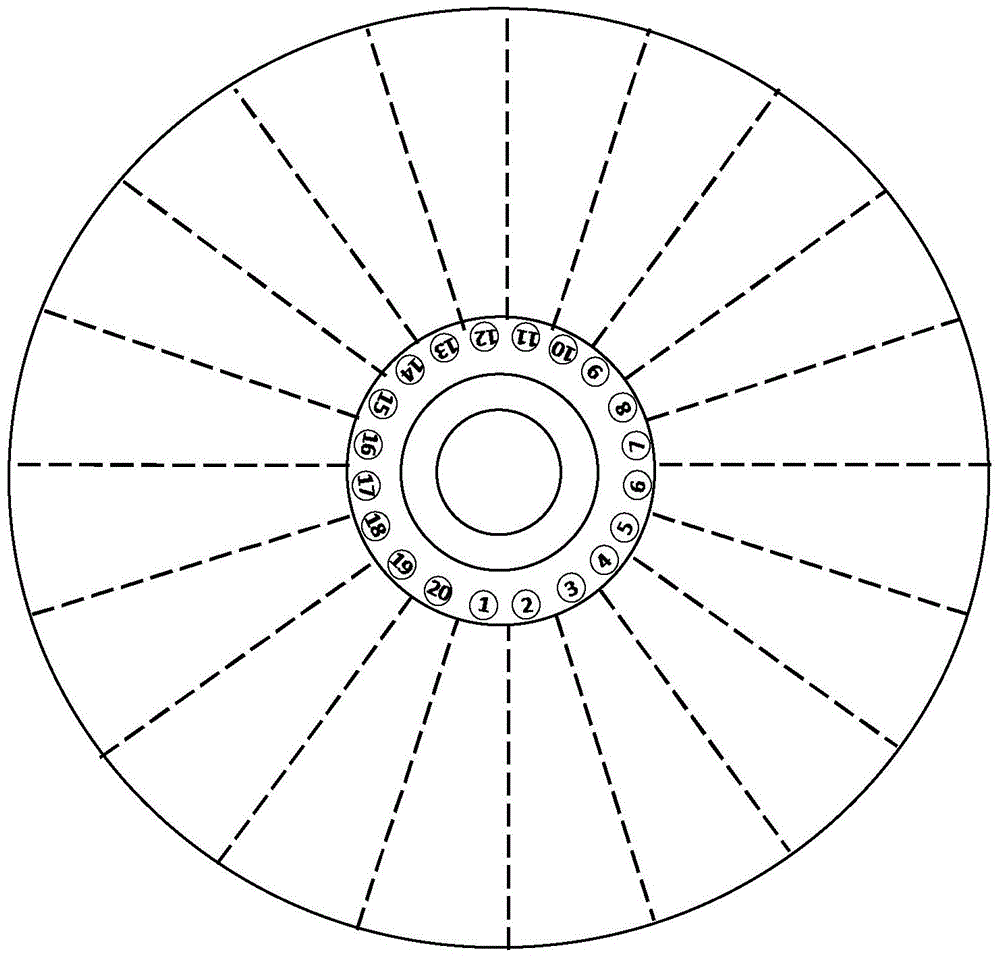
Microfluidics detecting chip based on nucleic acid amplification technology and preparation method thereof
ActiveCN105331528AReduce volumeEasy to carryBioreactor/fermenter combinationsBiological substance pretreatmentsTemperature controlNucleic acid amplification technique
The invention discloses a microfluidics detecting chip based on a nucleic acid amplification technology and a preparation method thereof. The chip has four layers, and is in a disc centrifugal design, one chip can be used for detecting 20 samples, and each sample has 12 parameters. Through an optical driving type centrifugal machine, temperature control equipment and nucleic acid amplification reagent consumable items which are matched with the microfluidics detecting chip, requirements on detection of transgenic ingredients in agricultural products and food, pathogenic bacteria, animal and plant species identification and the like are met, and the microfluidics detecting chip has the features of quickness, convenience, high flux and the like.
Owner:TIANJIN INSTITUE OF QUALITY STANDARD & TESTING OF AGRICULTUAL PRODS
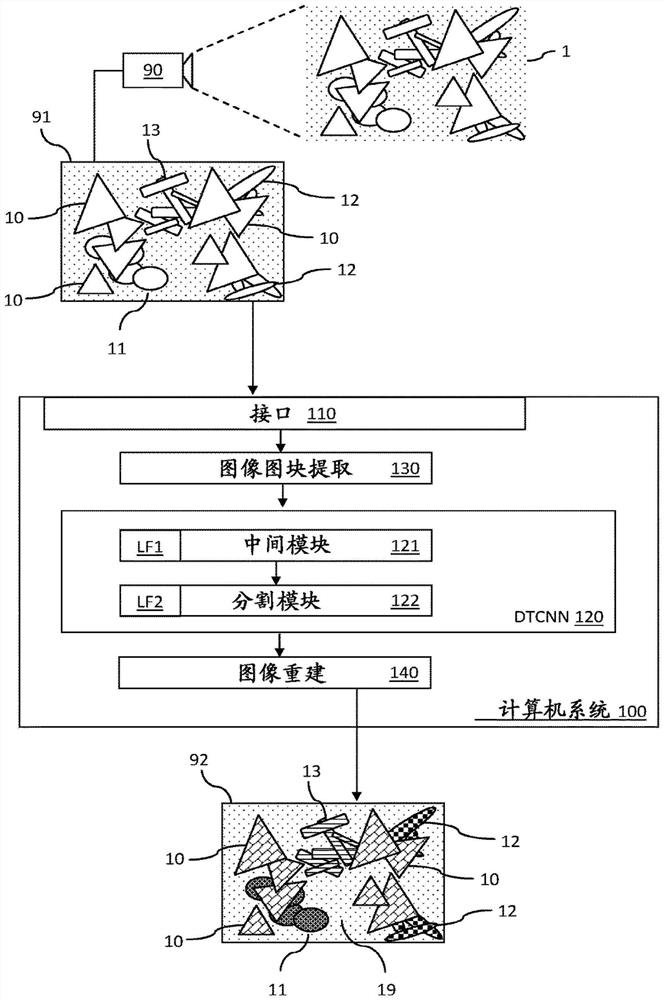
Systems and methods for plant species identification
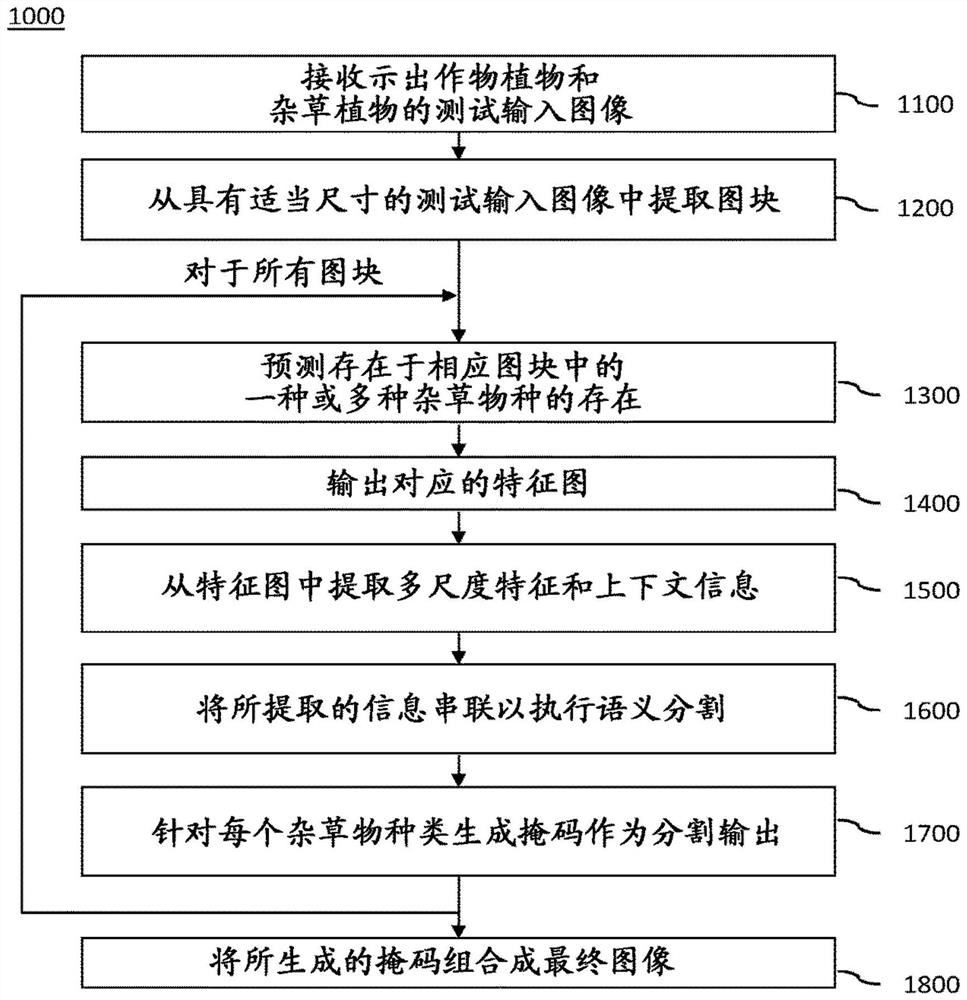
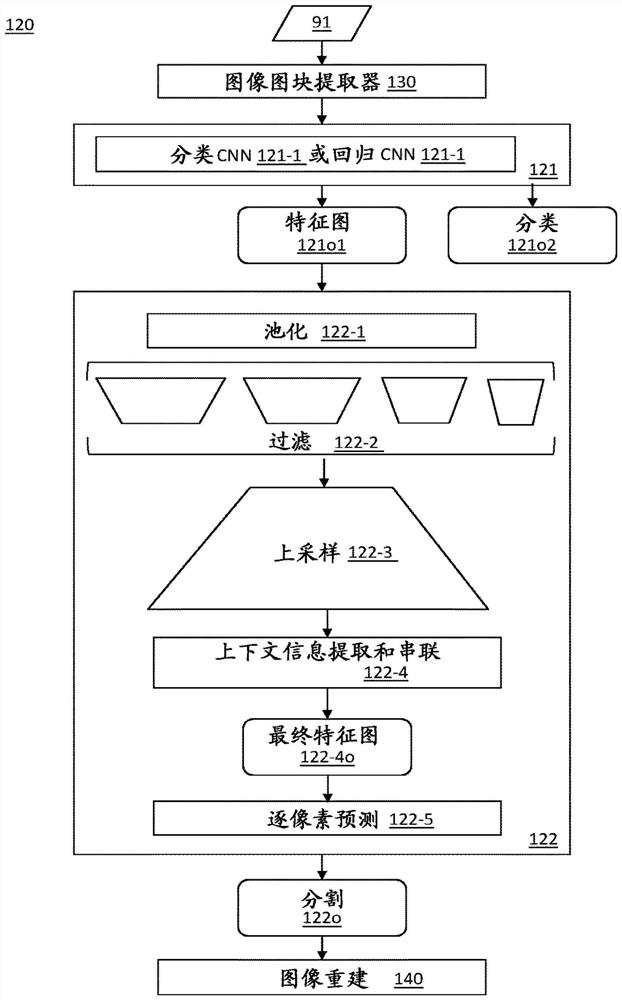
A computer-implemented method, computer program product and computer system (100) for identifying weeds in a farmland using a dual task convolutional neural network (120) having a topology of: an intermediate module (121) that performs a classification task associated with a first loss function (LF1); and a semantic segmentation module (122) that performs a segmentation task associated with a different second loss function (LF2). The intermediate module and the segmentation module are trained together in consideration of the first and second loss functions (LF1, LF2). The system performs a method comprising receiving a test input (91) comprising an image of a weed plant showing a crop plant of a crop species in a farmland and showing one or more weed species in the crop plant; predicting the presence of one or more weed species (11, 12, 13) present in the respective tiles; outputting the corresponding intermediate feature map to a segmentation module as the output of a classification task; generating a mask for each weed species as a segmentation output of the second task by extracting multi-scale features and context information from the intermediate feature map and serially connecting the extracted information to perform semantic segmentation; and generating a final image (92) indicating, for each pixel, whether the pixel belongs to a particular weed species, and if so, which weed species the pixel belongs to.
Owner:BASF AG
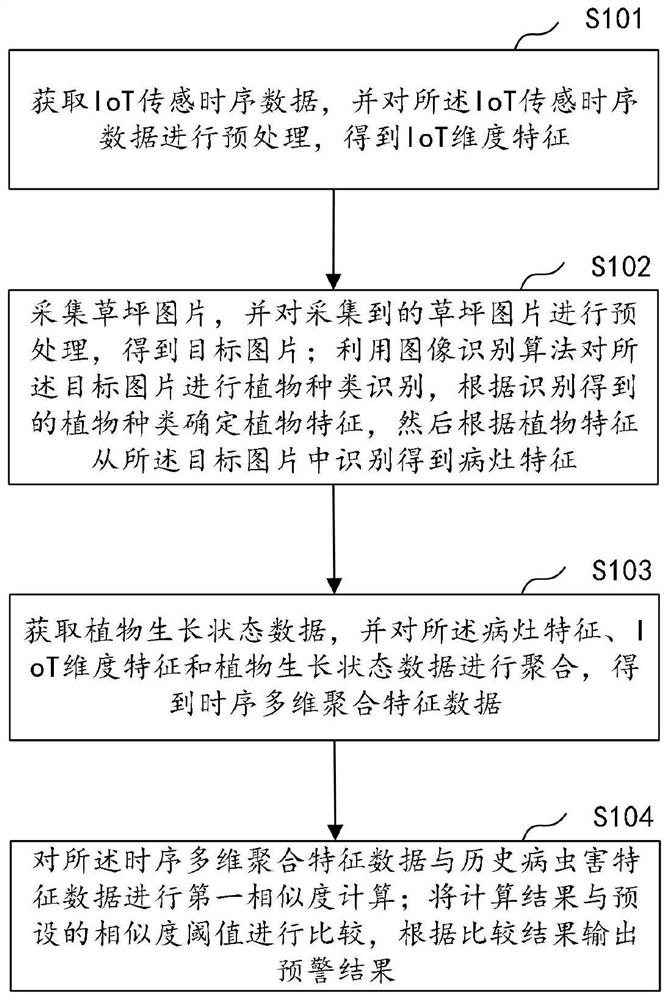
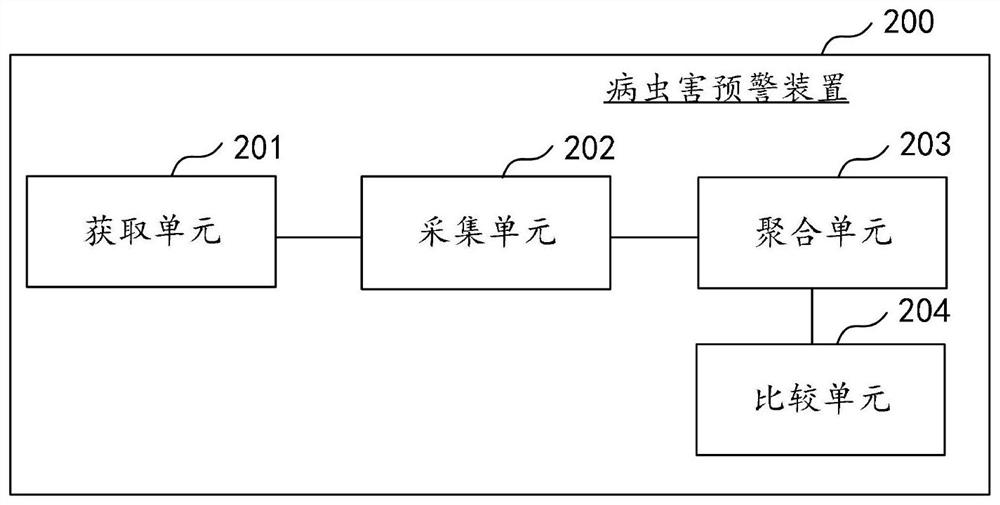
Disease and pest early warning method and device, computer equipment and storage medium
The invention discloses a disease and pest early warning method and device, computer equipment and a storage medium, and the method comprises the steps of obtaining IoT sensing time sequence data, andobtaining IoT dimension features; acquiring a lawn picture, and preprocessing the lawn picture to obtain a target picture; carrying out plant species identification on the target picture by utilizingan image identification algorithm, determining plant features according to the identified plant species, and then identifying focus features from the target picture according to the plant features; acquiring plant growth state data, and aggregating the focus characteristics, the IoT dimension characteristics and the plant growth state data to obtain time sequence multi-dimensional aggregation characteristic data; performing first similarity calculation on the time sequence multi-dimensional aggregation feature data and the historical pest and disease damage feature data; and comparing the calculation result with a preset similarity threshold, and outputting an early warning result according to a comparison result. According to the invention, the multi-dimensional time series data is predicted, so that effective early warning of outbreak of lawn diseases and insect pests is realized.
Owner:深圳市万物云科技有限公司
Plant identification method based on elliptical Fourier descriptors and weighted sparse representation
InactiveCN107392225AImprove robustnessImprove recognition rateImage analysisCharacter and pattern recognitionData setSparse representation classifier
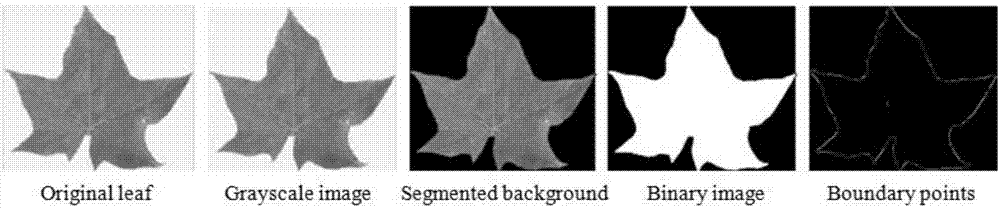
The invention provides a plant identification method based on elliptical Fourier descriptors and weighted sparse representation. The plant identification method is mainly and technically characterized by comprising the steps that leaf images are preprocessed, wherein all the colored leaf images are converted into grayscale images, the leaf images are separated from the background through an Otsu segmentation algorithm and converted into binary images, and small holes of the leaf images are eliminated through an erosion algorithm; edge detection is conducted by adopting a Canny edge detector; centroids of boundaries are calculated; the Fourier descriptors are calculated; a complete dictionary is constructured, wherein Fourier descriptor vectors of all leaf image data sets are divided into training sets and test sets, and the complete dictionary is composed of the Fourier descriptor vectors of all the training sets; and optimization is conducted by a weighted sparse representation classifier. According to the plant identification method, by adopting the elliptical Fourier descriptors, good robustness is achieved on noise and other factors, and by applying the weighted sparse representation classifier (WSRC) to plant species identification and particularly to a low-dimension space, the identification rate is obviously increased.
Owner:TIANJIN UNIV OF SCI & TECH
Analyzing and testing method for unearthed ancient lacquer film
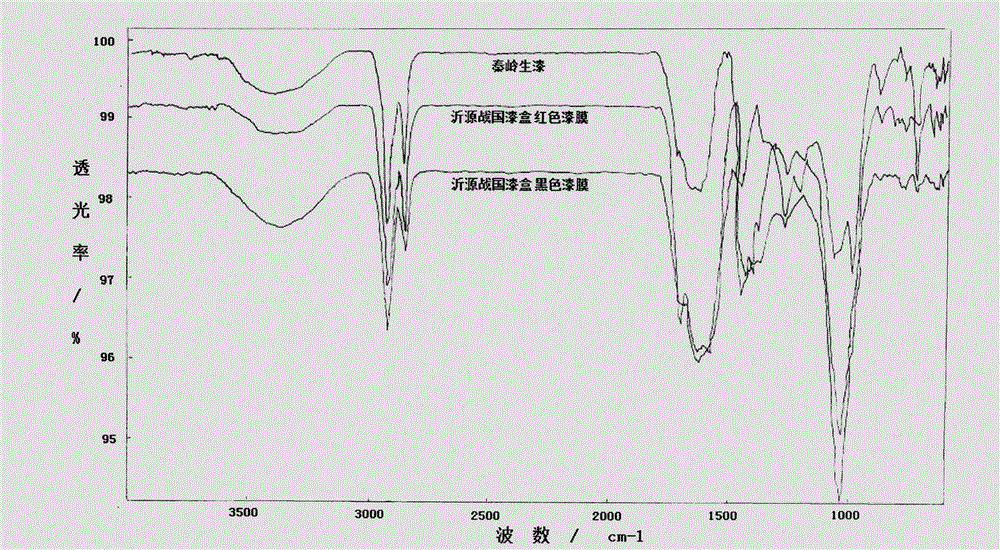
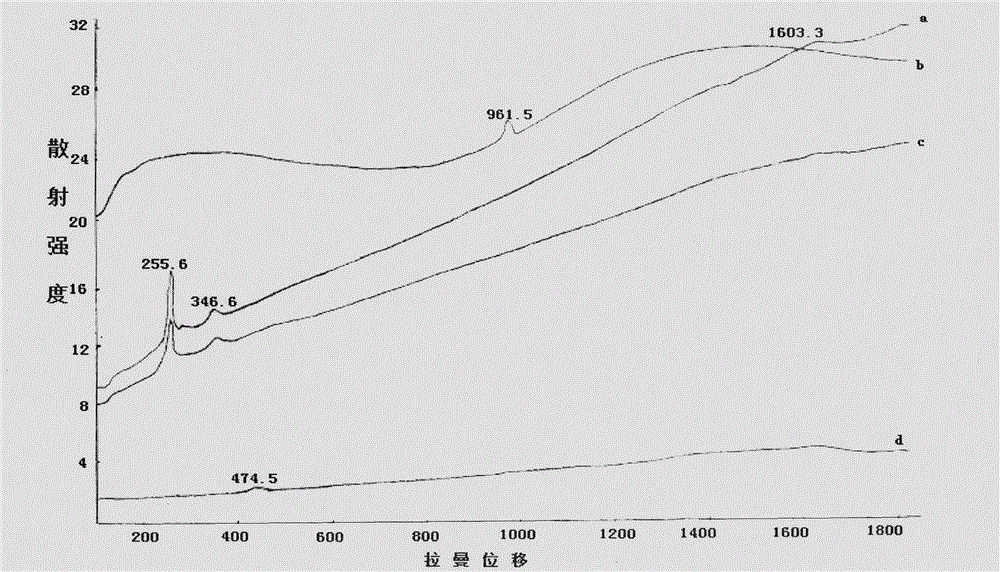
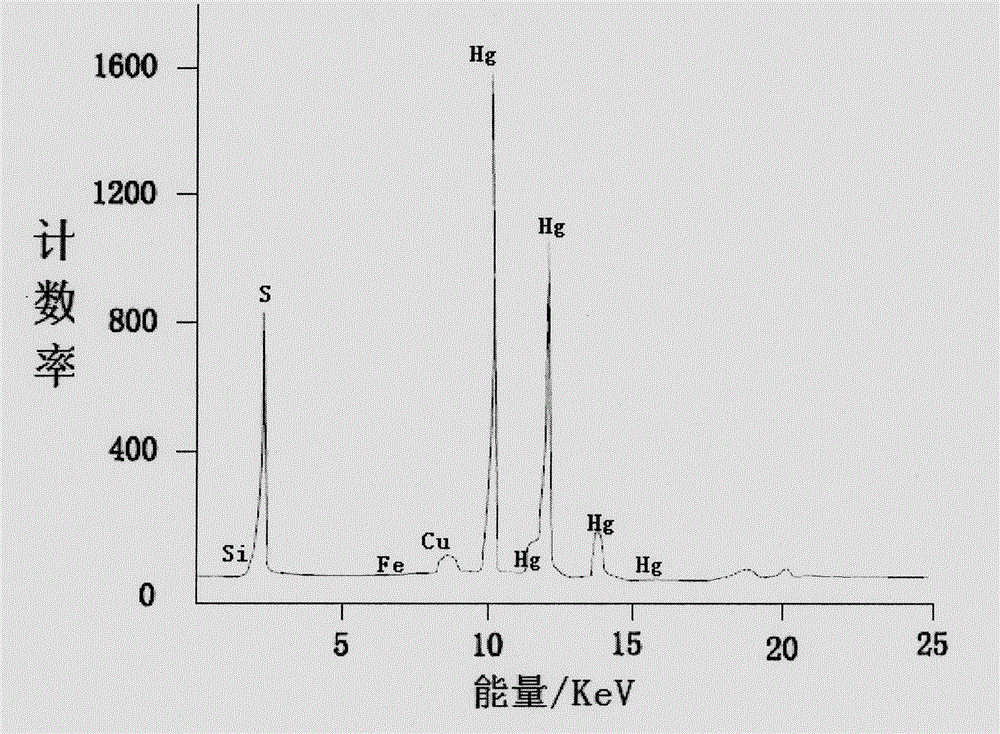
InactiveCN104807820AMaterial analysis by optical meansMaterial analysis using radiation diffractionChemical structureLacquer
The invention discloses an analyzing and testing method for an unearthed ancient lacquer film. Different analyzing methods are adopted for different coatings of the lacquer film; firstly, the overall structure (such as layering as well as the color, the thickness and the content condition of each layer) of the lacquer film is observed by the aid of a microscope, and then plant species identification is performed on a residual matrix and a fiber layer by the aid of the microscope; XRD (x-ray diffraction) analysis is performed on a lacquer ash layer, and main components and chemical structures of the main components can be detected; infrared spectroscopic analysis is performed on a raw lacquer layer, and the lacquer film is identified; the constitution structure of a pigment of a pigment layer is determined with an XRF (X-ray fluorescence) and XRD combined method. Besides, testing means are relatively flexible for analysis of additives in each layer, crystalline substances such as quartz and the like can be analyzed through XRD, non-crystalline substances can be analyzed with the Raman spectrum, a firm foundation is laid for further research on the lacquer film, a theoretical basis is provided for restoration of the ancient lacquer painting technology, and a valuable reference is provided for conservation and renovation work.
Owner:秦始皇帝陵博物院
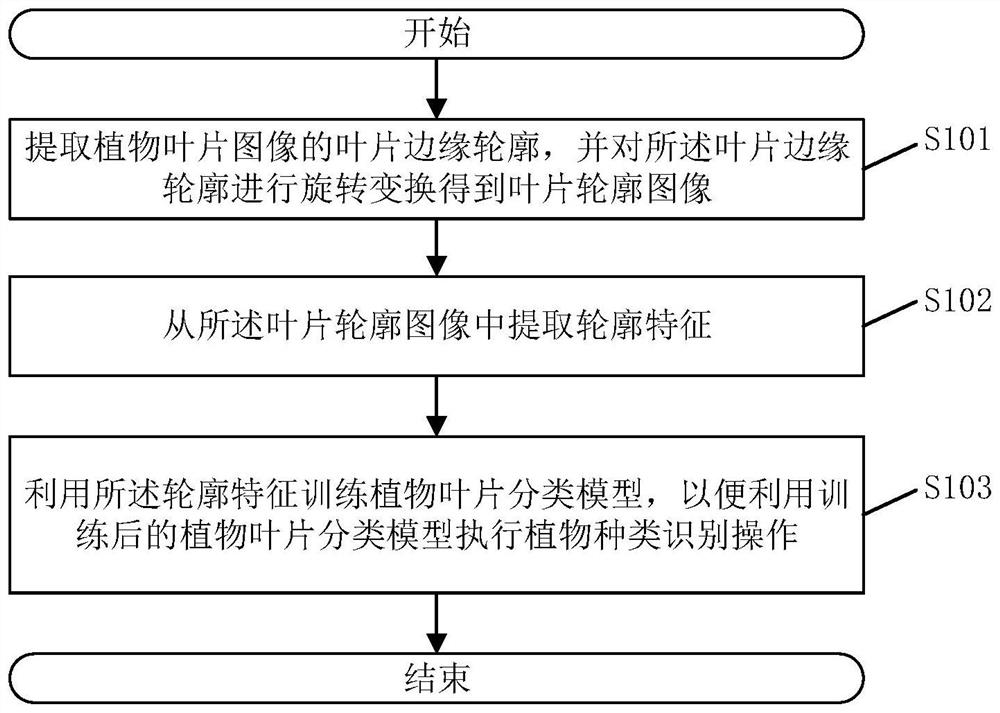
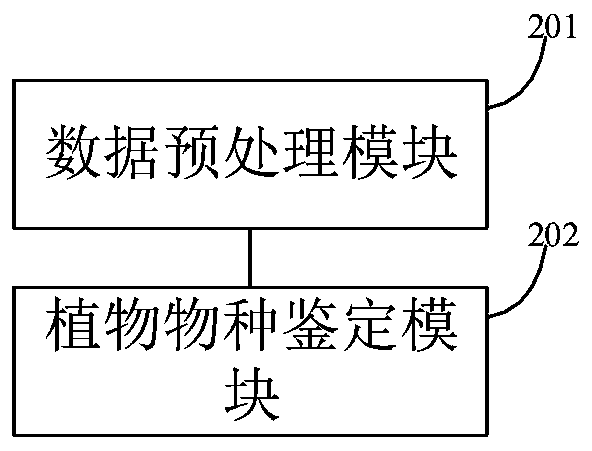
Plant species identification method and system, electronic equipment and storage medium
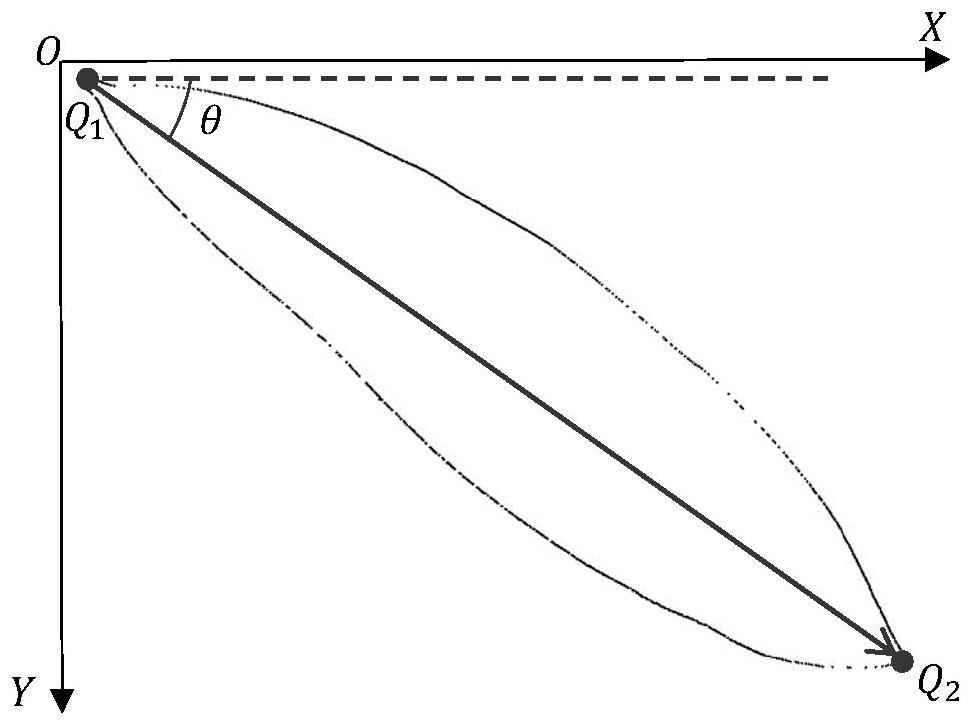
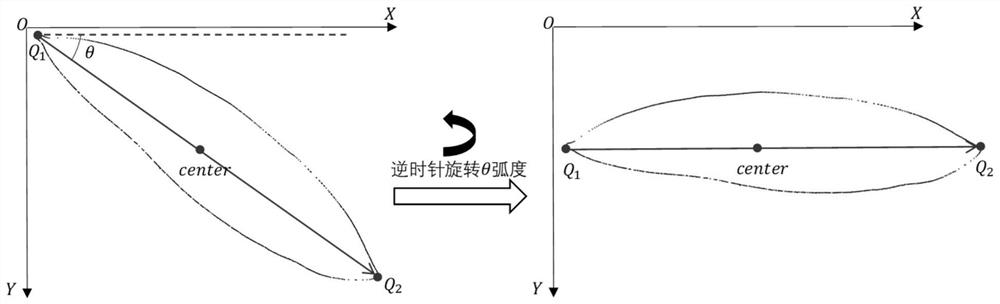
PendingCN113837037AGuaranteed feature invarianceImprove featuresCharacter and pattern recognitionMachine learningBiologyPlant species identification
The invention discloses a plant type identification method, and the method comprises the steps: extracting a leaf edge contour of a plant leaf image, and carrying out the rotation transformation of the leaf edge contour, and obtaining a leaf contour image; wherein the contour inclination angles of all the blade contour images are the same; extracting contour features from the blade contour image; wherein the contour features comprise any one or a combination of any several of a convex defect feature, a contour shape symmetry feature and a contour shape vector feature; and training a plant leaf classification model by using the contour features so as to execute a plant type identification operation by using the trained plant leaf classification model, thereby improving the accuracy of plant type identification. The invention also discloses a plant species identification system, electronic equipment and a storage medium, which have the above beneficial effects.
Owner:GUANGDONG INSPUR BIG DATA RES CO LTD
Method and equipment for identifying plant species based on ITS2
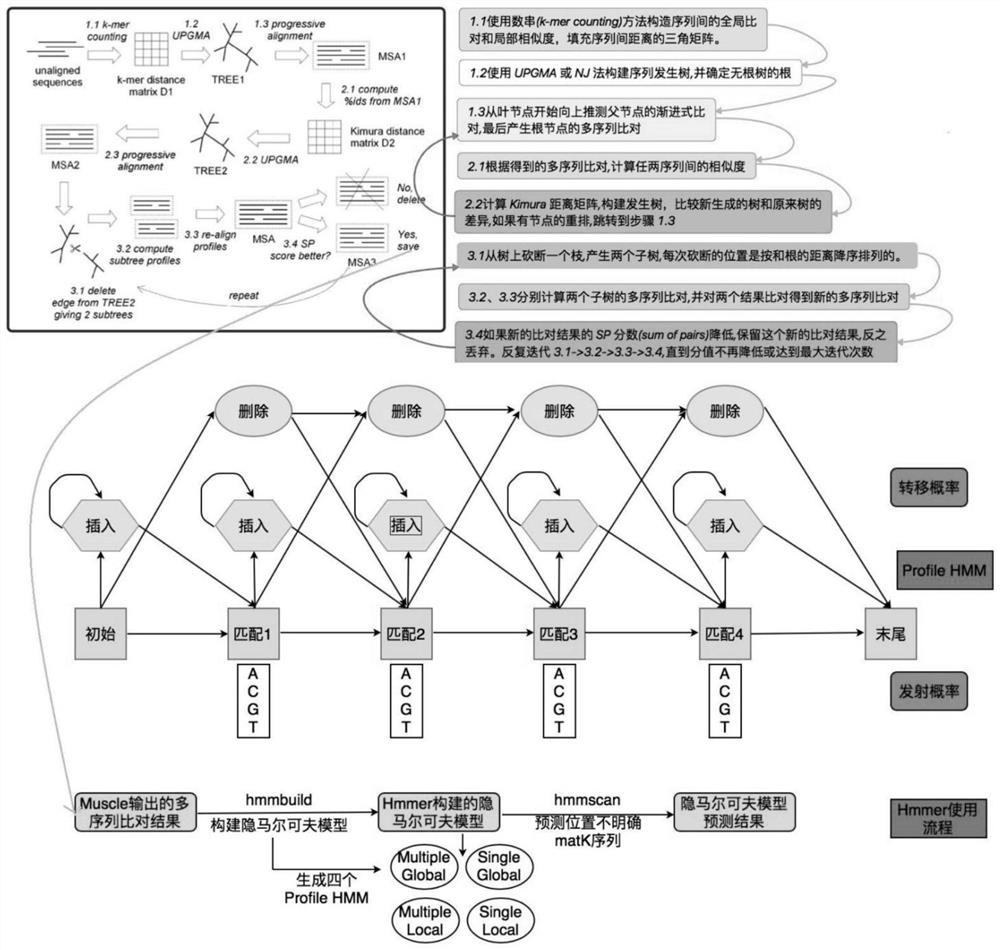
InactiveCN110111847AAchieve accurate identificationBiostatisticsSequence analysisClustered dataHide markov model
An embodiment of the invention provides a method and equipment for identifying plant species based on ITS2, wherein the method comprises the steps of acquiring taxology information and an ITS2 sequence of a to-be-identified plant, decomposing for obtaining a file for processing, extracting the ITS2 sequence and species note in the file for processing, by means of a hidden Markov model, positioningthe ITS2 sequence with the indefinite position, eliminating the ITS2 sequence which comprises more than three one-molecule nitrogenous base, and obtaining preprocessing data; clustering the preprocessing data, performing error elimination on the clustered data, obtaining final data, using the ITS2 sequence and the species note in the final data for constructing a database, and aligning the ITS2 sequence in the database for finishing plant species identification. The method and the equipment for identifying the plant species based on ITS2 according to the embodiment of the invention can realize accurate identification of the plant species in a relatively strict and complete plant species identification flow through high-flux sequencing technology.
Owner:EZHOU INST OF IND TECH HUAZHONG UNIV OF SCI & TECH +1
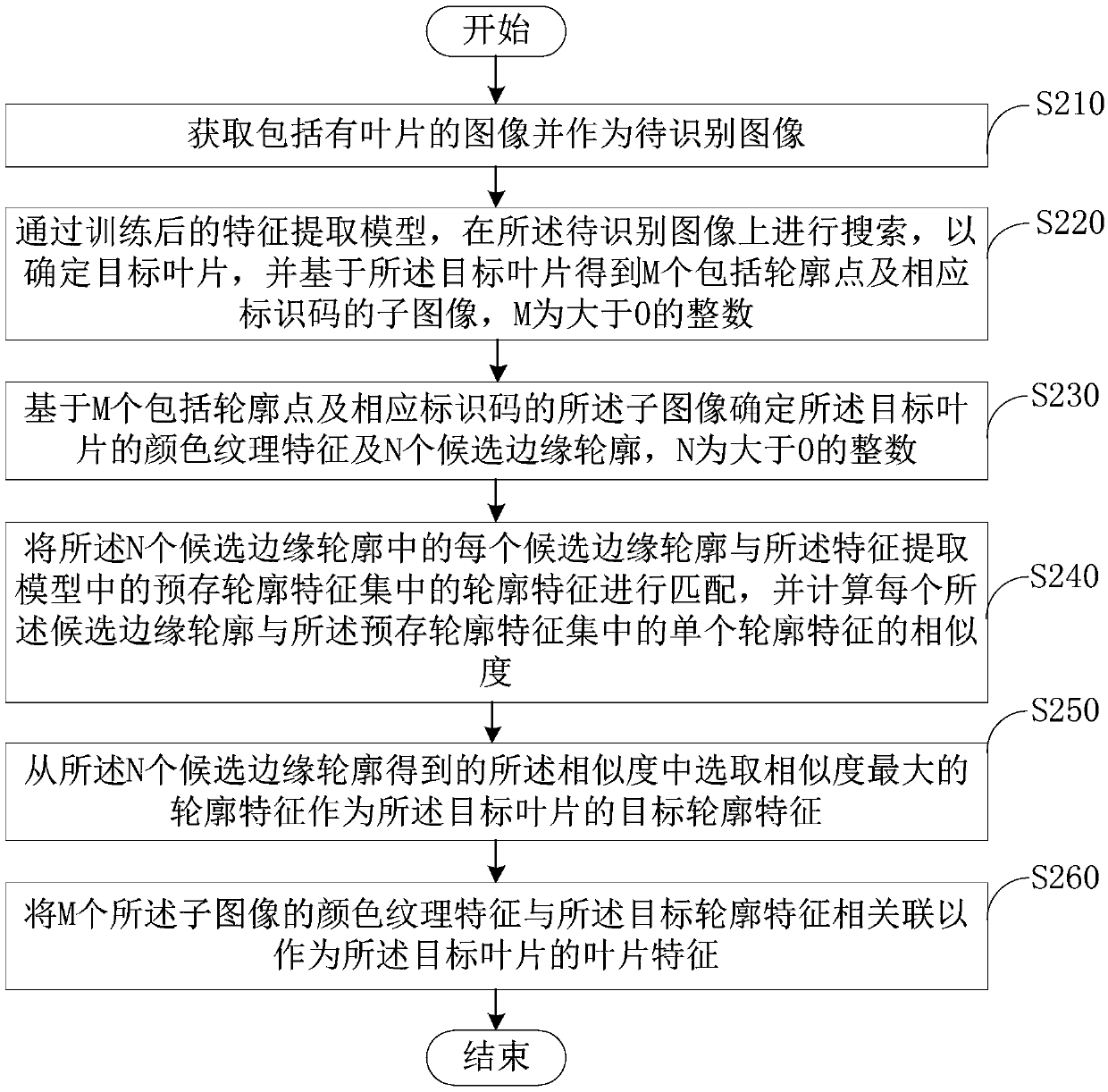
Blade feature extraction method and device
ActiveCN109558883AImprove the efficiency of extracting leaf featuresAccelerated identification of plant speciesCharacter and pattern recognitionNeural architecturesImage extractionFeature extraction
The invention provides a blade feature extraction method and device, and relates to the technical field of computer image processing. According to the method, a target leaf is decomposed into M contour point sub-images with identification codes, then all the sub-images are input into a feature extraction model, and color texture features and contour features of the target leaf are extracted basedon all the sub-images. According to the scheme, the feature extraction model based on deep learning is adopted, the blade features can be automatically extracted, manual operation is simplified, and therefore the efficiency of extracting the blade features is improved. In addition, the color texture features and the contour features are combined, so that the plant species identification accuracy can be improved by utilizing the extracted leaf features.
Owner:宁夏智启连山科技有限公司
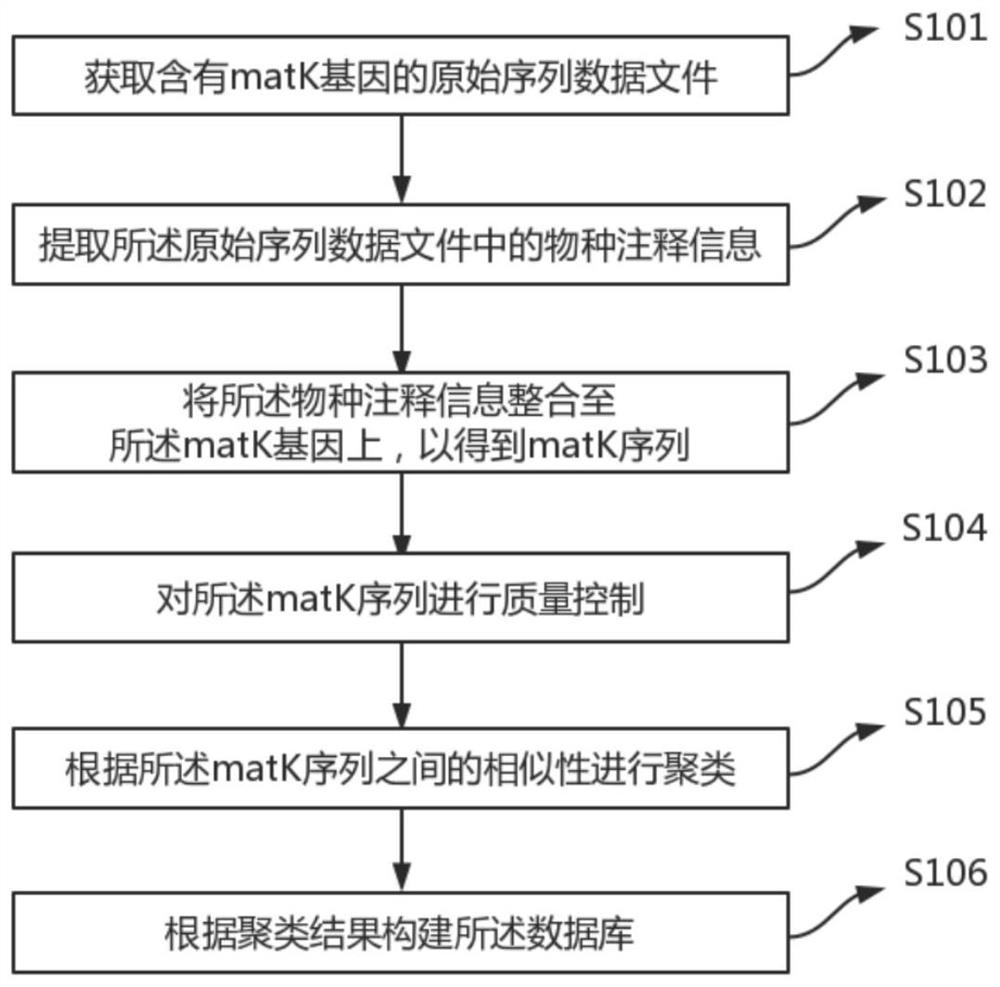
A construction method and database of unknown plant species identification database based on matk gene
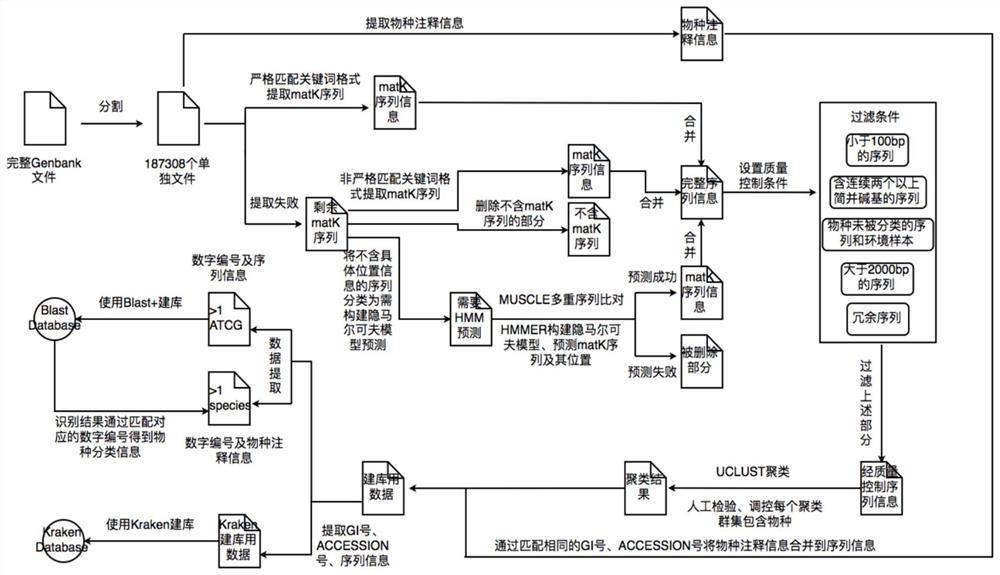
ActiveCN111681704BWide coverageImprove data qualityCharacter and pattern recognitionProteomicsData fileSpecies level
A construction method and database of an unknown plant species identification database based on the matk gene, the method comprising the steps of: obtaining an original sequence data file containing the matk gene; extracting species annotation information in the original sequence data file; The annotation information is integrated into the matK gene to obtain the matK sequence; quality control is performed on the matk sequence; clustering is performed according to the similarity between the matk sequences; and the database is constructed according to the clustering results. The construction method and database advantages of a kind of unknown plant species identification database based on matk gene provided by the application are: (1) high detection throughput; (2) high sensitivity and selectivity; (3) database coverage constructed by the method (4) Bioinformatics methods can be used to identify unknown species at the species level.
Owner:EZHOU INST OF IND TECH HUAZHONG UNIV OF SCI & TECH +1
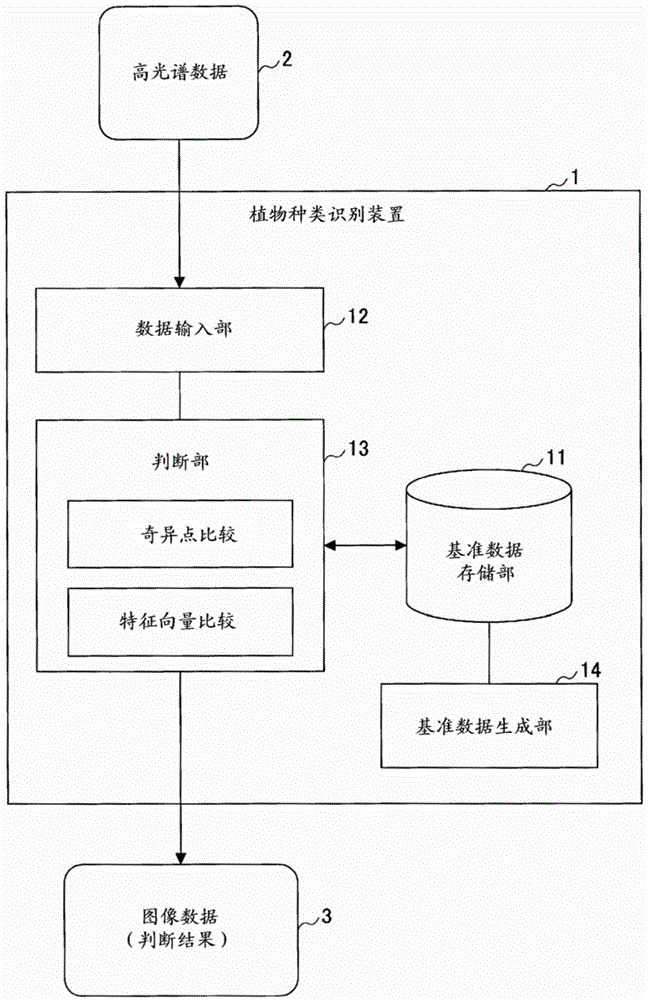
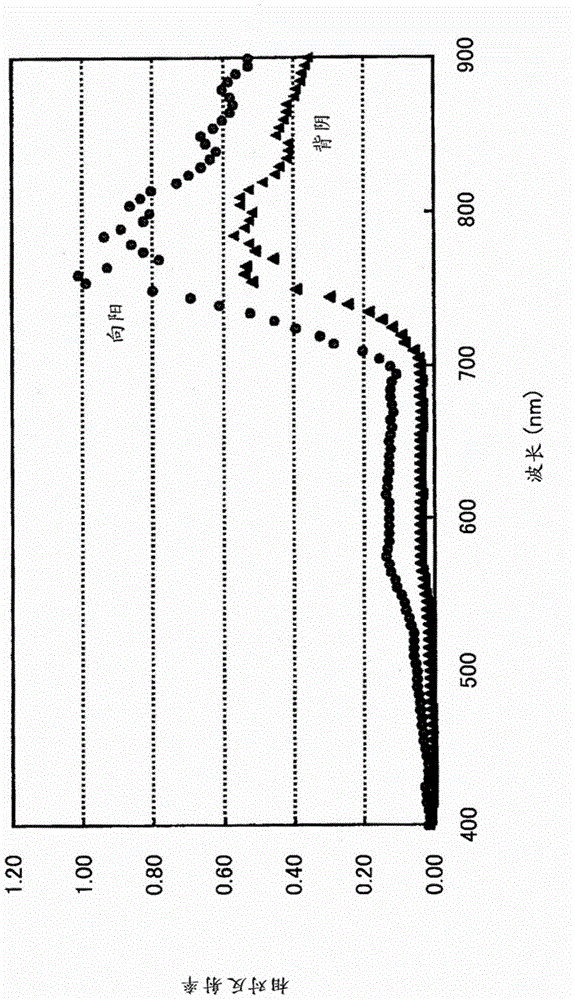
Plant species identification device and method
ActiveCN103620381BEasy to identifyInvestigation of vegetal materialCharacter and pattern recognitionBaseline dataPattern recognition
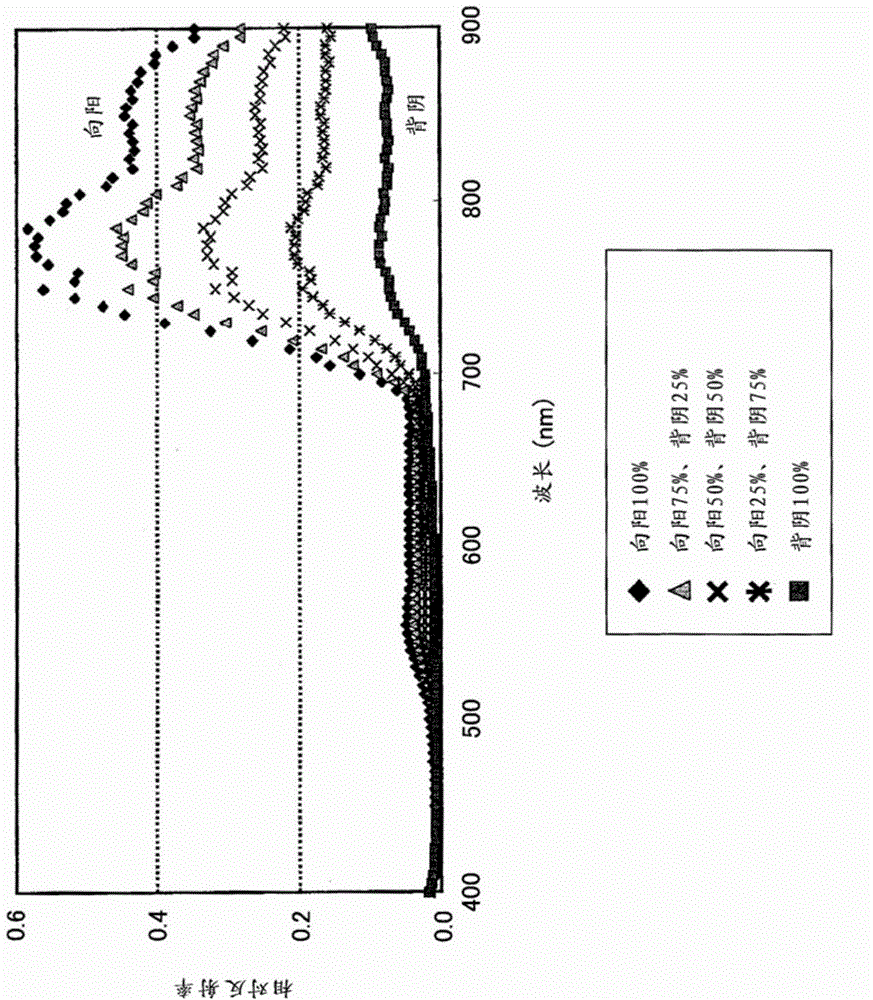
A plant species identification apparatus for identifying plant species is disclosed. A reference data storage part stores reference spectral data which indicate a reflectance spectral feature classified by area segments including a sunlit portion and a shaded portion in addition to the plant species. A data input part acquires hyperspectral data to be a target. A determination part specifies the reflectance spectral feature of a pixel for each of pixels of the hyperspectral data from the reference data storage part and to determine the plant species of the pixels based on a classification of the reference spectral data.
Owner:FUJITSU LTD
Plant species identification method based on digital image
ActiveCN102324038BEliminate the impact of recognition accuracyImprove accuracyCharacter and pattern recognitionFeature vectorTest sample
The invention provides a plant species identification method based on a digital image. The method comprises the following steps: collecting a plant organ digital image as a test sample, and extracting a characteristic vector; inputting the characteristic vector into a first stage classifier, and obtaining first n species in a vote number ranking, wherein n is larger than 3 and smaller than 10; the first stage classifier is obtained through the following modes: carrying out classifier training based on a characteristic set of all training samples; inputting the characteristic vector into a second stage classifier, and obtaining an identification result; the second stage classifier is obtained through the following modes: extracting a characteristic set corresponding to the n species from the characteristic set of all training samples to carry out classifier training. According to the invention, through a grading SVM classifier, sensitivity of the classifier to sample kind quantity is effectively reduced, influence of sample species increase on identification accuracy is eliminated, a problem of low accuracy of large sample size identification by the SVM classifier is overcome, and plant identification accuracy is raised.
Owner:BEIJING FORESTRY UNIVERSITY
Plant species identification method based on leaf hog characteristics and intelligent terminal platform
InactiveCN104036235BEfficient identificationImprove accuracyCharacter and pattern recognitionHistogram of oriented gradientsSvm classifier
Owner:TONGJI UNIV
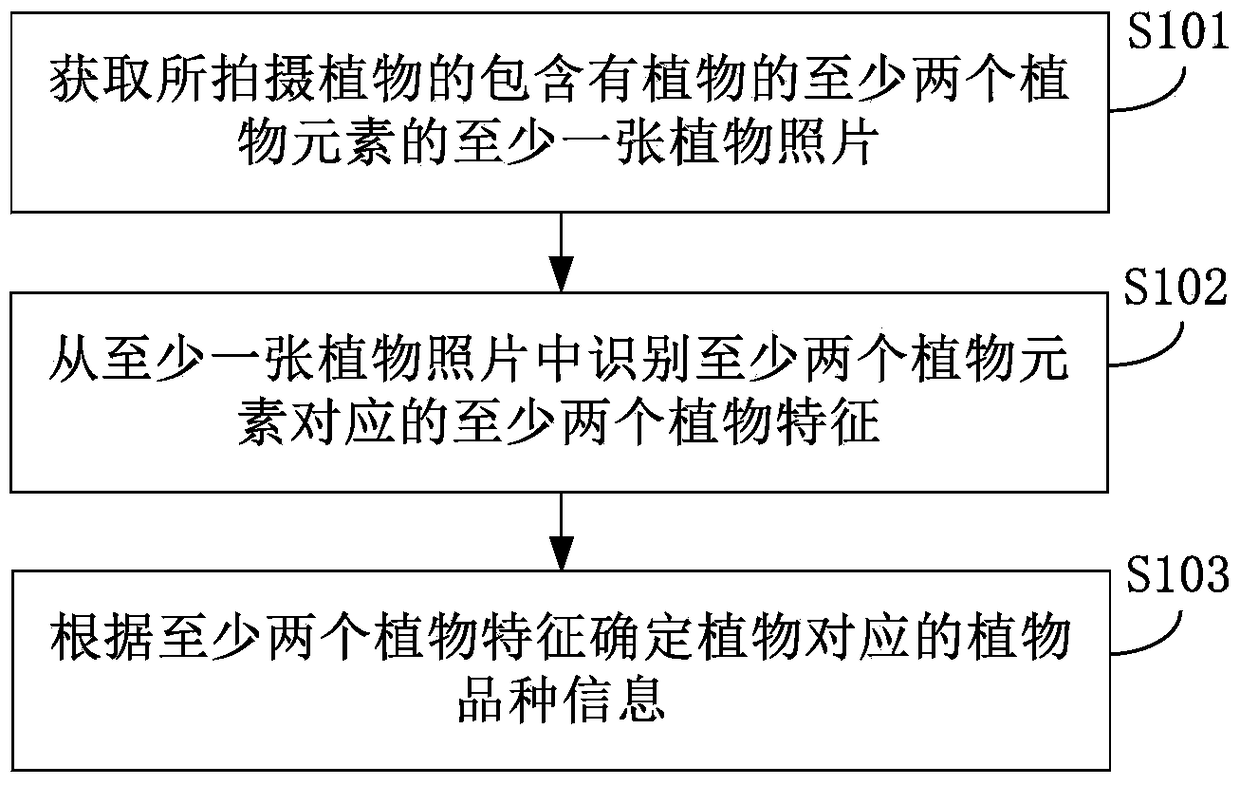
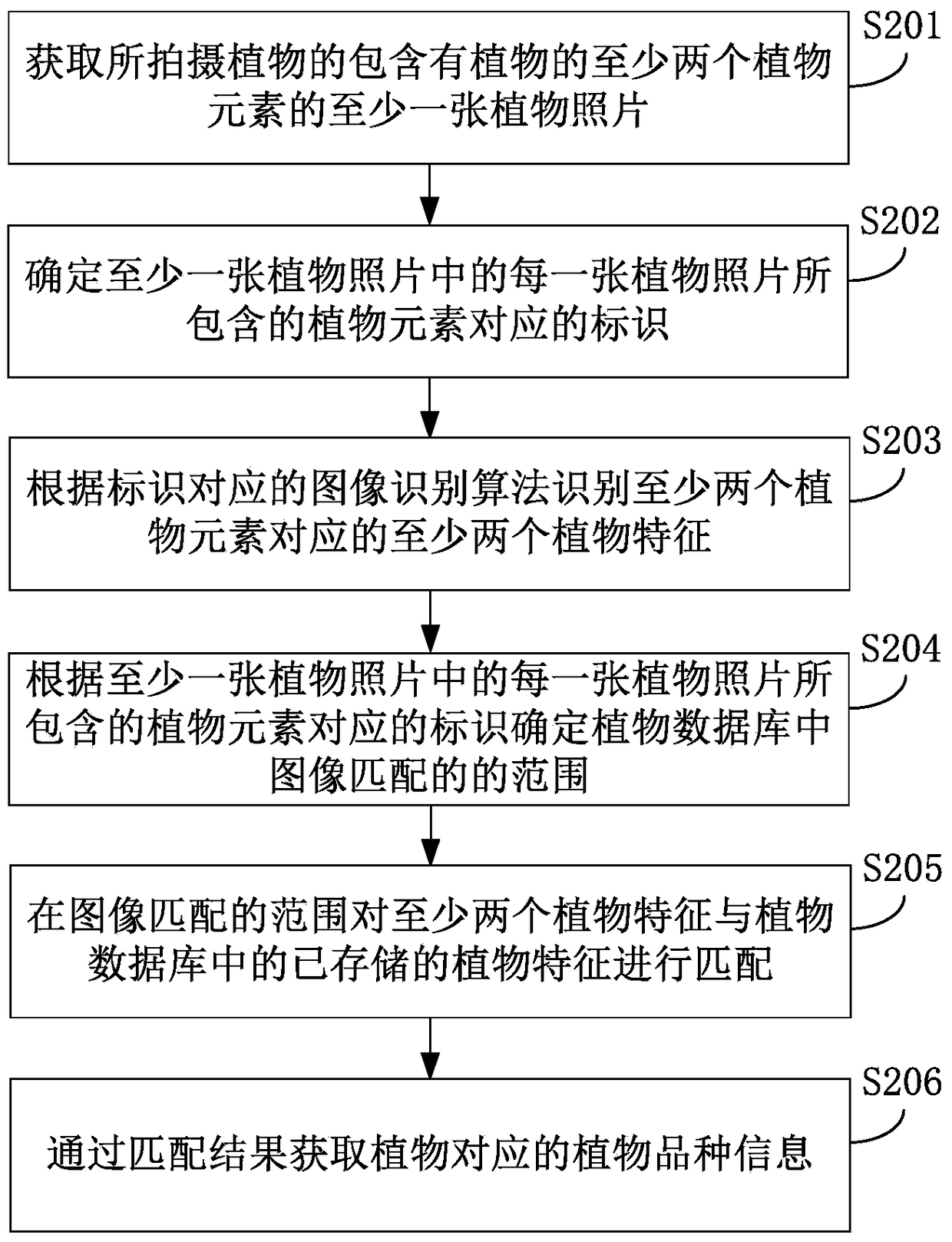
Plant variety identification method and device
ActiveCN105203456BReduce difficultyAdd reference informationMaterial analysis by optical meansPlant varietyPlant species identification
The invention discloses a plant species identification method and an apparatus thereof. The method comprises the following steps: acquiring at least one plant picture containing at least two plant elements of a shot plant; identifying at least two plant characteristics corresponding to the at least two plant elements from the at least one plant picture; and determining plant species information corresponding to the plant according to the at least two plant characteristics. The method and the apparatus allow a user to obtain the plant species information of the shot plant through shooting the plant, so plant reference information is increased in the plant identification process, the plant identification accuracy is improved, identification of the plant by the user in a subjective judgment mode is avoided, and the user's identification difficulty of the plant is reduced.
Owner:XIAOMI INC
Plant species identification method, system and device and storage medium
PendingCN114463629AAvoid missingImprove accuracyCharacter and pattern recognitionNeural architecturesBiologyNeutral network
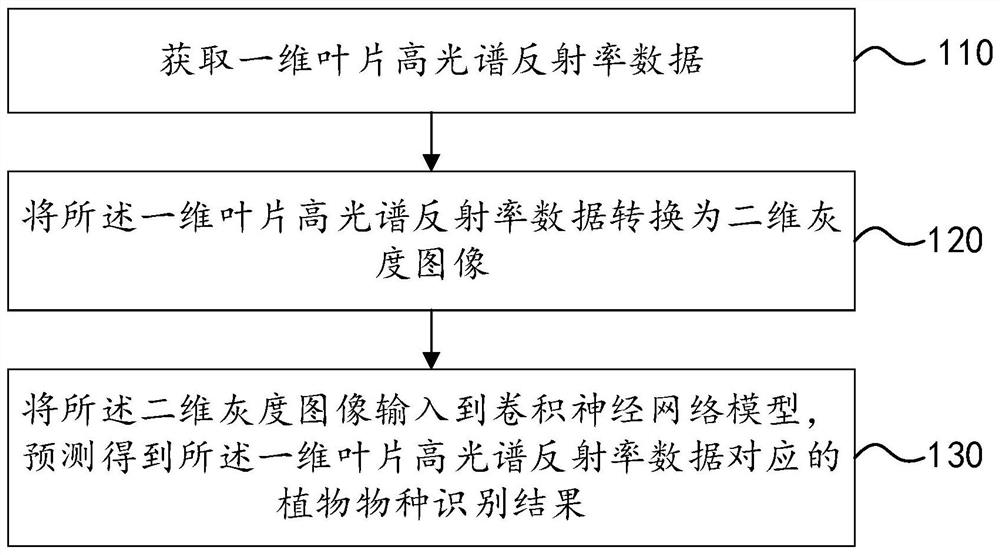
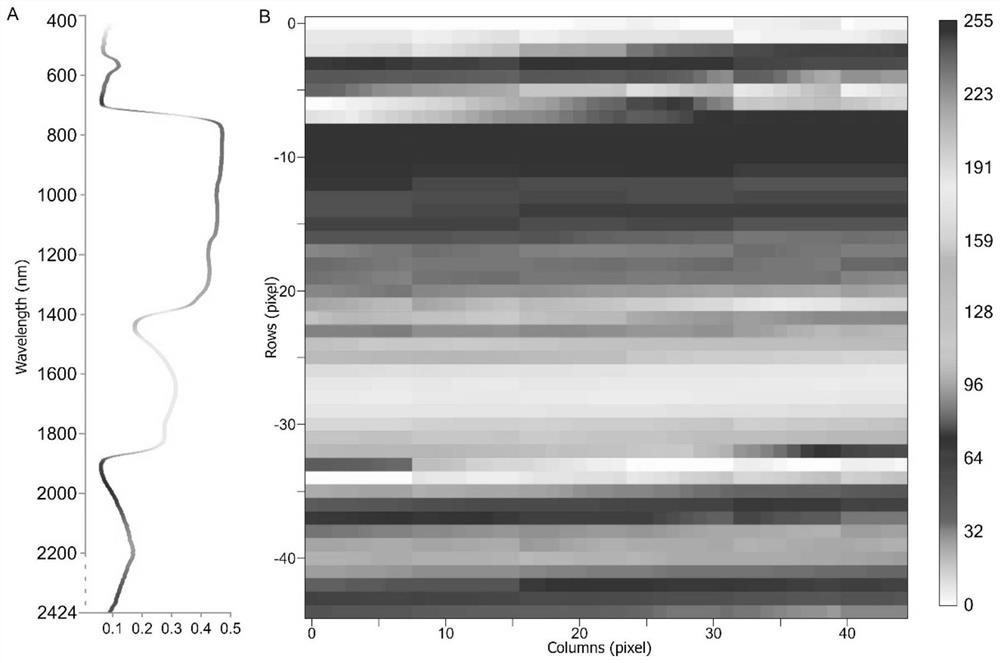
The invention discloses a plant species identification method, system and device and a storage medium, and can be applied to the technical field of species identification. The method comprises the following steps: acquiring one-dimensional leaf hyperspectral reflectivity data; converting the one-dimensional leaf hyperspectral reflectivity data into a two-dimensional grayscale image; and inputting the two-dimensional grayscale image into a convolutional neural network model, and predicting to obtain a plant species identification result corresponding to the one-dimensional leaf hyperspectral reflectivity data. According to the method, after all the acquired one-dimensional leaf hyperspectral reflectivity data are converted into the two-dimensional gray level image, all the hyperspectral data are fully utilized, useful information is prevented from being omitted, then the two-dimensional gray level image is input into the convolutional neural network model, and plant species identification is carried out by utilizing the convolutional neural network model, so that plant species identification efficiency is improved. Therefore, the accuracy of a plant species identification result is improved.
Owner:GUANGZHOU INST OF GEOGRAPHY GUANGDONG ACAD OF SCI
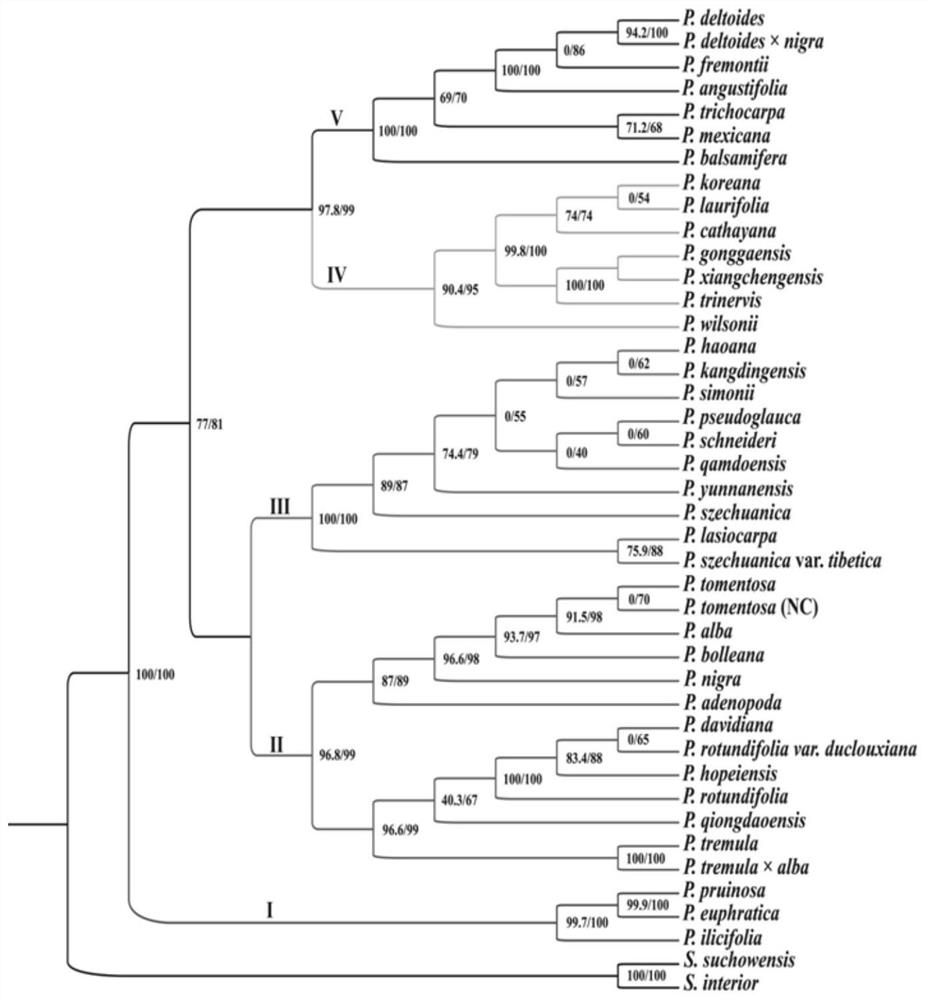
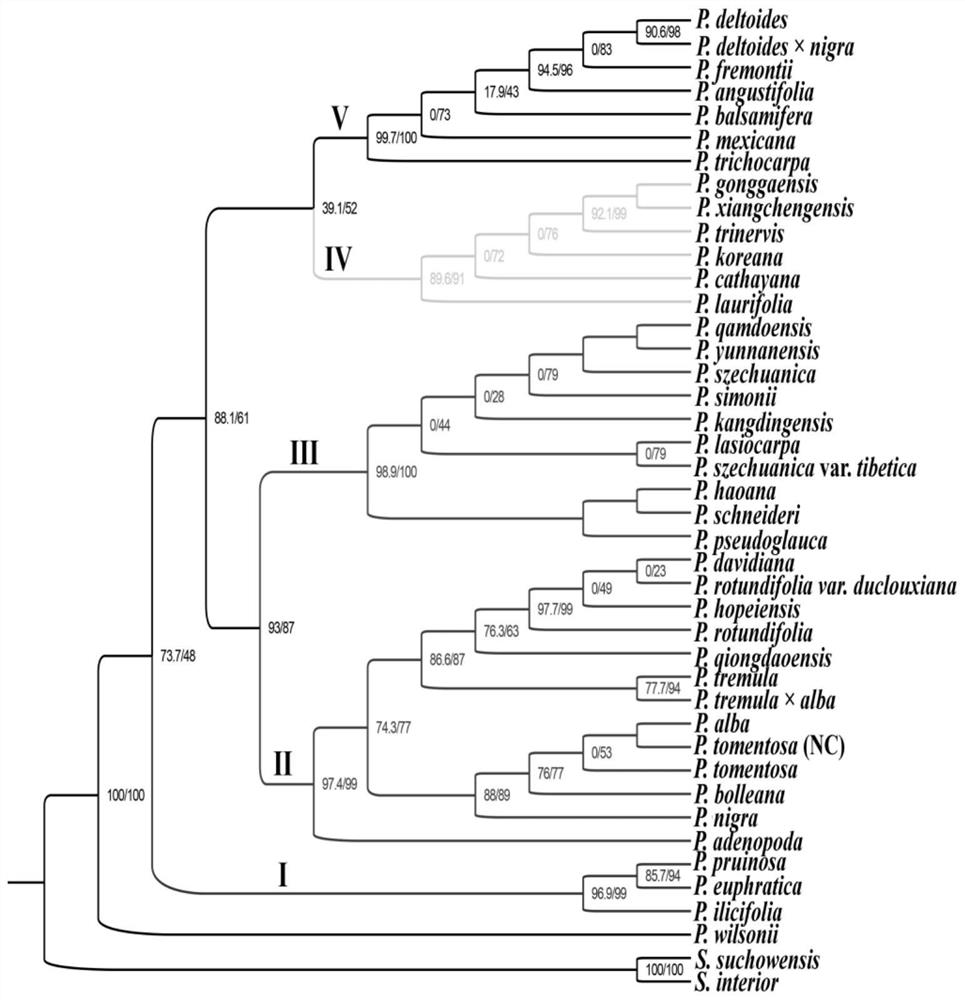
Chloroplast genome capable of identifying populus species as well as PCR amplification primer and application of chloroplast genome
ActiveCN112921111ARapid identificationAccurate identificationMicrobiological testing/measurementDNA/RNA fragmentationNdhFNucleotide
The invention belongs to the technical field of molecular biology, relates to the field of plant species identification, and discloses a chloroplast genome sequence capable of specifically identifying populus species and a specific PCR amplification primer of the chloroplast genome sequence, the specific primer comprises 9 hypervariable regions in the populus chloroplast genome, including ccsA-ndhD, ndhC-trnV, ndhF-ccsA, psbM-trnD, psbZ-trnfM, rpoB-petN, rps15-ycf1, ycf1 and trnK-psbK respectively, and the nucleotide sequences of the 9 hypervariable regions are shown in SEQ ID NO.1-9. The invention aims to provide the 9 hypervariable regions of the chloroplast genome suitable for identifying the populus species and the specific primer of the chloroplast genome, the 9 hypervariable region sequences are combined, the populus species can be rapidly and accurately identified, and an effective DNA bar code is provided for identifying the populus species.
Owner:SOUTHWEST FORESTRY UNIVERSITY
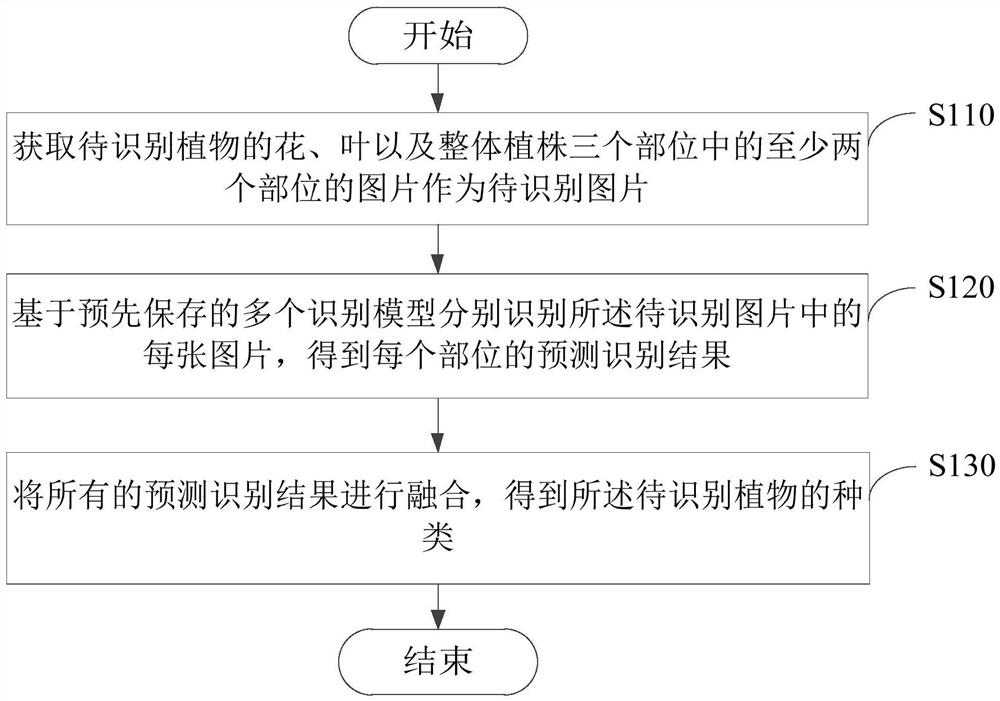
A plant species identification method and device
ActiveCN108256568BThe classification result is accurateImplement classificationCharacter and pattern recognitionBiotechnologyAlgorithm
The present invention provides a method and device for identifying plant species. The method obtains pictures of at least two parts of the plant to be identified, such as flowers, leaves, and the whole plant, as images to be identified; and based on multiple pre-stored The recognition model separately recognizes each of the pictures to be recognized, and obtains a predicted recognition result of each part; and then fuses all the predicted recognition results to obtain the type of the plant to be recognized. According to the overall information of leaves, flowers and plants, this method combines the overall and local information to perform fine identification and analysis on plants, so as to obtain accurate plant classification results, which can make the algorithm more robust. When users use this algorithm , enabling better plant classification than ever before.
Owner:宁夏智启连山科技有限公司
A microfluidic detection chip based on nucleic acid amplification technology and its preparation method
ActiveCN105331528BReduce volumeEasy to carryBioreactor/fermenter combinationsBiological substance pretreatmentsNucleic acid amplification techniquePlant species identification
The invention discloses a microfluidic detection chip based on nucleic acid amplification technology and a preparation method thereof. The chip is a 4-layer disc centrifugal design, and one chip can meet the detection work of 20 samples and 12 parameters of each sample. It is equipped with CD-ROM centrifuge, temperature control equipment and nucleic acid amplification reagent consumables, which can meet the detection needs of agricultural products, genetically modified ingredients in food, pathogenic bacteria, animal and plant species identification, etc., and has the characteristics of fast, convenient and high throughput.
Owner:TIANJIN INSTITUE OF QUALITY STANDARD & TESTING OF AGRICULTUAL PRODS
Genome-wide SNP and indel molecular marker method of large yellow croaker based on double enzyme digestion
ActiveCN105349675BReduce the scope of sequencingLow costMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyEnzyme digestion
The invention discloses a larimichthys crocea genome-wide SNP and InDel molecular marker method based on double enzyme digestion. The method comprises the following steps: 1) joint sequence design; 2) genome DNA enzyme digestion; 3) joint sequence connection; 4) sample mixing and PCR amplification; 5) high-throughput sequencing; 6) SNP locus excavation and analysis through sequencing data. According to the method, the modern molecular biology and the advanced high-throughput sequencing technology are combined, the method of combination of EcoRII and NlaIII double enzyme digestion is used, the genome-wide DNA molecules are subjected to enzyme digestion, DNA segments with certain lengths are obtained for carrying out library establishment sequencing and SNP molecular marker excavation, and the uniformly distributed genome-wide SNP locus information is obtained; the workload and cost of genome-wide marker analysis are greatly reduced, and the method is also applicable to the genome-wide marker analysis of other species; the SNP marker excavated through the method is applicable to the study fields such as animal and plant species identification, species genetic genealogy analysis, germplasm resource genetic diversity analysis and genetic breeding.
Owner:JIMEI UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com