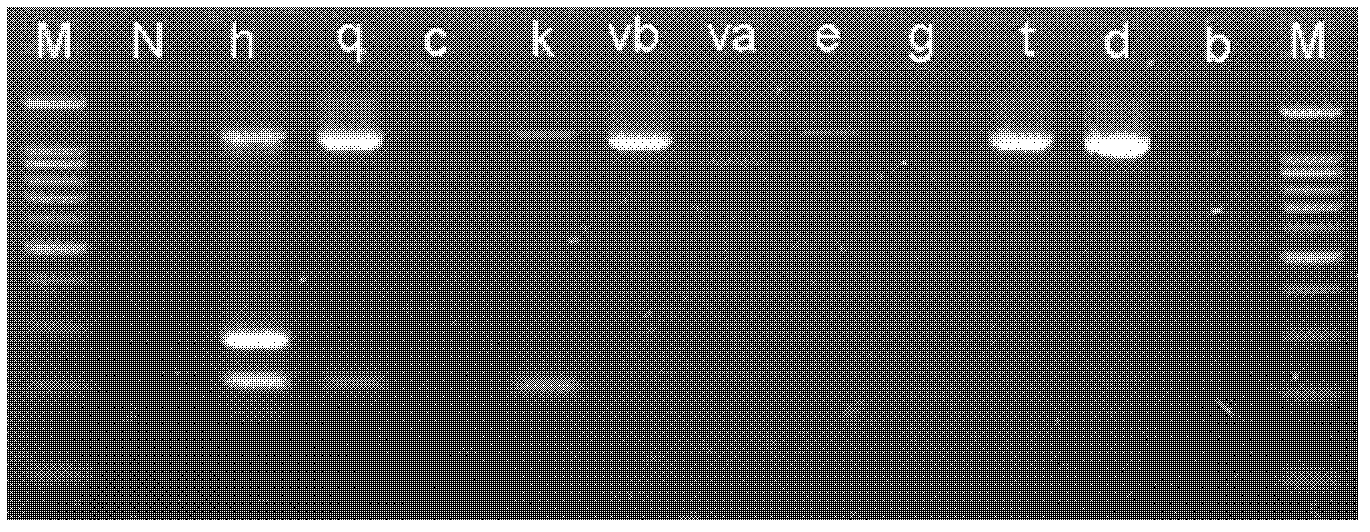Bartonella henselae PCR identification method
An identification method, the technology of the outer membrane protein gene, applied in the field of biological detection, can solve the problems of cumbersome operation, time-consuming, high cost, cumbersome analysis process, etc., and achieve the effect of increasing specificity and ensuring accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Embodiment 1 multiplex PCR primer screening
[0031] (1) RAPD-PCR
[0032] Three random primers S15: 5′-GGAGGGTGTT-3′ (SEQ ID No.16), S18: 5′-CCACAGCAGT-3′ (SEQ ID No.17) and S103: 5′-AGACGTCCAC-3′ (SEQ ID No. ID No.18) for RAPD-PCR amplification. The reaction system used 20 μL of QIAGEN Fast Cycling PCR Kit, 10 pmol of each of the above primers, and 50 ng of genomic DNA of Bartonella henselae (purchased from the American Type Culture Collection, ATCC49882). Amplification conditions were melting at 94°C for 5 minutes; denaturation at 94°C for 40 seconds, annealing at 40°C for 40 seconds, extension at 72°C for 1 minute, 35 cycles; extension at 72°C for 5 minutes.
[0033] (2) Cloning and sequencing of RAPD-PCR amplification products
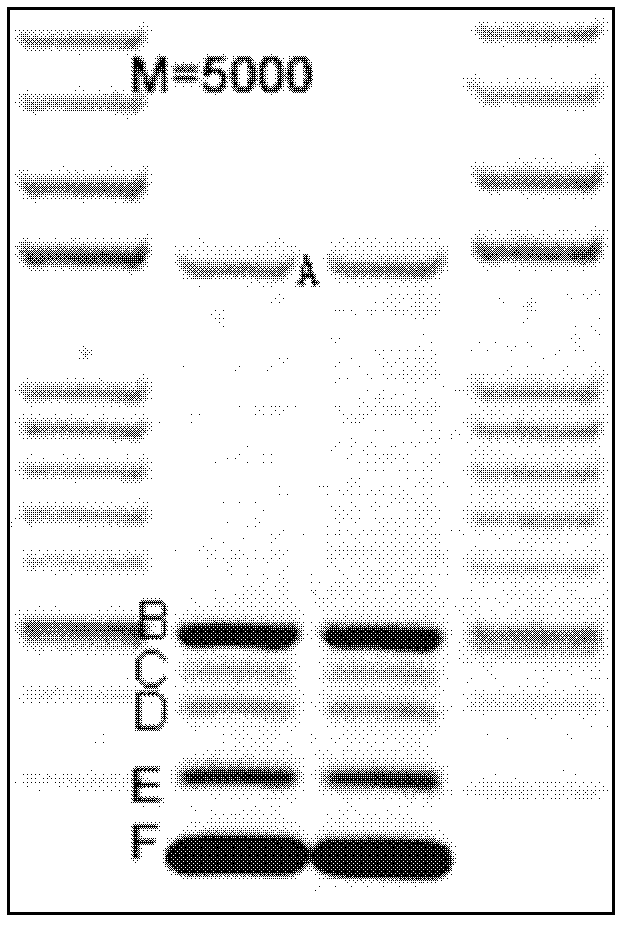
[0034] The Bartonella henselae RAPD-PCR amplification bands A, B, C, D, E and F ( figure 1 ) were gel-cut and purified, and cloned into plasmids for sequencing. Sequence analysis showed that the amplified bands A, B, C, D, E and F were...
Embodiment 2
[0040] Embodiment 2PCR reaction condition optimization
[0041] 1. Primer concentration
[0042] Template: A commercially available nucleic acid extraction kit was used to extract the genomic DNA of the Bartonella henselae strain.
[0043] Reaction system: 50 μL amplification system, 10 μL of 10×Ex Taq buffer from Treasure Biotech, TaKaRa Ex Taq 5U, 5 mM dNTP mixture, 10 pmol each of primers BhAF-BhAR, Cpf-Cpr and 6f1-6f2 or primers BhAF-BhAR and Cpf- 10 pmol each of Cpr, 15 pmol of primers 6f1-6f2, 1 μL of DNA template, and deionized water to make up to 50 μL.
[0044] PCR amplification parameters: denaturation at 94°C for 30s, annealing at 54°C for 30s, extension at 72°C for 1min, a total of 25 cycles.
[0045] Gel electrophoresis and result observation: Take 5 μL of PCR product and 1 μL of 6× loading buffer, mix thoroughly, and place on 1% agarose gel for electrophoresis. The voltage was 5v / cm, the electrophoresis buffer was 0.5×TBE, and the results were observed and pho...
Embodiment 3
[0051] Embodiment 3 specificity detection experiment
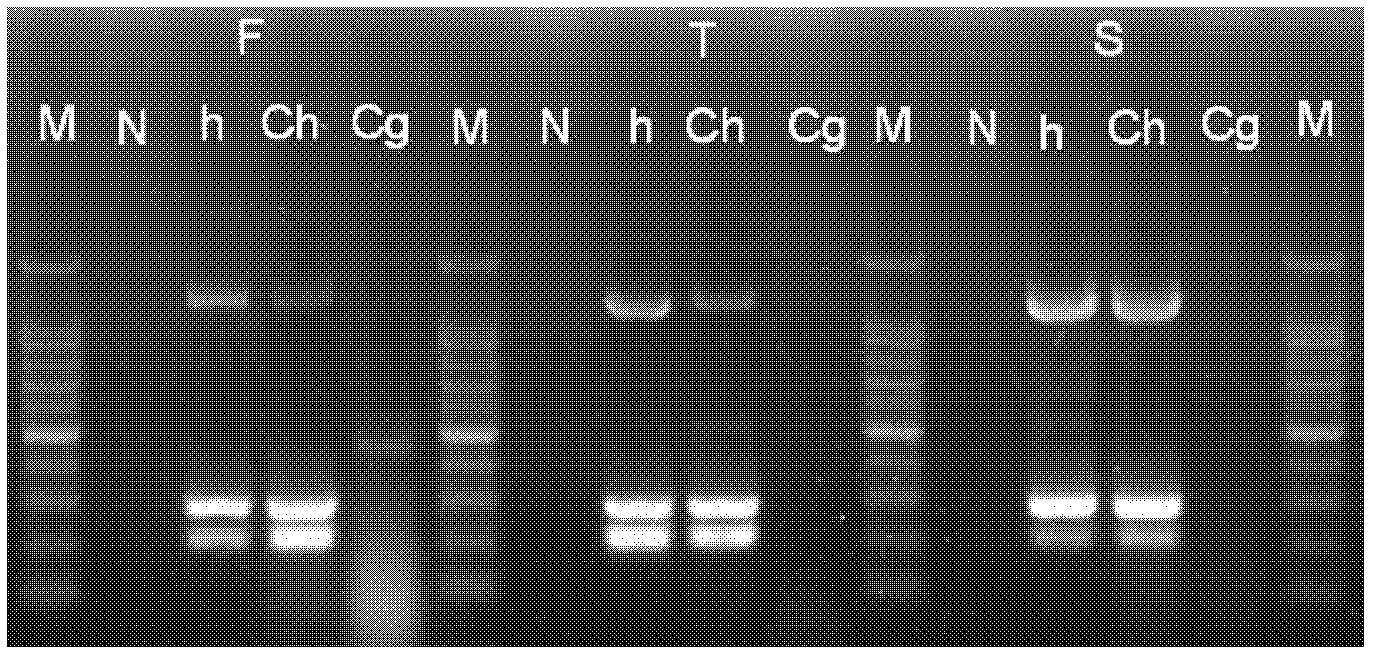
[0052] Using the system optimized in Example 2 to amplify Bartonella quintana, Bartonella keshii (B.clarridgeiae), Bartonella keele (B.koehlerae), Bartonella vinsonii Boghof subspecies (B.vinsonii subsp.berkhoffii), Bartonella Vinsonii subsp.arupensis (B.vinsonii subsp.arupensis), Bartonella Elizabeth (B.elizabethae), Bartonella Graham (B.grahamii), Bartonella Tribochi ( B.tribocorum), B.doshiae, and B.bacilliformis did not have the three amplification bands A, C, and F at the same time. The A and F bands were amplified throughout the body, but the C band did not appear ( image 3 ). The results showed that amplifying the C band alone could also specifically identify Bartonella henselae. Amplifies Agrobacterium tumefaciens, Brucella, Rickettsia, Escherichia coli, Shigella, Vibrio cholerae, Staphylococcus aureus, Anaplasma, Campylobacter jejuni, Legionella, Acinetobacter baumannii, Hook 27 kinds of bacteria such as term...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com