Chloroplast genome capable of identifying populus species as well as PCR amplification primer and application of chloroplast genome
A technology of chloroplast genome and tree species, which is applied in the field of plant species identification and molecular biology, and can solve the problems of difficult amplification, sequencing and assembly, high identification rate, and cumbersome methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
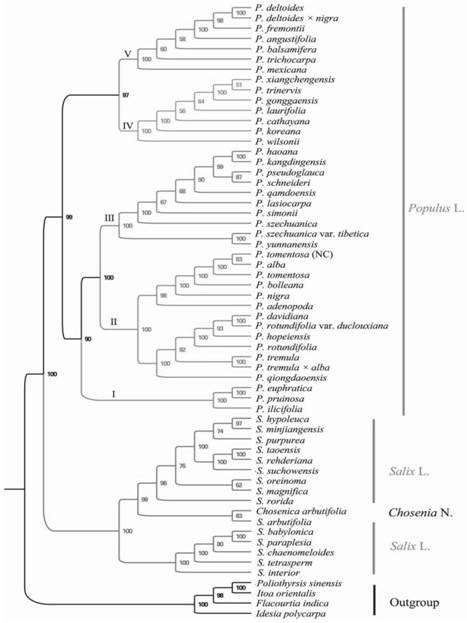
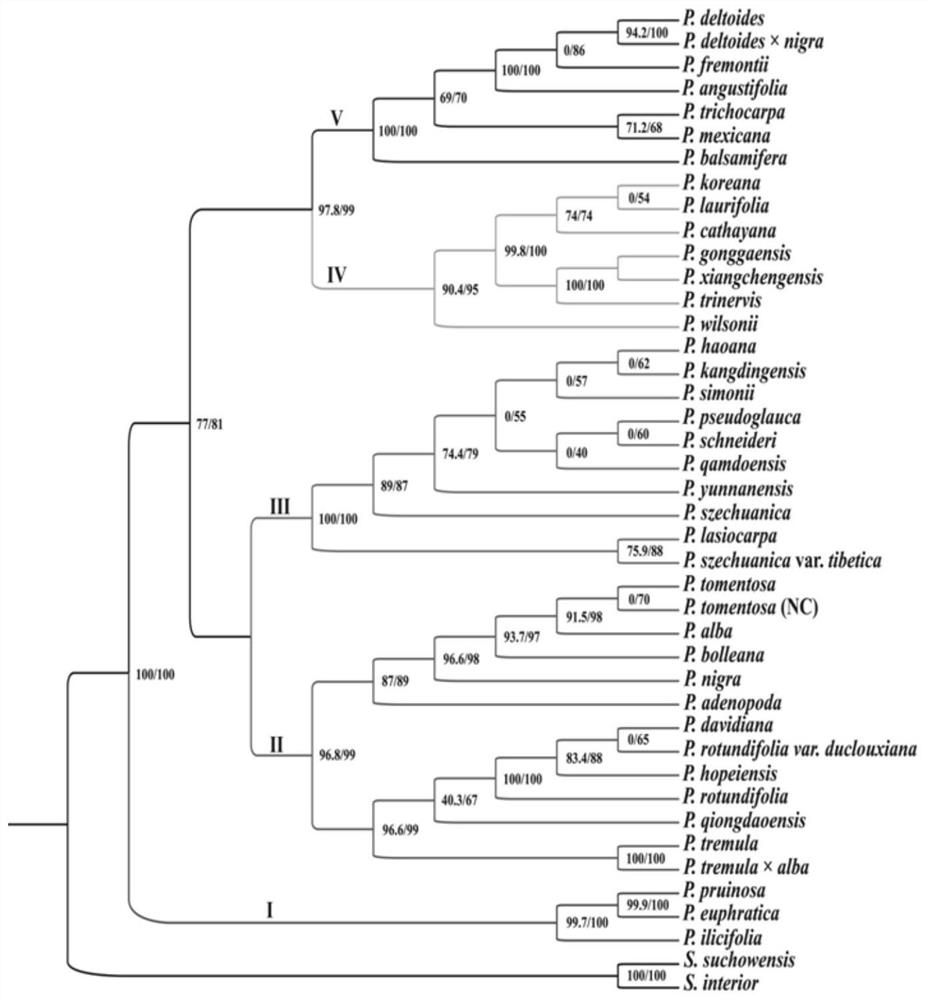
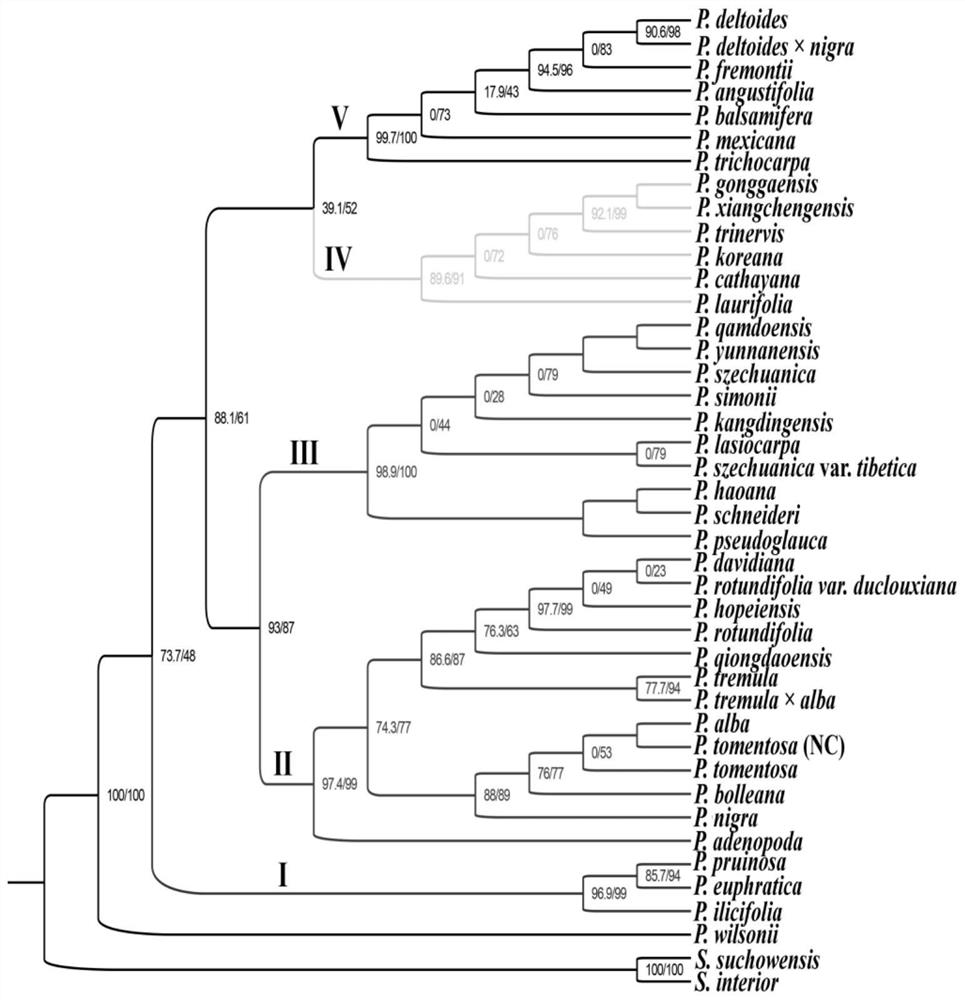
[0020] The present invention will be further described below in conjunction with the accompanying drawings and specific embodiments.
[0021] 1. Materials
[0022] The young leaves of different species of Populus were collected, stored in a ziplock bag filled with silica gel for rapid drying, and brought back to the laboratory for later use. The materials used are shown in Table 1, including:
[0023] Table 1 Sample Source Table
[0024]
[0025] 2. Genomic DNA extraction
[0026] Weigh 100 mg of poplar leaves dried with color-changing silica gel into a 2 mL centrifuge tube, add an appropriate amount of PVP powder, and grind with a freeze-mixing ball mill. The ground samples are extracted from the total genomic DNA according to the Ezup Column Plant Genomic DNA Extraction Kit.
[0027] 3. PCR amplification
[0028] The 9 hypervariable regions were amplified by PCR. The total volume of the PCR reaction was 25 μL, the DNA template was 1.0 μL, the 10×buffer Mix was 12.5 μL...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com