Patents
Literature
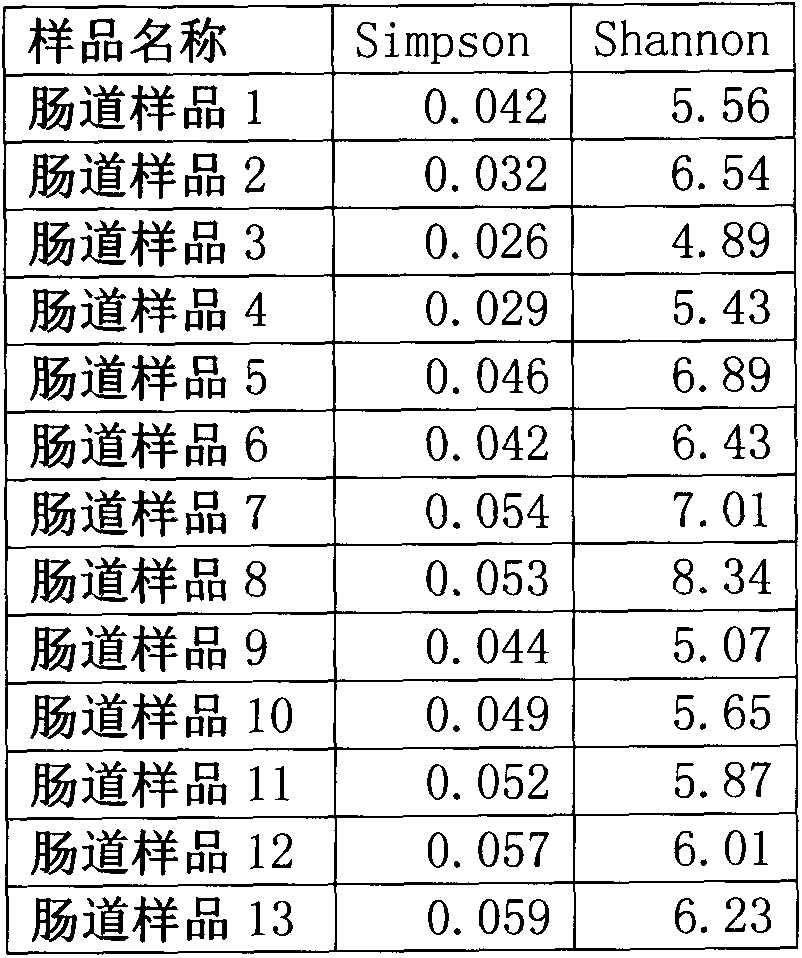
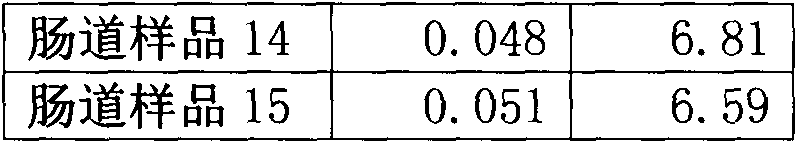
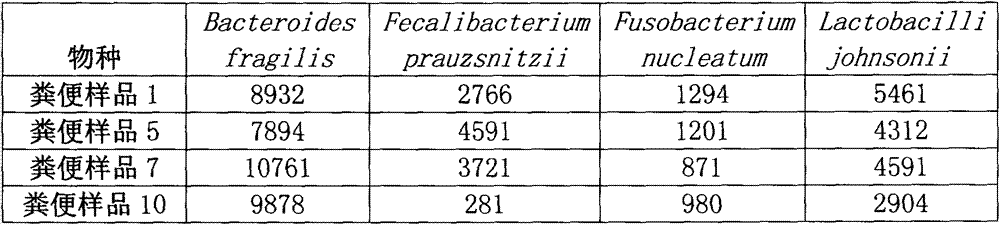
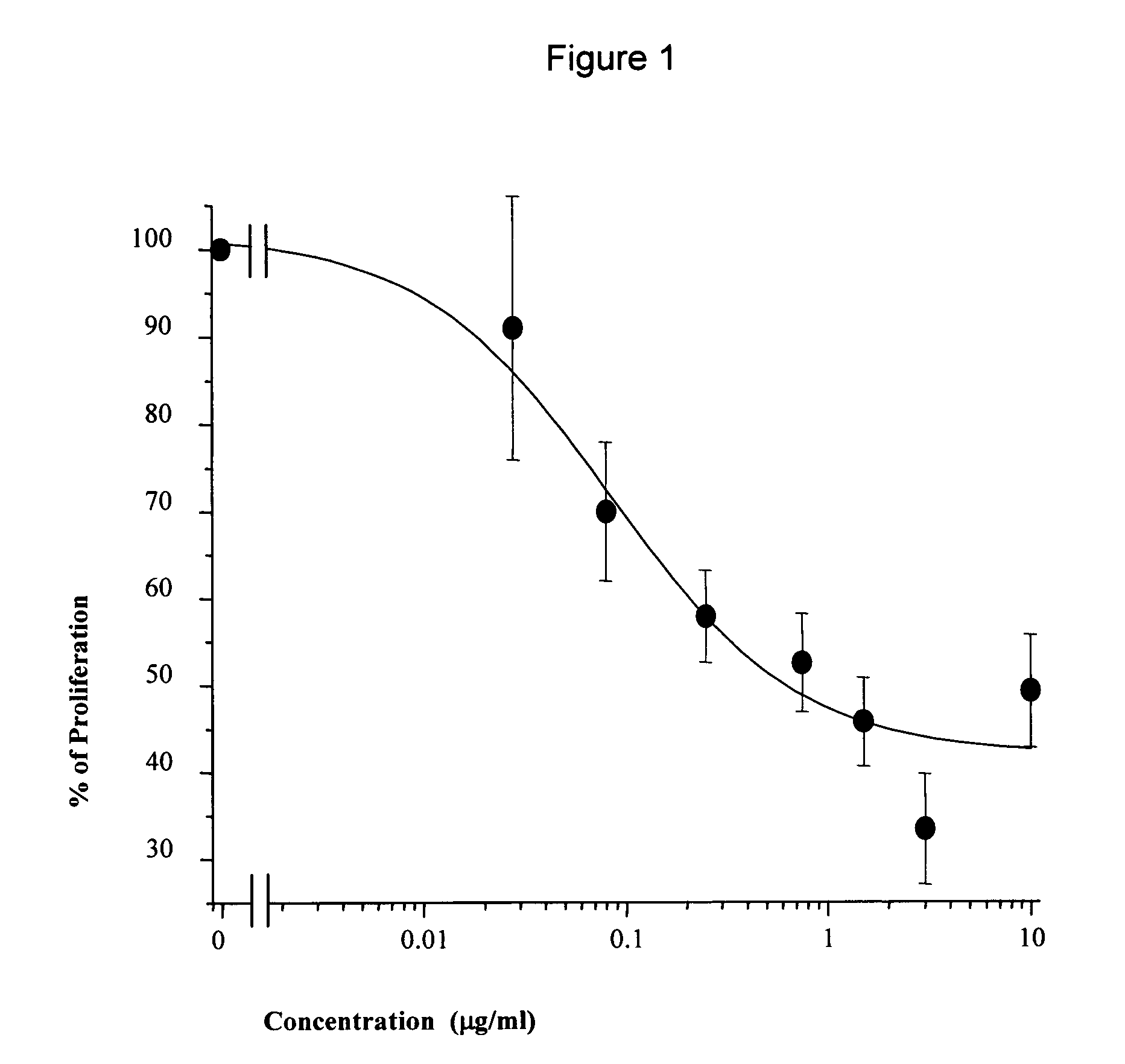
146 results about "Hypervariable region" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
A hypervariable region (HVR) is a location within nuclear DNA or the D-loop of mitochondrial DNA in which base pairs of nucleotides repeat (in the case of nuclear DNA) or have substitutions (in the case of mitochondrial DNA). Changes or repeats in the hypervariable region are highly polymorphic.
Binding polypeptides with diversified and consensus vh/vl hypervariable sequences
ActiveUS20070160598A1Raise the possibilitySmall sizeAnimal cellsSugar derivativesAntibody hypervariable regionBioinformatics
The invention provides variant hypervariable regions comprising selected amino acid sequence diversity. Libraries comprising a plurality of these polypeptides are also provided. In addition, methods of and compositions for generating and using these polypeptides and libraries are provided.
Owner:GENENTECH INC
Antibody variants
Antibody variants of parent antibodies are disclosed which have one or more amino acids inserted in a hypervariable region of the parent antibody and a binding affinity for a target antigen which is at least about two fold stronger than the binding affinity of the parent antibody for the antigen.
Owner:CHEN YVONNE M +2
Binding polypeptides with diversified and consensus VH/VL hypervariable sequences
ActiveUS8679490B2Small sizeImprove efficiencyAnimal cellsSugar derivativesAntibody hypervariable regionAmino acid
The invention provides variant hypervariable regions comprising selected amino acid sequence diversity. Libraries comprising a plurality of these polypeptides are also provided. In addition, methods of and compositions for generating and using these polypeptides and libraries are provided.
Owner:GENENTECH INC
Metagenome 16S hypervariable region V3 based classification method and device thereof
InactiveCN102517392AReduce manual laborSave moneyBioreactor/fermenter combinationsBiological substance pretreatmentsClassification methodsAntibody hypervariable region
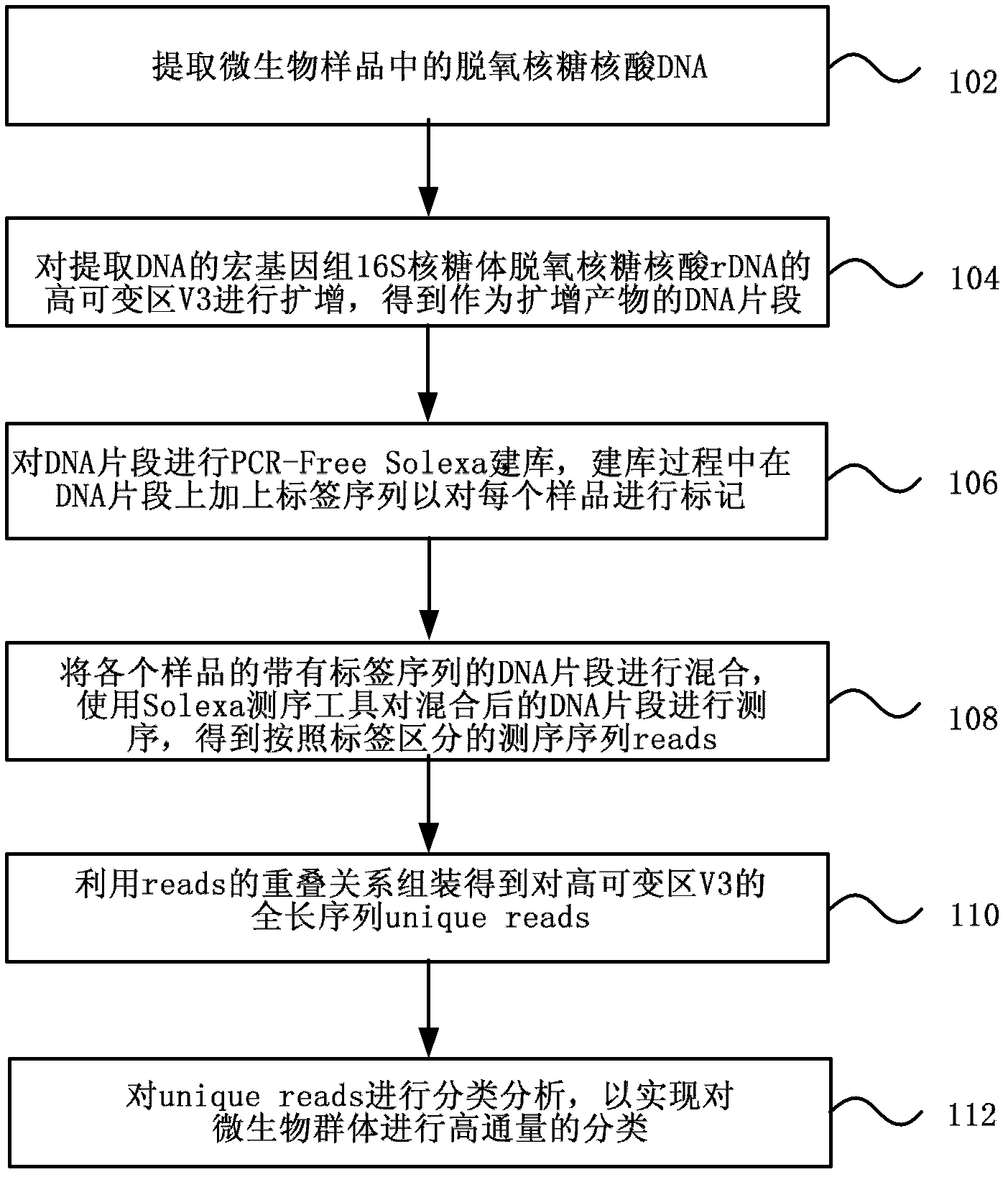
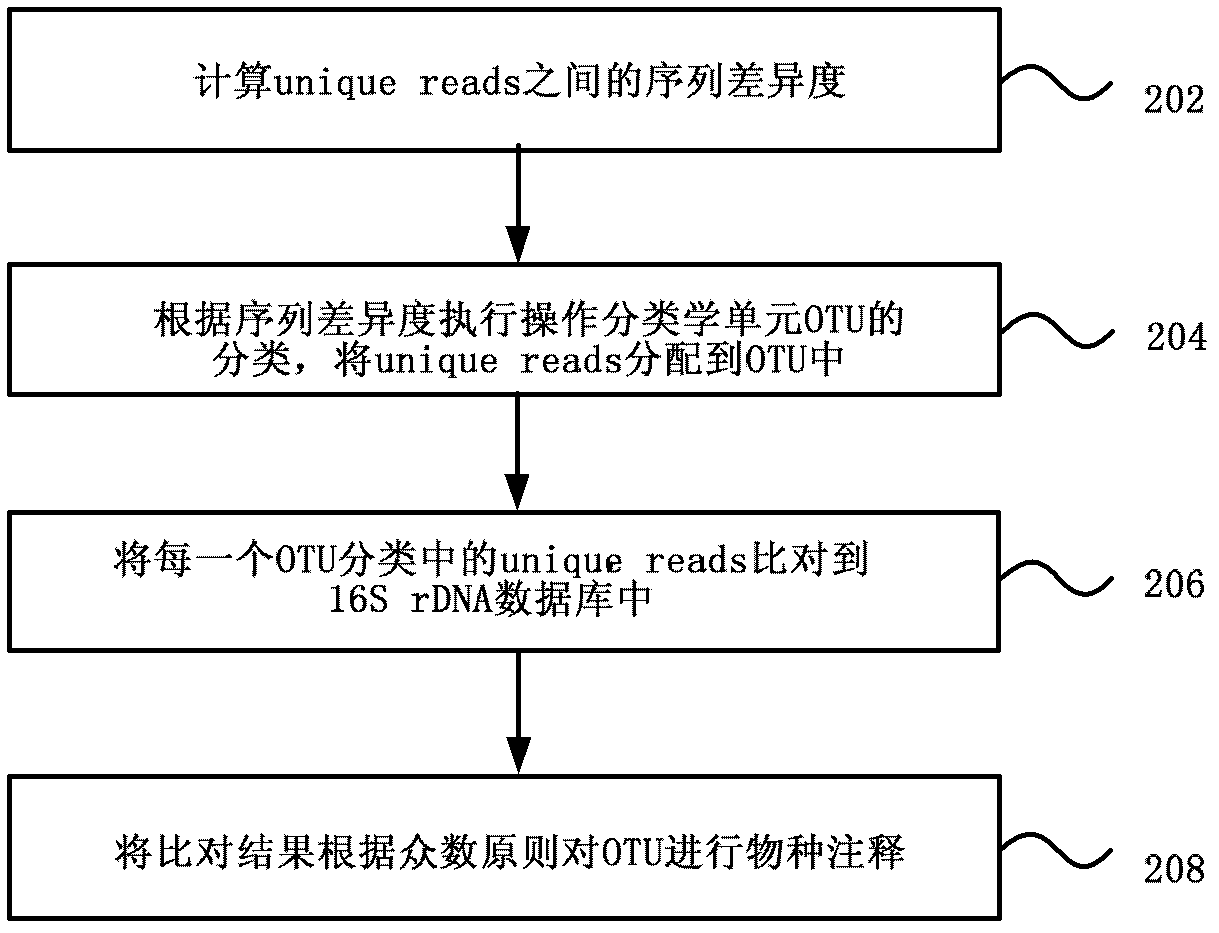
The invention discloses a metagenome 16S hypervariable region V3 based classification method and a device thereof. The method contains the following steps of: extracting DNA in microbial samples; carrying out amplification of metagenome 16S rDNA hypervariable region V3, carrying out Solexa database construction on amplification products, and simultaneously marking each sample by adding a connector with a label sequence in the process of database construction; mixing different samples with label sequences, sequencing by the use of a Solexa sequencing tool after mixing to obtain original sequencing sequences reads distinguished by labels; assembling by the use of reads overlapping relations to obtain hypervariable region V3 full-length sequences unique reads; and carrying out classification analysis on unique reads to accomplish classification of microbial population. By the adoption of the method and the device provided by the invention, classification of microbial population is accurate and sequencing cost is greatly reduced.
Owner:SHENZHEN HUADA GENE INST +1
Monoclonal antibodies
The invention provides heterochimeric antibodies and / or fragments thereof comprising (i) hypervariable region sequences wholly or substantially corresponding to sequences found in antibodies from a donor species; (ii) constant region sequences wholly or substantially corresponding to sequences found in antibodies from a target species which is different from the donor species; and (iii) heavy and / or light chain variable framework sequences which contain at least three non-CDR residues corresponding to sequences found in antibodies from a target species and at least three contiguous non-CDR residues corresponding to sequences found in antibodies from a donor species. The invention further provides antibody to canine or feline or equine antigens, e.g., CD20 or CD52, and methods of making and using antibodies as described.
Owner:ARATANA THERAPEUTICS
Analysis method for diversity of microbes and abundance of species in coal seam water
InactiveCN103981259AEfficient detectionEasy to operateMicrobiological testing/measurementSequence analysisOriginal data
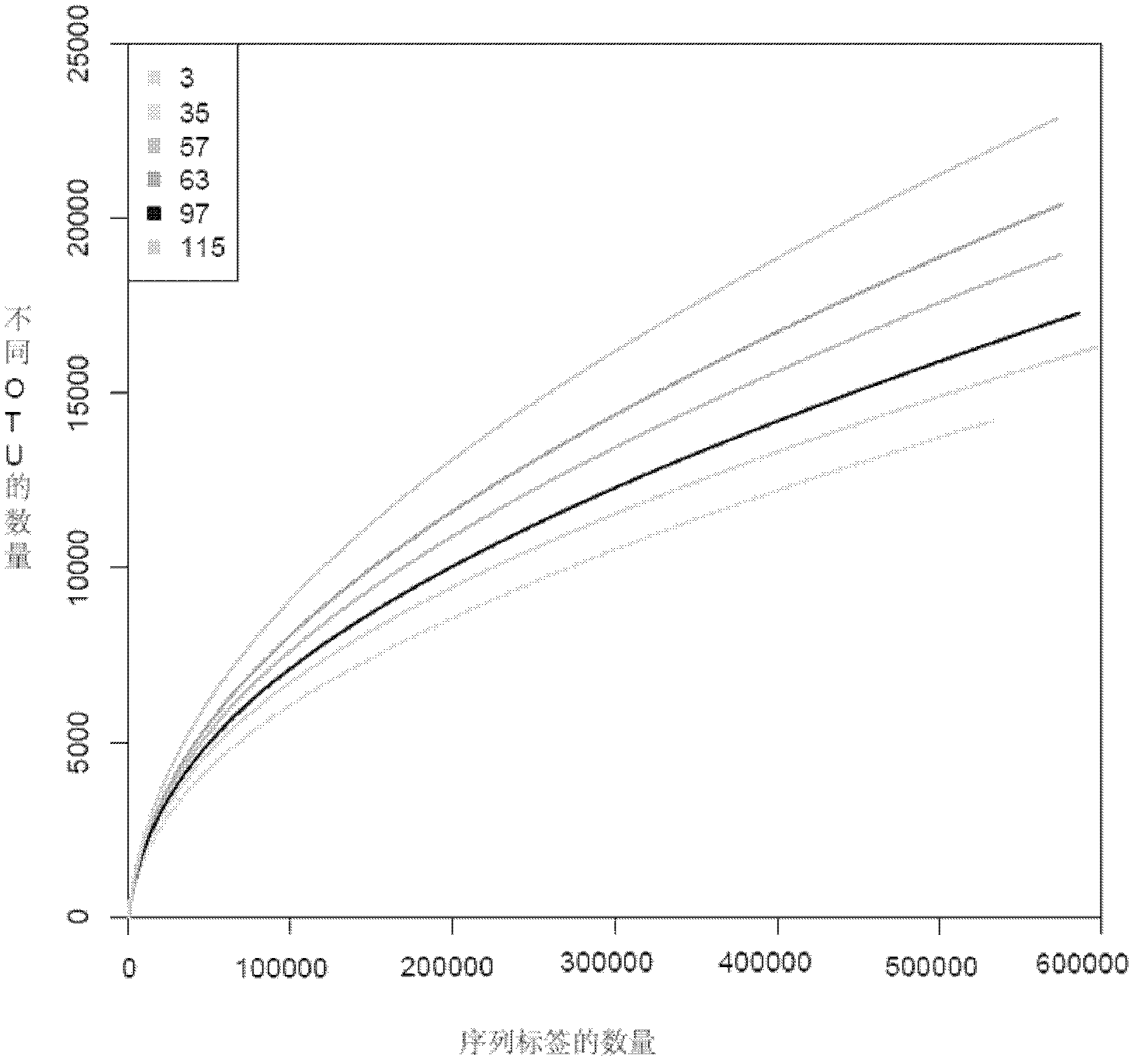
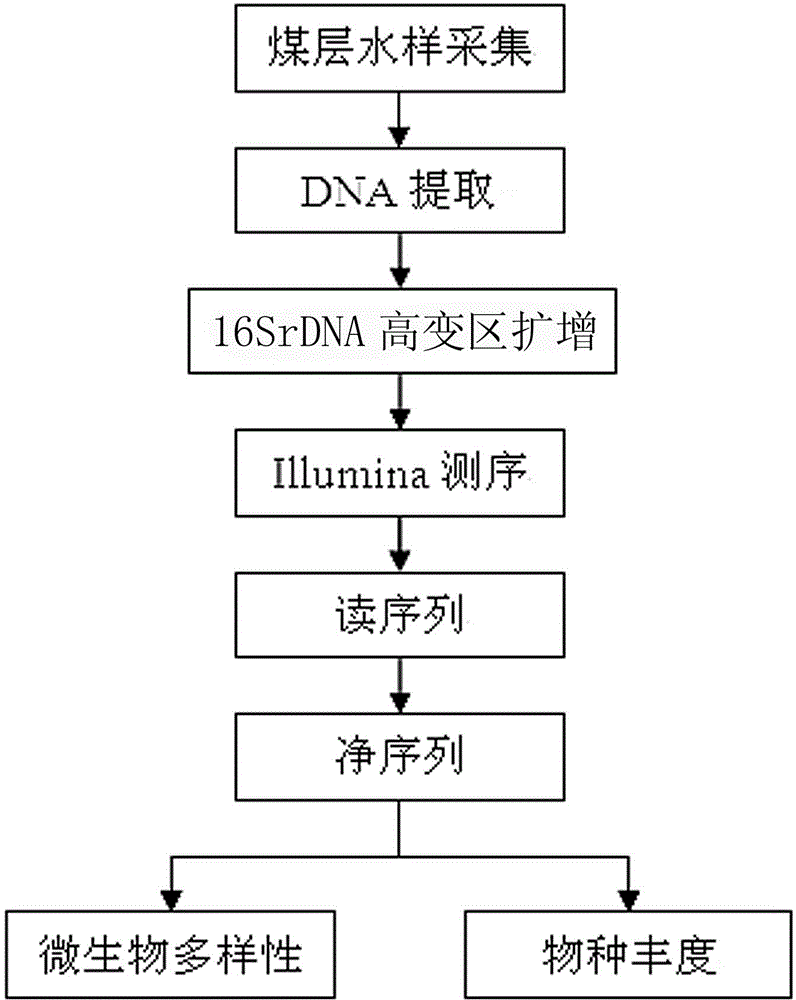
The invention belongs to the field of research on microbes in stratum water and provides an analysis method for diversity of microbes and abundance of species in coal seam water to overcome the problems that research on diversity of microbes in an environment has great limitation, species with low abundance in a sample cannot be effectively detected, species with high similarity in the sample are difficult to distinguish, only a minority of microbe species in the sample can be revealed, etc. The method comprises the following steps: acquisition of a coal seam water sample; enrichment of microbial thalli in the water sample and extraction of total DNA of the microbial thalli; amplification of the hypervariable region of 16SrDNA with specific primers; sequencing analysis with an Illumina sequencing platform; and processing of original data of sequencing so as to obtain diversity and abundance of microbes in the water sample. The method can effectively detect species with low abundance in the sample, identifies more than 400 genuses of bacteria in coal seam water, has obvious superiority in research on species and abundance of microbes in coal seam water and is applicable to analysis of diversity of microbes and abundance of species in environments like coal seams and oil gas or shale gas strata.
Owner:SHANXI JINCHENG ANTHRACITE COAL MINING GRP CO LTD
Therapeutic Binding Molecules
InactiveUS20110076270A1Immunoglobulins against cell receptors/antigens/surface-determinantsAntibody ingredientsMedicinePharmaceutical drug
A molecule comprising at least one antigen binding site, comprising in sequence the hypervariable regions CDR1, CDR2 and CDR3, said CDR1 having the amino acid sequence Asn-Tyr-Ile-Ile-His (NYIIH), said CDR2 having the amino acid sequence Tyr-Phe-Asn-Pro-Tyr-Asn-His-Gly-Thr-Lys-Tyr-Asn-Glu-Lys-Phe-Lys-Gly (YFNPYNHGTKYNEKFKG) and said CDR3 having the amino acid sequence Ser-Gly-Pro-Tyr-Ala-Trp-Phe-Asp-Thr (SGPYAWFDT); e.g. further comprising in sequence the hypervariable regions CDR1′, CDR2′ and CDR3′, CDR1′ having the amino acid sequence Arg-Ala-Ser-Gln-Asn-Ile-Gly-Thr-Ser-Ile-Gln (RASQNIGTSIQ), CDR2′ having the amino acid sequence Ser-Ser-Ser-Glu-Ser-Ile-Ser (SSSESIS) and CDR3′ having the amino acid sequence Gln-Gln-Ser-Asn-Thr-Trp-Pro-Phe-Thr (QQSNTWPFT), e.g. a chimeric or humanised antibody, useful as a pharmaceutical.
Owner:NOVARTIS AG
Plant endophyte 16S rRNA gene amplification method and application
ActiveCN106282165ABig amount of dataThe test result is completeMicrobiological testing/measurementDNA preparationSequence analysisAmplicon sequencing
The invention discloses a plant endophyte 16S rRNA gene amplification method and application. The amplification method includes the following steps of plant pretreating; plant-sample total DNA extracting; sample 16S rRNA gene amplifying, wherein sample 16S rRNA gene amplifying comprises 16S rRNA gene total-length emulsion PCR amplifying and 16S rRNA gene hypervariable-region / conserved-region amplifying sub-amplifying; high-throughput sequencing and biological information analyzing based on a Illumina platform, wherein high-throughput sequencing and biological information analyzing based on the Illumina platform comprises plant endophyte 16S rRNA gene amplicon purifying and recycling, amplicon sequencing library establishing, Illumina HiSeq sequencing and sequencing data bioinformatics analysis. The gene amplification method is used for high-throughput-sequencing plant disease detection and phytophagous animal enteric microorganism detection. Sequencing analysis of plant endophytic bacteria is carried out with the high-throughput sequencing technology, the data size is larger, the detection result is more complete, pollution of plant hosts is reduced to the maximum degree, the result multiformity is higher, and the cost is low.
Owner:成都罗宁生物科技有限公司
Remedies for mammary cancer
InactiveUS20050037061A1Broad specificityPrevent proliferationImmunoglobulins against cell receptors/antigens/surface-determinantsAntibody ingredientsTreatment effectHeavy chain
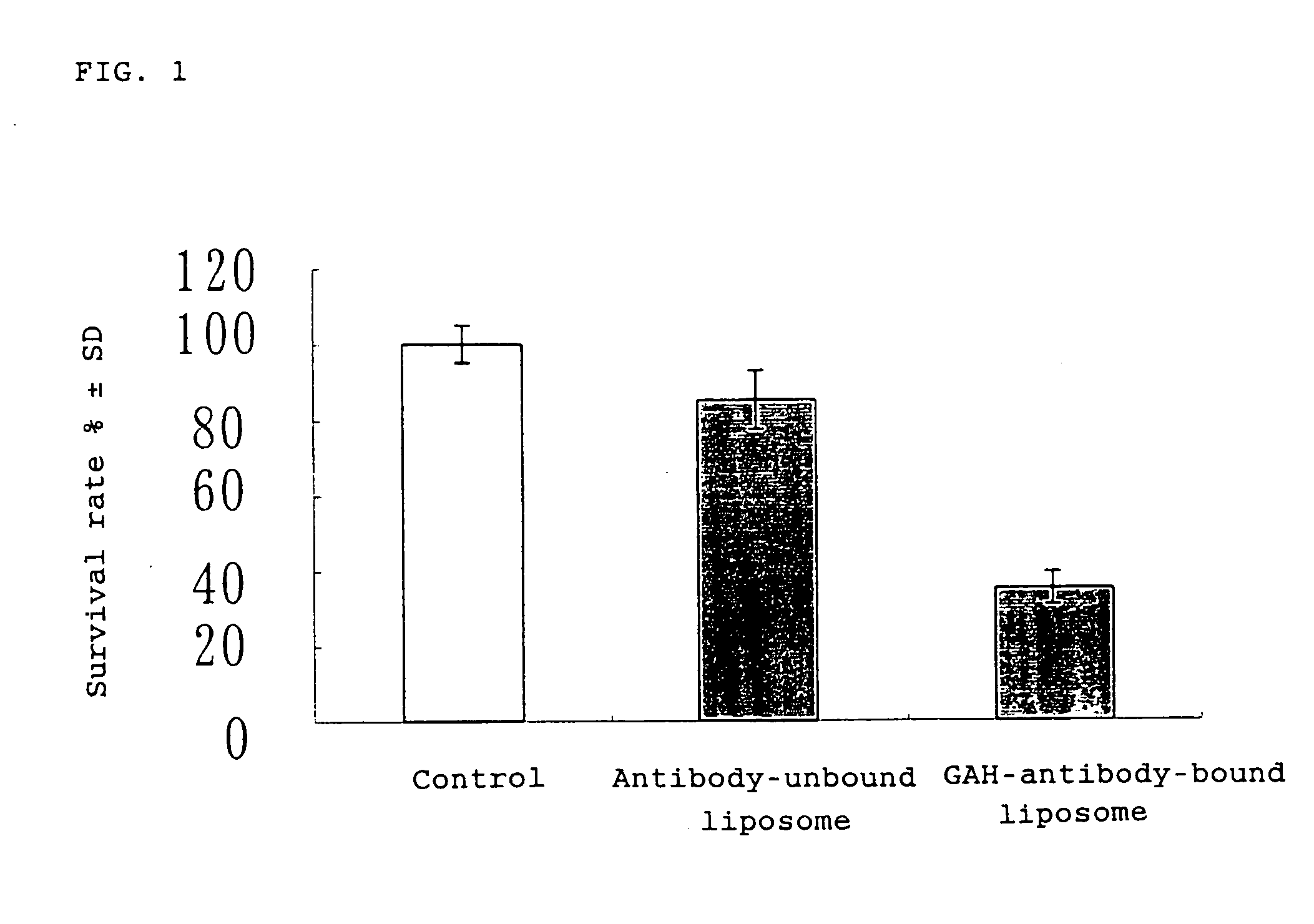
Provided is a remedy for cancer which remedy is specific to cancer tissue and has therapeutic effects on mammary cancer. The remedy is available by associating an antitumor substance with a human monoclonal antibody having amino acid sequences of SEQ. ID. NOS. 1, 2 and 3 of Sequence Listing in hypervariable regions of a heavy chain and amino acid sequences of SEQ. ID. NOS. 4, 5 and 6 of Sequence Listing in hypervariable regions of a light chain by attaching the antibody to a liposome having the antitumor substance encapsulated therein.
Owner:MITSUBISHI TANABE PHARMA CORP
Microbial community composition method and device based on high-throughput sequencing
InactiveCN107292123AReduce manual laborSave moneySpecial data processing applicationsMolecular structuresNatural abundanceCommunity composition
The invention discloses a microbial community composition method and device based on 16S rRAN gene hypervariable regions V1 / V2. The method comprises the steps of extracting DNAs in microbe samples and carrying out PCR (Polymerase Chain Reaction) amplification on the hypervariable regions V1 / V2 of the 16S rRANs of the DNSs in the samples; carrying out Solexa database establishment on amplified products and marking each sample through junctions with label sequences in a database establishment process; mixing different samples with the label sequences, carrying out sequencing on the mixed samples through utilization of a Solexa sequencing tool, thereby obtaining original sequencing sequence reads distinguished according to labels; carrying out assembling through utilization of overlapping relationships of the reads, thereby obtaining unique reads of the hypervariable regions V1 / V2 or V6; and carrying out sequence similarity comparison analysis on the unique reads, thereby classifying microbes in the samples and realizing relatively abundant calculation. According to the method and the device provided by the invention, the microbial community composition is analyzed accurately.
Owner:SUZHOU PRECISION BIOTECH CO LTD
Nucleotide and deduced amino acid sequences of hypervariable region 1 of the envelope 2 gene of isolates of hepatitis C virus and the use of reagents derived from these hypervariable sequences in diagnostic methods and vaccines
The nucleotide and deduced amino acid sequences of hypervariable region 1 of the envelope 2 gene of 49 isolates of hepatitis C are disclosed. The invention relates to the use of these sequences to design proteins and nucleic acid sequences useful in diagnostic methods and vaccines.
Owner:SEC DEPT OF HEALTH & HUMAN SERVICES
Therapeutic binding molecules
InactiveUS7825222B2Hybrid immunoglobulinsImmunoglobulins against cell receptors/antigens/surface-determinantsHumanized antibodyAmino acid
A molecule comprising at least one antigen binding site, comprising in sequence the hypervariable regions CDR1, CDR2 and CDR3, said CDR1 having the amino acid sequence Asn-Tyr-Ile-Ile-His (NYIIH), said CDR2 having the amino acid sequence Tyr-Phe-Asn-Pro-Tyr-Asn-His-Gly-Thr-Lys-Tyr-Asn-Glu-Lys-Phe-Lys-Gly (YFNPYNHGTKYNEKFKG) and said CDR3 having the amino acid sequence Ser-Gly-Pro-Tyr-Ala-Trp-Phe-Asp-Thr (SGPYAWFDT); e.g. further comprising in sequence the hypervariable regions CDR1′, CDR2′ and CDR3′, CDR1′ having the amino acid sequence Arg-Ala-Ser-Gln-Asn-Ile-Gly-Thr-Ser-Ile-Gln (RASQNIGTSIQ), CDR2′ having the amino acid sequence Ser-Ser-Ser-Glu-Ser-Ile-Ser (SSSESIS) and CDR3′ having the amino acid sequence Gln-Gln-Ser-Asn-Thr-Trp-Pro-Phe-Thr (QQSNTWPFT), e.g. a chimeric or humanised antibody, useful as a pharmaceutical.
Owner:NOVARTIS AG
Monoclonal antibodies
The invention provides heterochimeric antibodies and / or fragments thereof comprising (i) hypervariable region sequences wholly or substantially corresponding to sequences found in antibodies from a donor species; (ii) constant region sequences wholly or substantially corresponding to sequences found in antibodies from a target species which is different from the donor species; and (iii) heavy and / or light chain variable framework sequences which contain at least three non-CDR residues corresponding to sequences found in antibodies from a target species and at least three contiguous non-CDR residues corresponding to sequences found in antibodies from a donor species. The invention further provides antibody to canine or feline or equine antigens, e.g., CD20 or CD52, and methods of making and using antibodies as described.
Owner:ARATANA THERAPEUTICS
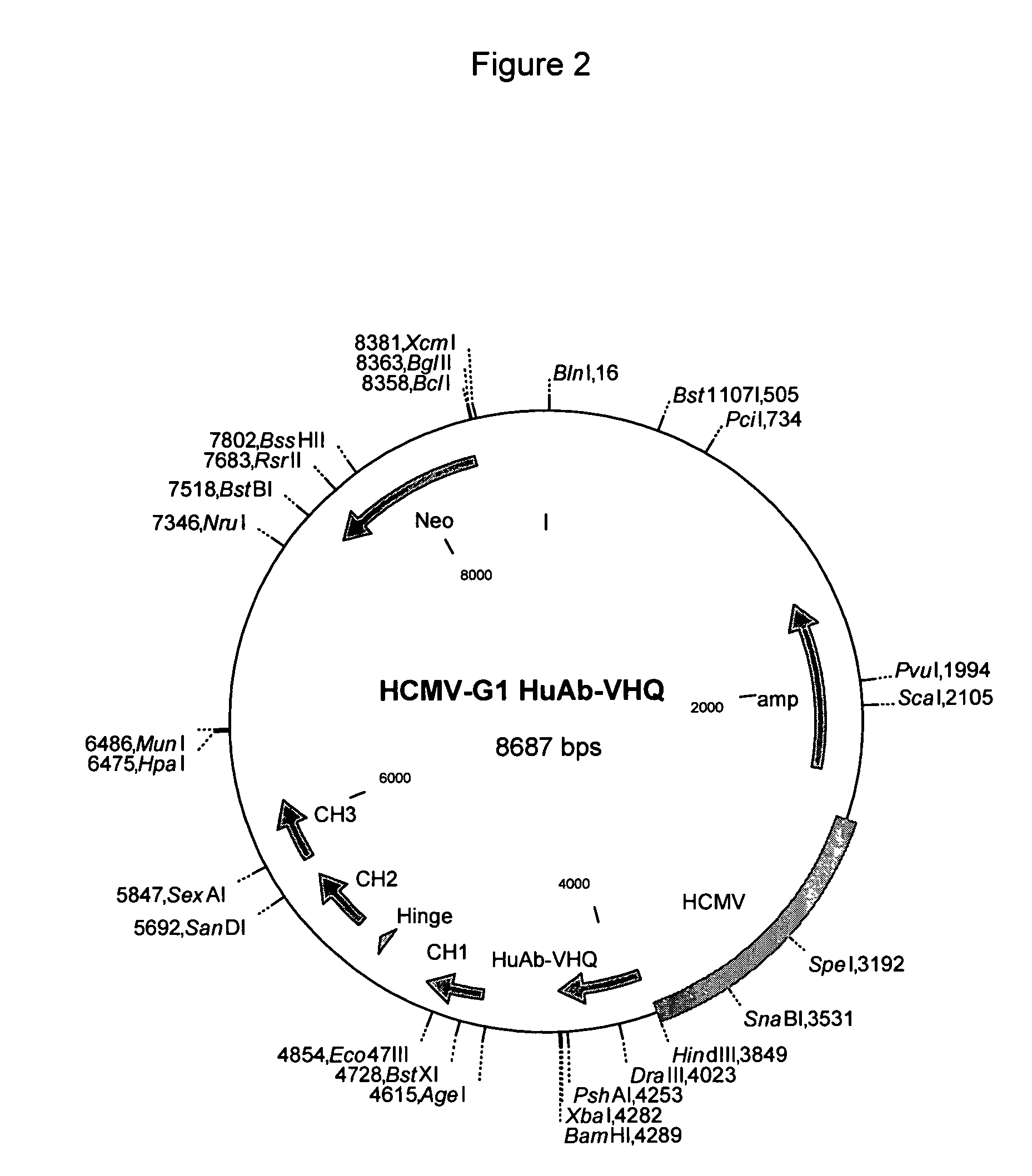
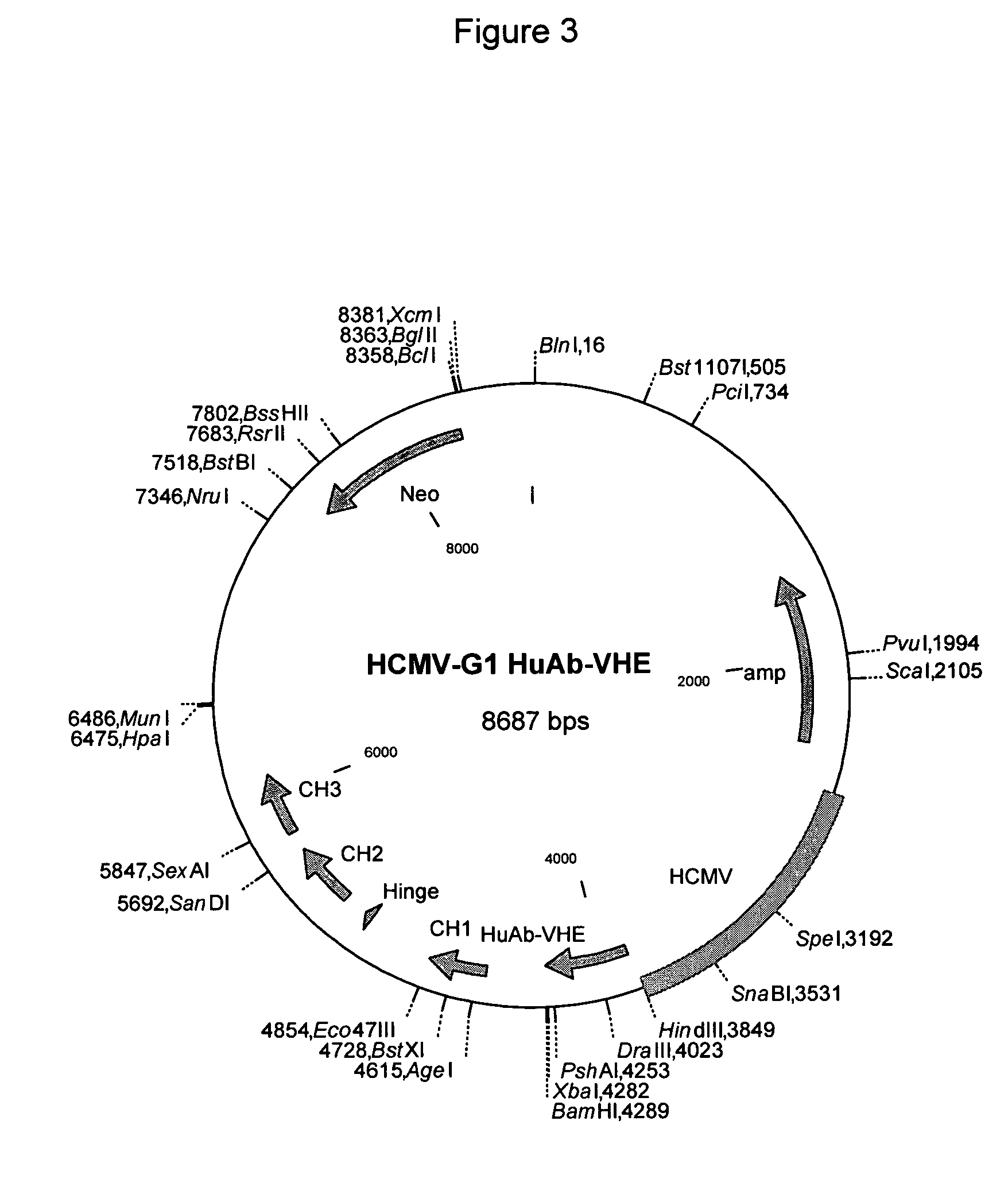
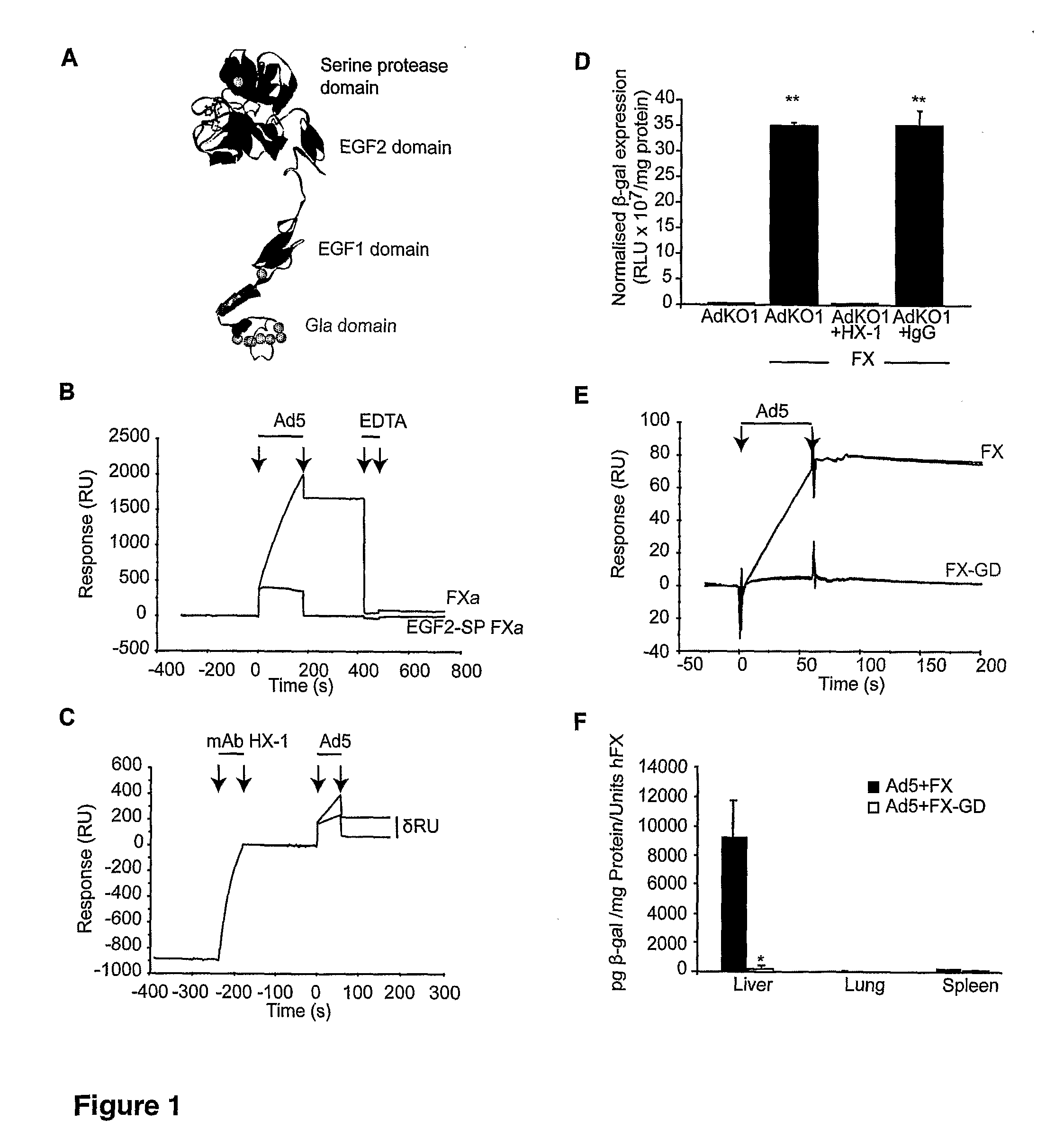
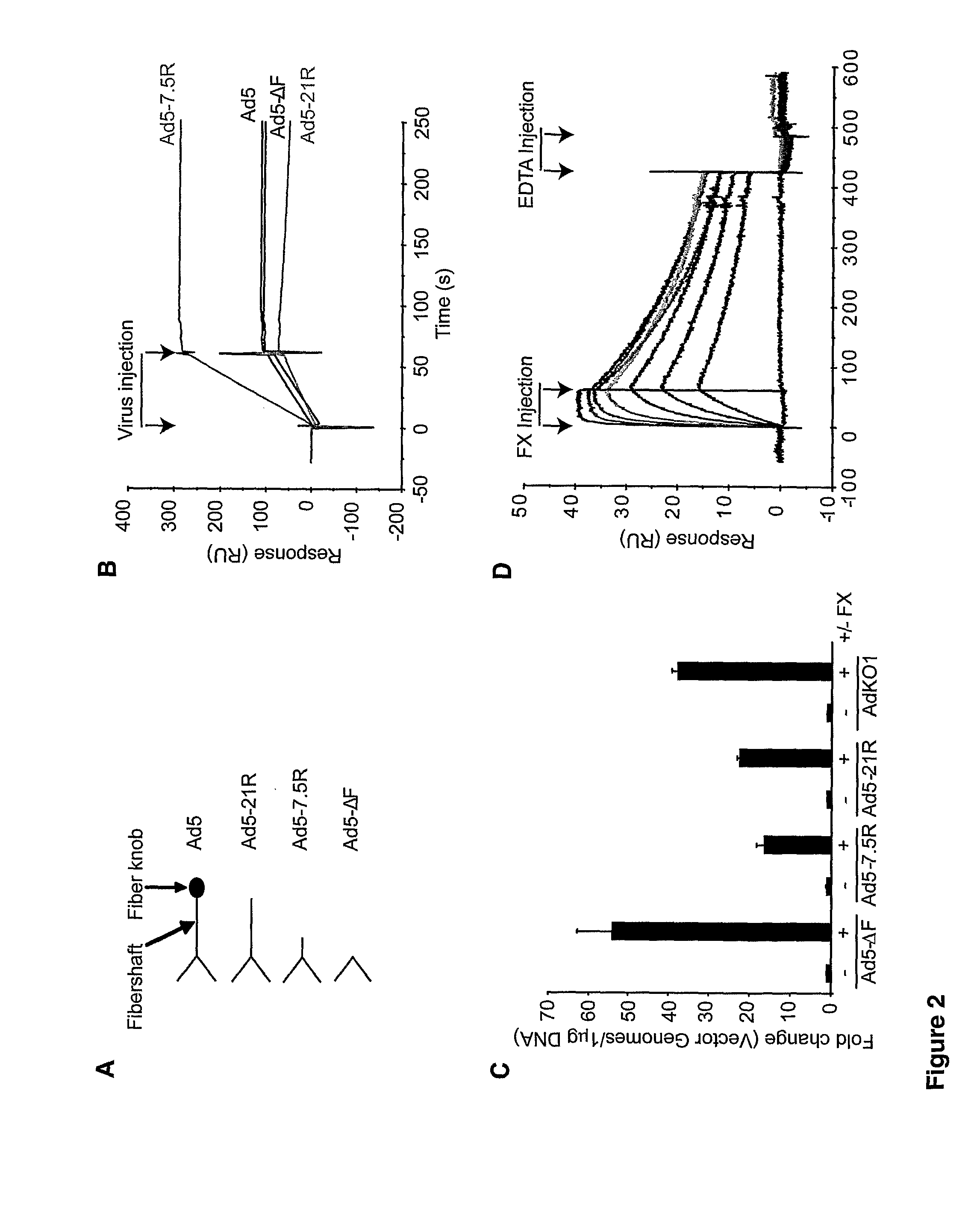
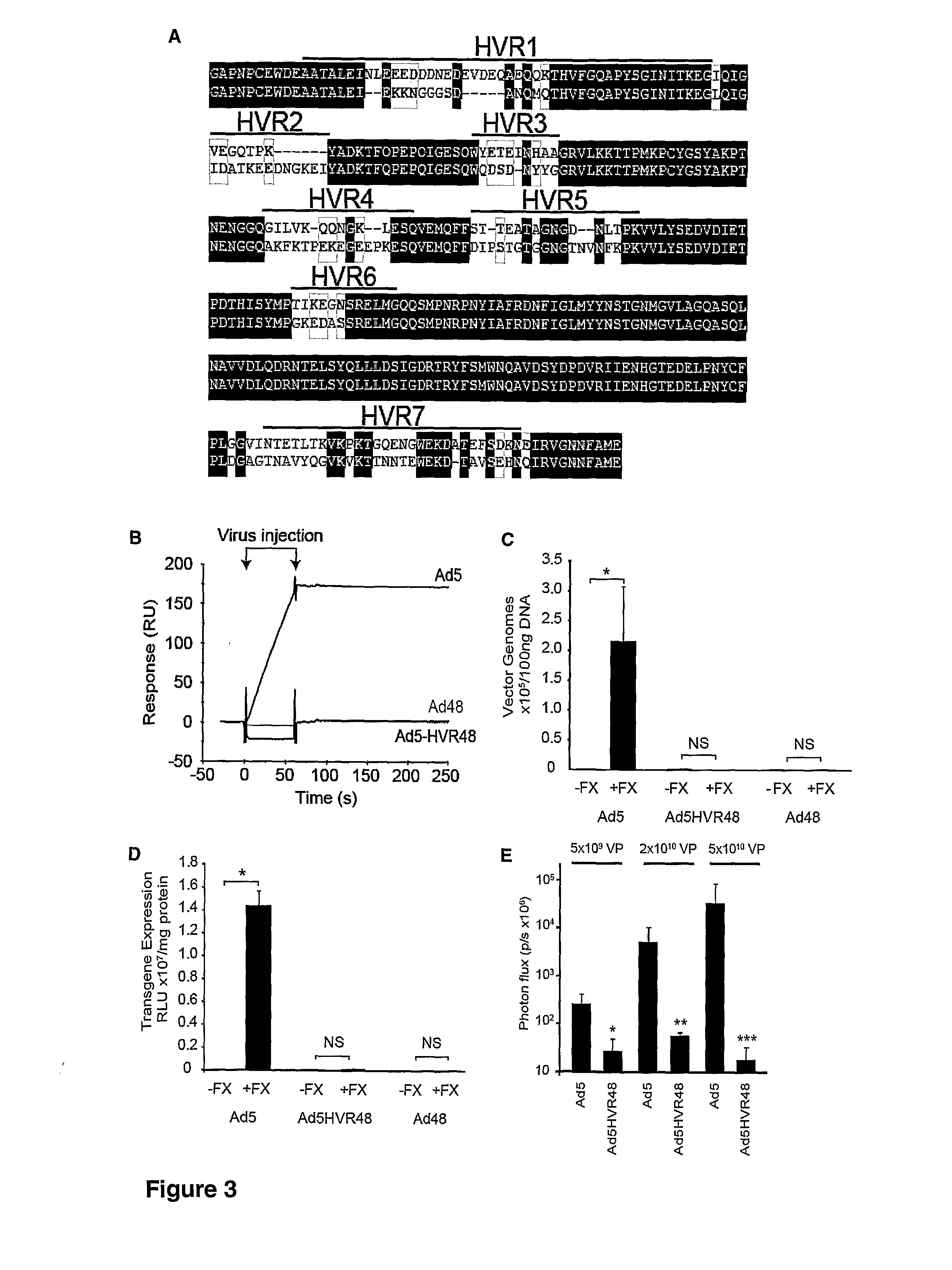
Modulation of Adenoviral Tropism
InactiveUS20110104788A1Increase opportunitiesHelp studyCompound screeningApoptosis detectionProviding materialTissues types
The invention provides materials and methods for modulating adenoviral tropism for hepatocytes and other cell types such as splenocytes. It relates to the findings that hypervariable regions (HVRs) of the viral hexon protein interact with the Gla domain of the blood clotting factor FX as part of the infective process in vivo. The invention provides means to disrupt the interaction between hexon and FX, thus reducing infection of hepatocytes and splenocytes, as well as use of targeting agents comprising the Gla domain or a fragment thereof to direct adenoviral vectors to desired target cell or tissue types.
Owner:BAKER ANDREW +2
Humanized anti-CD79b antibodies and immunoconjugates and methods of use
ActiveUS9845355B2Immunoglobulins against cell receptors/antigens/surface-determinantsTissue cultureVariable domainAntibody hypervariable region
Owner:GENENTECH INC
Method for predicting health and immunity levels of introduced milk cows based on intestinal flora
The invention relates to the technical field of biotechnology, in particular to a method for extracting and analyzing the DNA of introduced milk cow feces samples to predict the health and immunity levels of the introduced milk cows. The method comprises the processes such as sample selection, 16S amplicon sequencing, sequencing data analysis, intestinal flora diversity and abundance analysis, user report push and the like. According to the method, a monitoring index of intestinal flora diversity and abundance analysis is linked with the health and immunity levels of the introduced milk cows,after experiment studies and data analysis, fresh introduced (fewer than half a year) and early introduced (more than half a year) milk cow feces samples are collected, a hypervariable region is amplified, and the method for predicting the health and immunity levels of the introduced milk cows is realized. The samples are easy to collect, the analysis speed is rapid, the cost is low, and the assessment results are consistent with the existing studies, making the method for predicting the health and immunity levels of the introduced milk cows based on the intestinal flora possible.
Owner:DALIAN UNIV
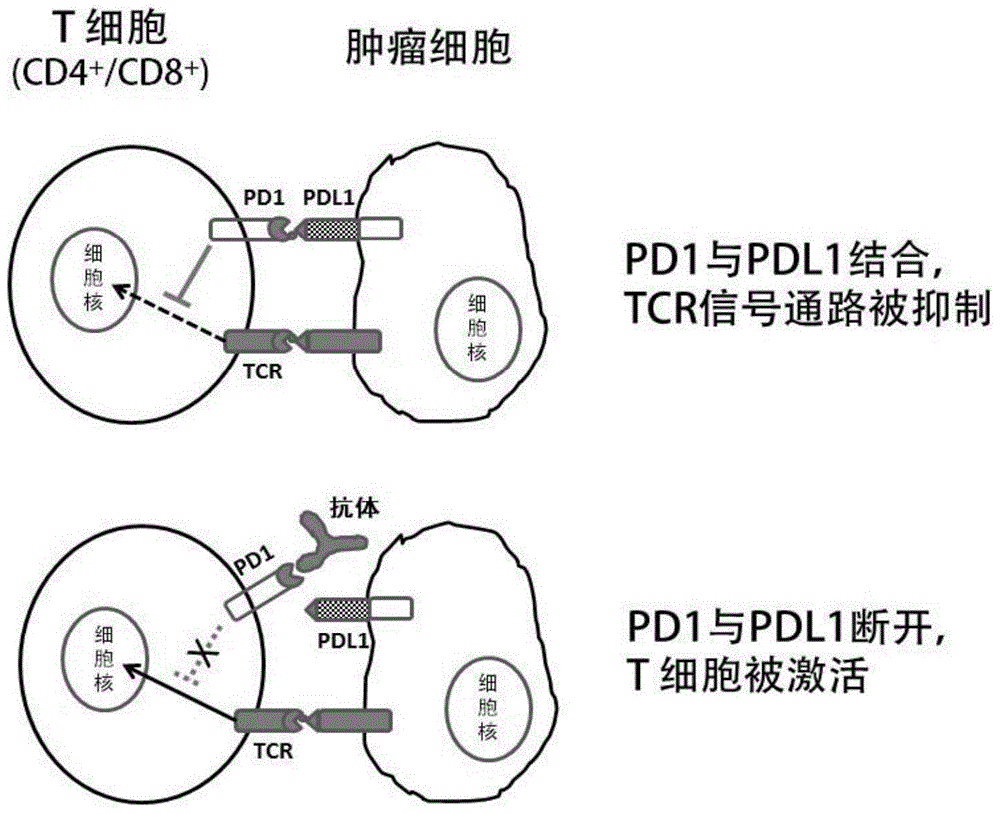
Anti-human PD-1 protein antibody, and coding gene and application thereof
InactiveCN106336460AEffective treatmentEffective preventionImmunoglobulins against cell receptors/antigens/surface-determinantsAntibody ingredientsSingle-Chain AntibodiesHeavy chain
The invention discloses an anti-human PD-1 protein antibody, and a coding gene and application thereof. The antibody comprises a heavy-chain variable region and a light-chain variable region; the amino acid sequences of three hypervariable regions, i.e., CDRH1, CDRH2 and CDRH3, of the heavy-chain variable region are GGSFSGYYWS, EINHSGSTNYNPSLKS and GSPDSSRARGYYMDV, respectively; and the amino acid sequences of three hypervariable regions, i.e., CDRL1, CDRL2 and CDRL3, of the light-chain variable region are RASQGIRNDLG, AASSLQS and LQHNSYPL, respectively. According to the invention, the anti-human PD-1 protein antibody prepared by constructing a human single-chain antibody library and screening single-chain antibodies by using ribosome displaying technology can specifically bind to human PD-1 protein, has high affinity, is applicable to detection of cells expressing PD-1 ad applicable as a fully human-derived anti-human PD-1 protein antibody for treating human diseases, and effective prevents and treats tumor diseases.
Owner:杭州贝颐药业有限公司
Method for detecting rhizosphere soil prokaryotic microorganisms of various soybeans based on 16SrDNA deep sequencing
InactiveCN105525025AComprehensive detectionImprove throughputMicrobiological testing/measurementMicroorganismSoil microbiology
The invention belongs to the technical field of soil microbiology, and in particular relates to a method for detecting rhizosphere soil prokaryotic microorganisms of various soybeans based on 16SrDNA deep sequencing. The method comprises the following steps: 1. collecting root shook-off soil and rhizosphere soil of various soybeans in different development stages; 2. extracting microorganism metagenome DNA from the soil; 3. performing PCR amplification on a 16S rDNA fourth hypervariable region in the DNA by virtue of a dual-tag primer so as to construct a library; 4. simultaneously synthesizing and sequencing the qualified library by virtue of a Illumina Miseq platform in a mode of 250 nucleotides at dual ends, so that pure READS is obtained; 5. splicing: clustering at least 38000 effective tags generated from each sample into an operable classifying unit; 6. conducting significance analysis on species composition, structure, diversity and relative abundance difference; and 7. by taking the root shook-off soil as a control group of the system, accurately determining the composition, structure, diversity and relative abundance of a rhizosphere soil prokaryotic microorganism colony, and comparing the various soybeans.
Owner:NANJING UNIV
Neutralizing monoclonal antibody in human adenovirus 7 and preparation method and application thereof
ActiveCN104086650AEffective displayEnhance immune responseMicroorganism based processesImmunoglobulins against virusesResponse effectInfective disorder
The invention discloses a method for preparing a neutralizing monoclonal antibody in a human adenovirus 7. The hexon fifth hypervariable region of a human adenovirus 3 vector is substituted by a hexon fifth hypervariable region gene of the human adenovirus 7 to construct a chimeric virus which is taken as an immunogen. A murine human adenovirus 7 neutralizing monoclonal antibody and a hybridoma cell strain 5MH5 / 3G5 prepared by using the method are further provided. A humanized and chimeric monoclonal antibody prepared by using the method and a medicinal composition comprising the humanized or human-mouse chimeric monoclonal antibody serving as an effective component are further provided. An exogenous neutralizing epitope is embedded into the hypervariable region of the human adenovirus 3 vector to construct a recombinant adenovirus showing the exogenous neutralizing epitope. By using the recombinant adenovirus as an immunogen, the exogenous neutralizing epitope can be shown effectively, the immune response effect is enhanced, and the research of a monoclonal antibody for treating infectious diseases is facilitated.
Owner:广州瑞发一号健康投资中心(有限合伙)
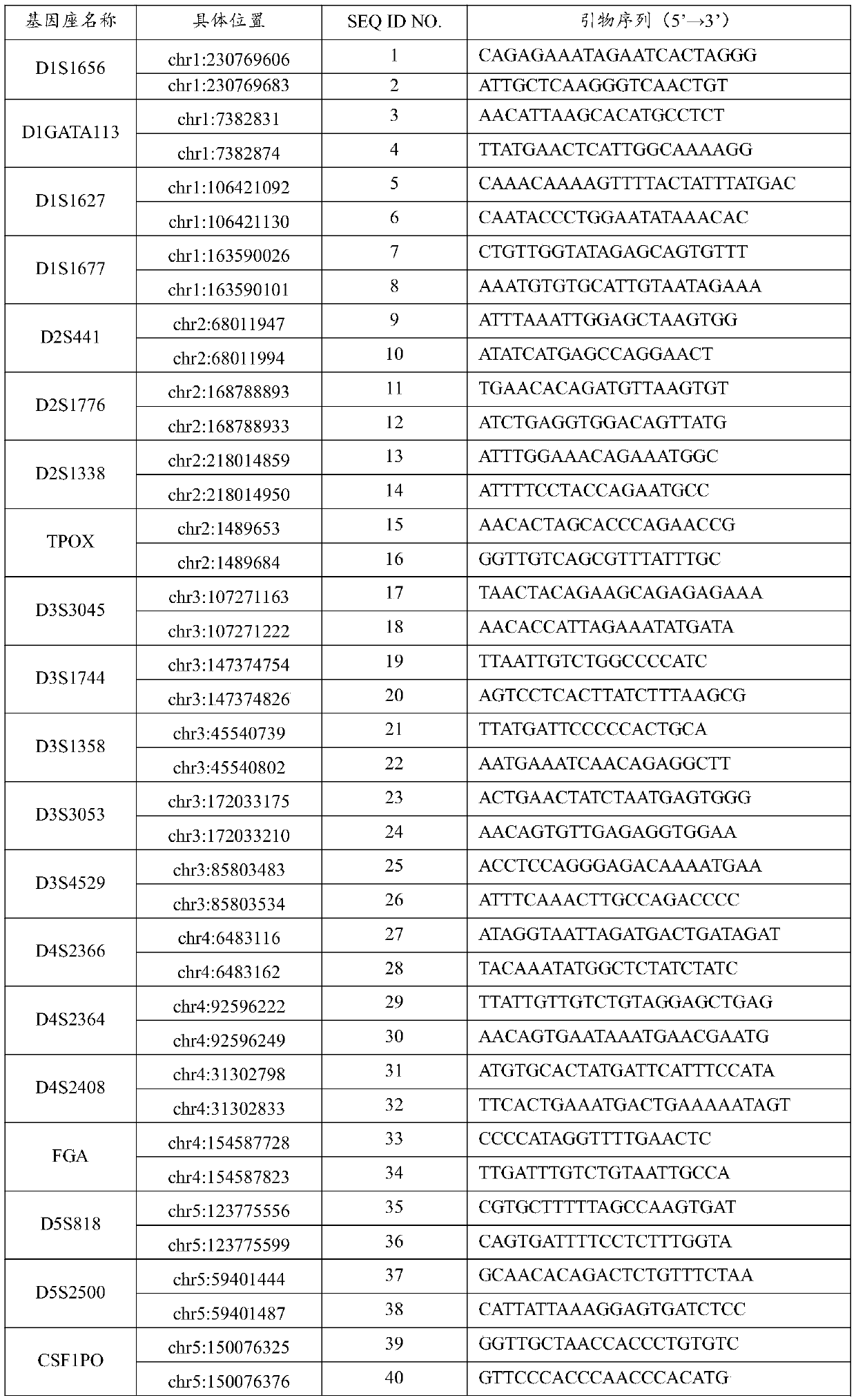
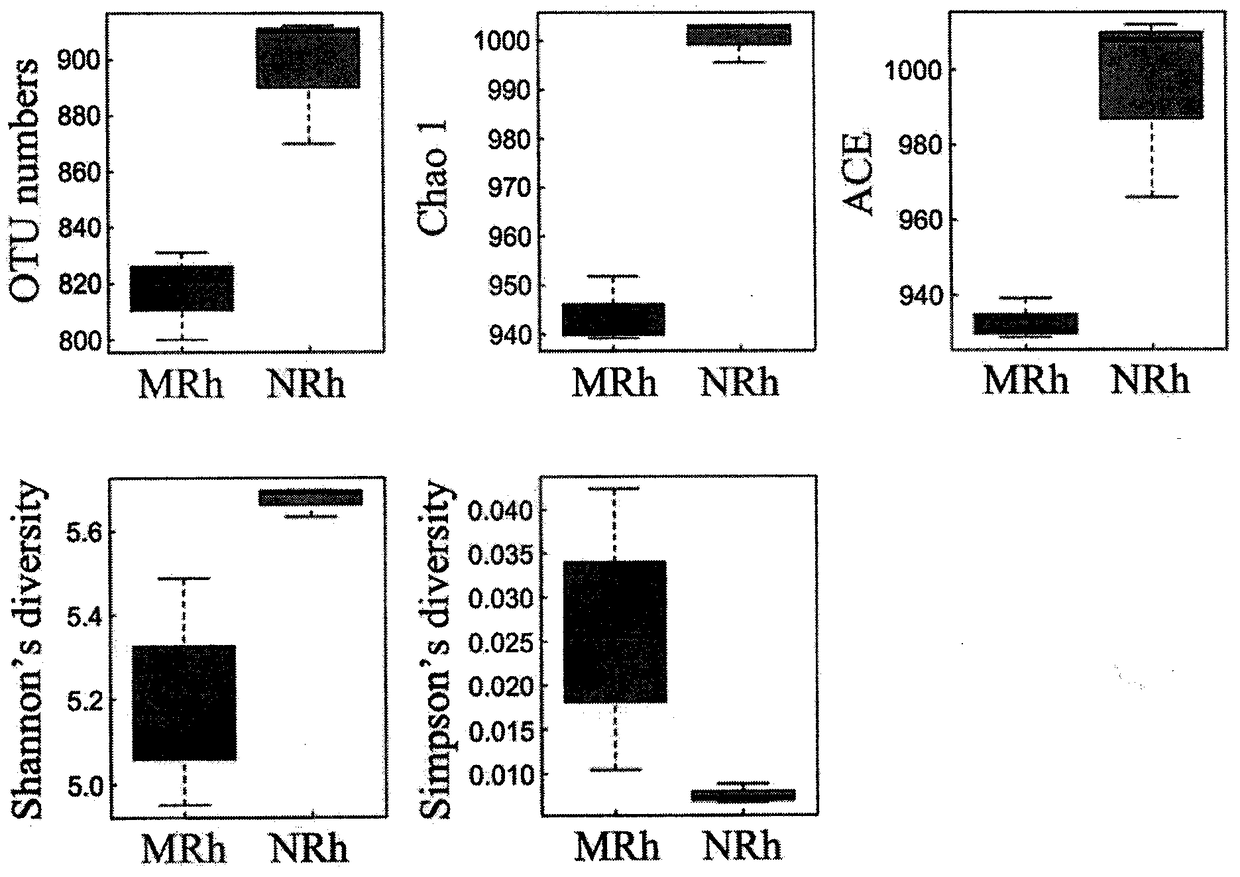
Accurate human DNA typing method, reagent and application
PendingCN110863056AHigh resolutionImprove individual recognitionMicrobiological testing/measurementDNA/RNA fragmentationHuman DNA sequencingGenomic data
The application discloses an accurate human DNA typing method, a reagent and application. According to the accurate human DNA typing method, a short tandem repeat sequence, polynucleotide polymorphicsites covering a whole genome, a mitochondrial DNA hypervariable region I, a mitochondrial DNA hypervariable region II and an Amel enamel gene are detected simultaneously to obtain accurate typing andbase sequences of all the sites, thereby realizing accurate typing of human DNAs. The disclosed method having high resolution ratio is capable of carrying out efficient and accurate individual recognition and has the extremely high individual recognition capacity for difficult detection materials and highly-degraded detection materials; the non-parent exclusion rate is larger than 99.999999% in paternity identification and the accuracy is high; the compatibility is high, the existing individual recognition detection kit can be covered, and the method can be used for analyzing race groups in different regions or countries by combining human genome data. Besides, the method disclosed by the invention is relatively high in adaptability of detection materials and the human DNA extracted by various experimental methods can be detected.
Owner:BGI FORENSIC TECH (SHENZHEN) CO LTD
Method for detecting prokaryotic microorganisms in crop rhizosphere based on high-throughput sequencing
The invention relates to a method for detecting prokaryotic microorganisms in rhizosphere soil of different crops based on 16S rDNA high-throughput sequencing. The method comprises the following stepsof 1, collecting shaking-off soil (as a system control) of different genotype crops outside root zones or at root systems and the rhizosphere soil; 2, extracting total microbial DNA from each samplesoil; 3, using a double-label primer method for PCR amplification of V5-V7 hypervariable region fragments of 16S rDNA in the total DNA to construct a library; 4, for a qualified library, simultaneously conducting synthesis and sequencing of a double-end 300 nucleotide pattern by using an Illumina Miseq platform; 5, splicing after pure paired READS is obtained, and generating 40,000 effective TAGsfrom each sample for clustering into operable classification units; 6, according to the system control, accurately determining the composition, structure, diversity and relative abundance saliency analysis of prokaryotic microbial communities in the rhizosphere soil of each sample, and comparing similarities and differences between different crops.
Owner:NANJING UNIV
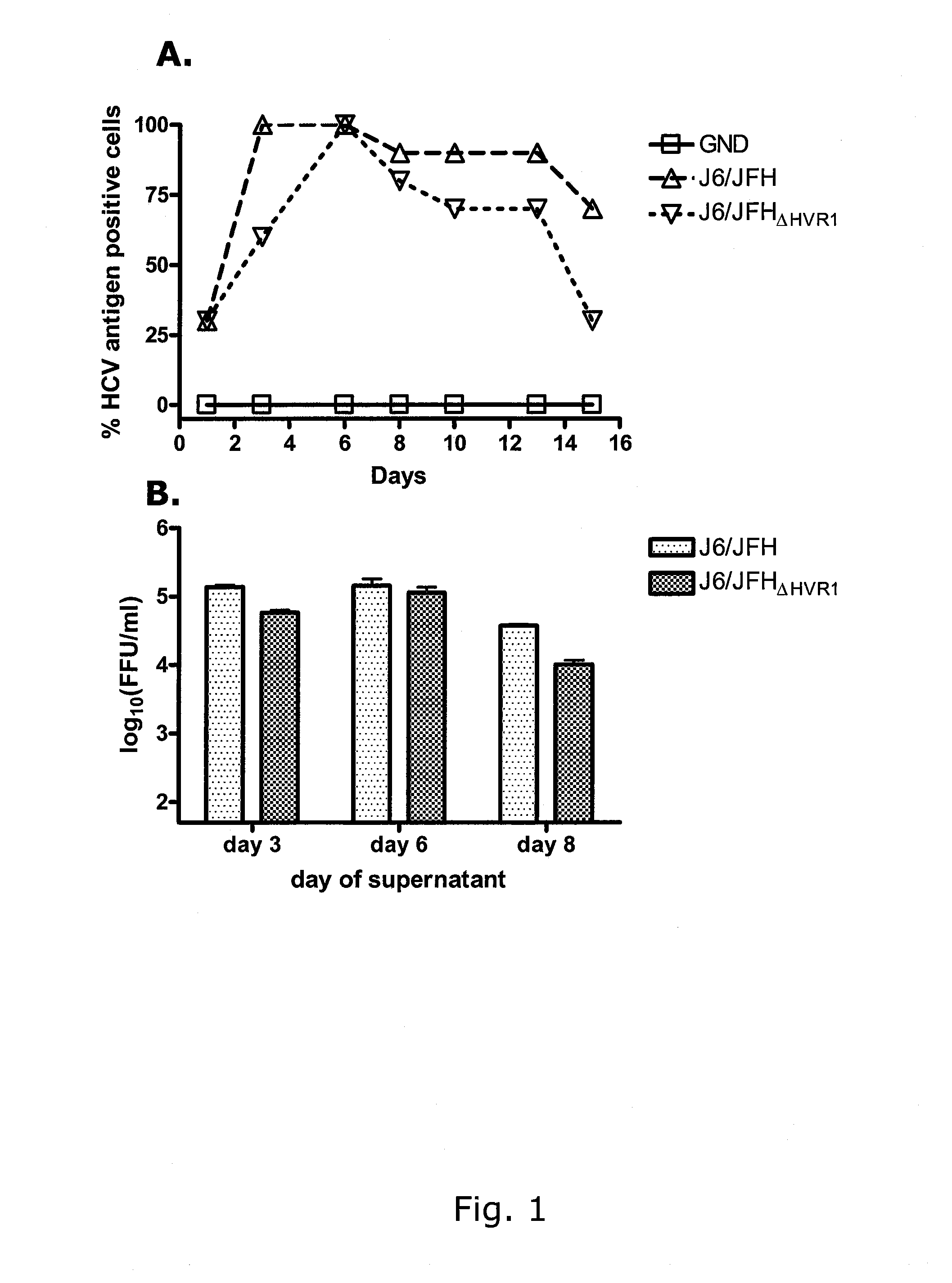
INFECTIOUS GENOTYPE 1a, 1b, 2a, 2b, 3a, 5a, 6a and 7a HEPATITIS C VIRUS LACKING THE HYPERVARIABLE REGION 1 (HVR1)
The present inventors used the previously developed H77 / JFH1T27OOC,A4O8OT (1a / 2a), J4 / JFH1T2996C,A4827T,ΔHVRI (1b / 2a), J6 / JFH1ΔHVRI (2a / 2a), J8 / JFH1ΔHVRI (2b / 2a), S52 / JFH1T27i8G,τ7i6oc (3a / 2a), SA13 / JFH1C34O5G,A3696G (5a / 2a) and HK6a / JFH1T1389c,A1590G (6a / 2a) constructs for the deletion of Hypervariable Region 1 (HVR1) to construct viable, JFH1 (geno-type 2a) based, genomes. The present inventors serially passaged the viruses in cell culture obtaining relatively high HCV RNA titers and infectivity titers. Sequence analysis of the viruses identified mutations adapting H77 / JFH1T27OOC,A4O8OT,ΔHVR1 (1a / 2a), J8 / JFH1ΔHVR1 (2b / 2a), S52 / JFH1T2718G,T716OC,ΔHVR1 (3a / 2a) and J4 / JFH1T2996C,A4827T,ΔHVR1 (1b / 2a) to the HVR1 deletion.
Owner:HVIDOVRE HOSPITAL +1
Recombination human rabies viruses resisting antibody
InactiveCN101235086ANo pollution problemEasy to purifyFungiImmunoglobulins against animals/humansHeavy chainNeutralizing antibody
The invention relates to a production method of recombination human antibody, in particular to a production method of recombination human anti rabies virus antibody, which is characterized in that the recombination antibody is neutralizing antibody specially combined with rabies virus, which is determined by the specific gene sequence in hypervariable region (CDRs) of antibody heavy chain and light chain variable region and can be expressed in procaryotic cell and eukaryotic cell. The CDR area, part or all gene of the antibody can generate the antibody in any expression system of procaryotic cell and eukaryotic cell, to prevent and treat rabies clinically.
Owner:吉林圣元科技有限责任公司
Primer for amplifying molecules of microbes in pit mud of Luzhou-flavor liquor and detection method
ActiveCN101974519AReflect the characteristics of the structureAbundant stripsMicrobiological testing/measurementDNA preparationMicroorganismCulture independent
The invention belongs to the technical field of wine making, which particularly relates to a primer for detecting molecules of microbes in pit mud of Luzhou-flavor liquor and a molecule detection method for microbes in pit mud of Luzhou-flavor liquor. The method is characterized in that protogenome of prokaryotic microbes in the microbes in pit mud is extracted by a culture-independent approach; the hypervariable regions V6-V8 of a microbe 16S rDNA are amplified through PCR (Polymerase Chain Reaction), and are processed by denaturing gradient gel electrophoresis (DGGE); and the diversity of the microbes and the variation of microbe structures can be analyzed through an electrophoretogram. By using the primer and the method, the 16S rDNA of the microbes in different kinds of pit mud in the process of making Luzhou-flavor liquor can be amplified quickly and accurately, the difference of microbes in different kinds of pit mud in the process of making Luzhou-flavor liquor can be further detected by DGGE, and thus realizing in-time monitoring on variations in the fermentation process of microbes in pit mud for making wine.
Owner:LUZHOU PINCHUANG TECH CO LTD
Anthropogenic H5N1-resisting hemagglutinin protein broad-spectrum neutralising antibody and application thereof
The invention provides an anthropogenic H5N1-resisting hemagglutinin protein broad-spectrum neutralising antibody. The antibody is obtained by screening through a phage display technology. The sequence of the hypervariable region (CDRs) determining the specificities of the antibody is represented by figure 1. The antibody of the invention specifically identifies H5N1 highly pathogenic avian influenza virus particle antigen, has obvious fluorescence immune reaction and enzyme-coupled immune reaction with the avian influenza virus H5N1 aiming at avian influenza virus hemagglutinin protein HA, and has the neutralising activity function of resisting the avian influenza virus H5N1. The antibody of the invention can be made into specific antibody medicines clinically used for preventing and treating human avian influenza that is caused by the H5N1 avian influenza virus, so as to clinically prevent or treat human avian influenza that is caused by the H5N1 avian influenza virus.
Owner:STATION OF VIRUS PREVENTION & CONTROL CHINA DISEASES PREVENTION & CONTROL CENT
Anti-human papilloma virus L1 protein antibody, and coding gene and application thereof
ActiveCN103694345AEfficient detectionDetects HPV infection and can also be used for effective treatmentImmunoglobulins against virusesAntiviralsDiseaseHeavy chain
The invention discloses an anti-human papilloma virus L1 protein antibody, and a coding gene and application thereof. The anti-human papilloma virus L1 protein antibody comprises a heavy chain variable region and a light chain variable region. The anti-human papilloma virus L1 protein antibody is characterized in that amino acid sequences of three hypervariable regions CDRH1, CDRH2 and CDRH3 of the heavy chain variable region are respectively disclosed as SEQ ID NO.5-7; and amino acid sequences of three hypervariable regions CDRL1, CDRL2 and CDRL3 of the light chain variable region are respectively disclosed as SEQ ID NO.8-10. The invention also discloses application of the anti-human papilloma virus L1 protein antibody in preparing reagents and medicines for detecting, preventing and treating human papilloma virus related diseases. The anti-human papilloma virus L1 protein antibody has high affinity with human papilloma virus L1 protein, and has favorable specificity.
Owner:HANGZHOU DALTON BIOSCI
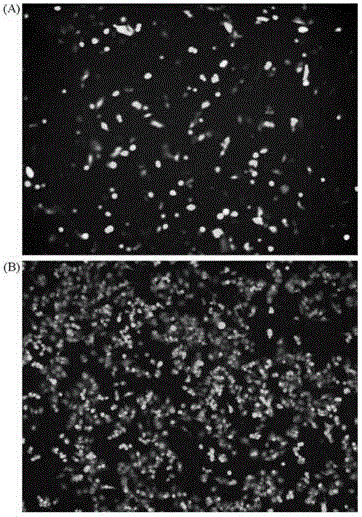
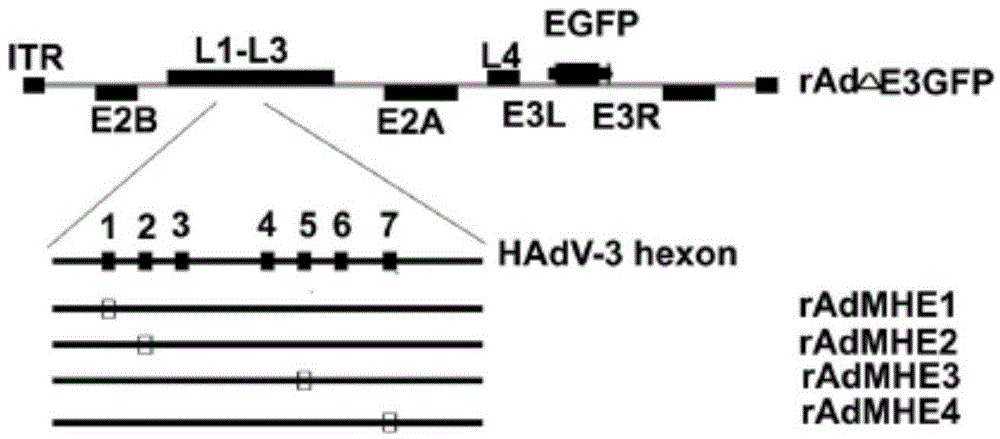
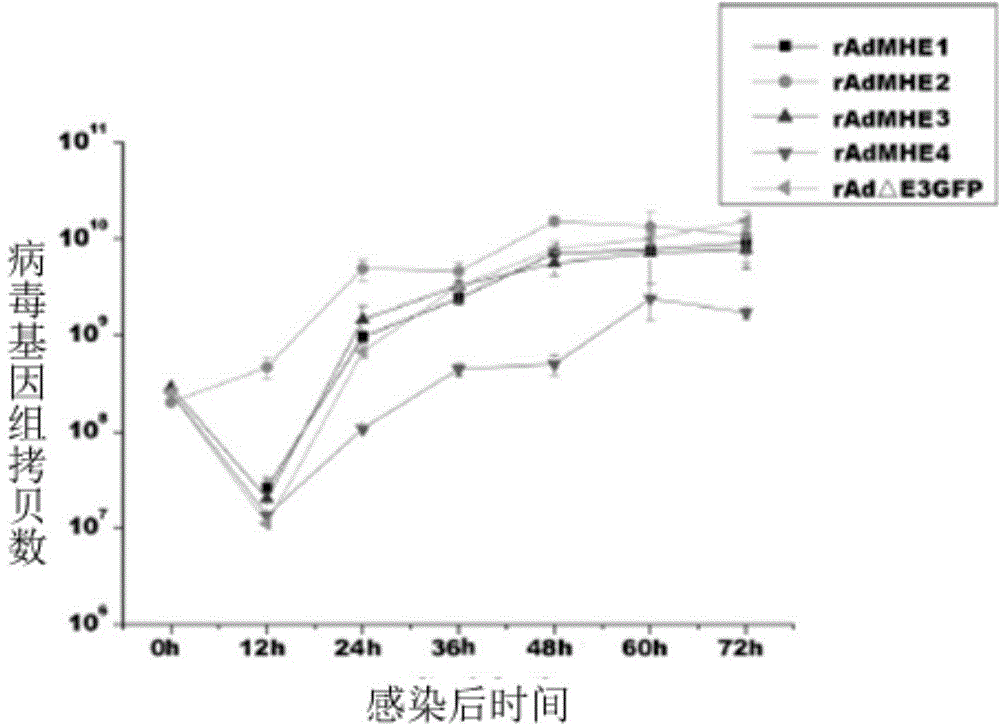
Modifiable loca of human type 3 adenovirus hexon and application thereof
The invention provides modifiable amino acid loca in hypervariable regions of human type 3 adenovirus hexon. The modifiable loca are positioned in the hypervariable regions of the hexon and have at least one amino acid locus in a sequence shown as SEQ ID NO:1-4. The amino acid loca are positioned in hypervariable regions 1, 2, 5, and 7 of the human type 3 adenovirus hexon. The invention also provides the application of the modifiable amino acid loca to the show of extrinsic protein and the preparation of immunogen, medicaments and detection reagents. The modifiable amino acid loca lay the foundation for a virus show technology platform which is researched and developed by using a technology platform of modifiable extrinsic polypeptides in the hypervariable regions of the human type 3 adenovirus hexon as vaccines or antigens.
Owner:GUANGZHOU INST OF RESPIRATORY DISEASE
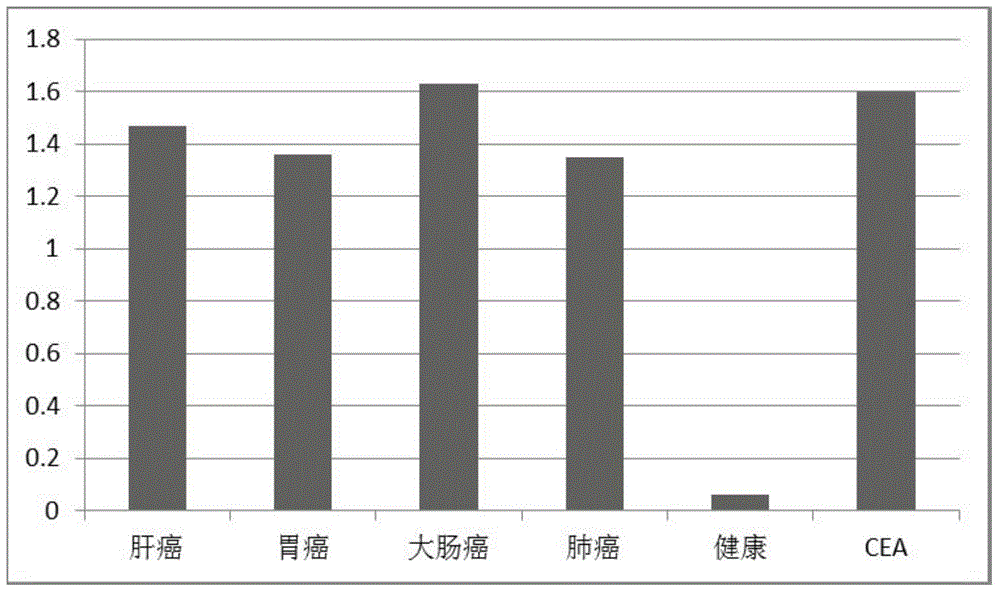
Anti-human carcino-embryonic antigen antibody as well as coding gene and application thereof
InactiveCN104628859AHigh affinityEfficient detectionImmunoglobulins against cell receptors/antigens/surface-determinantsIn-vivo testing preparationsDiseaseCarcinoembryonic Antigen Antibody
The invention discloses an anti-human carcino-embryonic antigen antibody as well as a coding gene and application thereof. The anti-human carcino-embryonic antigen antibody comprises a heavy chain variable region and a light chain variable region, wherein the amino acid sequences of three hypervariable regions CDRLH1, CDRH2 and CDRH3 of the heavy chain variable region are GFAVSSN, HRSGN and VPGFSRGQYEESWYFDL respectively; the amino acid sequences of three hypervariable regions CDRL1, CDRL2 and CDRL3 of the light chain variable region are RASQSIDSYLN, GASNLRN and HQAYSPFT respectively. The anti-human carcino-embryonic antigen antibody disclosed by the invention is a fully humanized anti-human carcino-embryonic antigen antibody, has high affinity with a human carcino-embryonic antigen and can be used for detecting and treating the cancer disease of a human body.
Owner:宁波盛华达科技有限公司
Bartonella henselae PCR identification method
InactiveCN102312006AAccurate detectionSensitive detectionMicrobiological testing/measurementDNA/RNA fragmentationBartonellaBartonella henselae
The invention provides a bartonella henselae PCR identification method. Specific primers are designed according to a sequence hypervariable region of a bartonella henselae outer membrane protein gene BH13010 and specific amplification is performed by using tested sample DNA or bacterial cultures as a template. If a specific band can be obtained by amplification, bartonella henselae is identified.The sequences of the primers in the method are shown as SEQIDNo.6 and 7. The invention has advantages of good specificity of primers, high accuracy of identification method, simple operation, low requirement of equipments and low cost.
Owner:ICDC CHINA CDC
Adenoviral vector-based dengue fever vaccine
The invention relates to a replication-deficient adenoviral vector comprising two or more nucleic acid sequences encoding Dengue virus antigens and a chimeric hexon protein. The chimeric hexon protein comprises a first portion and a second portion. The first portion comprises at least 10 contiguous amino acid residues from a first adenovirus serotype (e.g., serotype 5 adenovirus hexon protein), optionally with one amino acid substitution. The second portion comprises (a) at least one hypervariable region (HVR) of a hexon protein of an adenovirus of a second adenovirus serotype, or (b) at least one synthetic hypervariable region (HVR) that is not present in the hexon protein of the wild-type adenovirus of the first adenovirus serotype.
Owner:GEN VEC INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com