InDel molecular markers for assisted selection of early heading genes of rice and application of InDel molecule marker
A molecular marker, rice technology, applied in recombinant DNA technology, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problems of long growth period, affecting popularization and application, and not suitable for late production and planting, etc. achieve the effect of speeding up the cultivation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] 1. Identification of heading date of chromosome segment substitution line
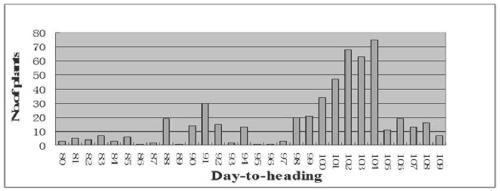
[0029] Through the identification of the heading date of 113 chromosomal fragment substitution lines with indica rice 9311 as the genetic background, it was found that a substitution line CL33 headed 15-20 days earlier than the recipient parent 9311. figure 1 shown.
[0030] 2. Genetic analysis of CL33 heading stage genes
[0031] Pass to 9311 / CL33 F 2 524 isolated individual plants were investigated for heading date, and the results were as follows figure 2 As shown, their variation range is 80-109 days, and the segregation ratio of early and late heading is 127:394, which is in line with the segregation ratio of 1:3. Genetic analysis shows that the early heading of CL33 is controlled by a pair of recessive genes, of which late heading is controlled by a pair of recessive genes. Heading is dominant to early heading.
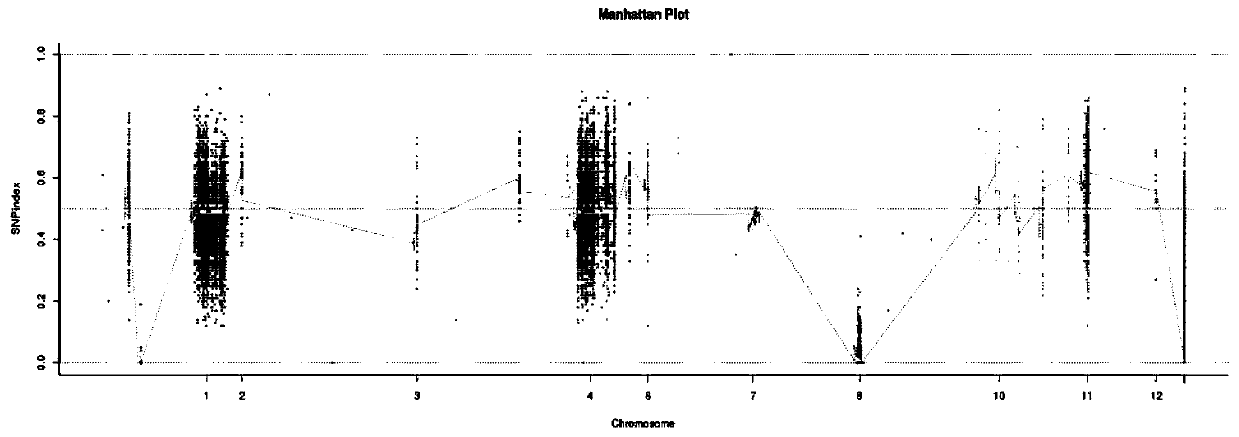
[0032] 3. Whole gene resequencing and BSA analysis based on CL33, 9311 a...
Embodiment 2
[0058] (1) Rice samples
[0059] Early and late heading parents CL33 and 9311, 9311 / CL33 hybrid F 2 In the genetic segregating population, 31 plants showed early heading (80-85 days), 2 plants showed late heading (105-106 days) and 1 plant with a heading period of 94 days.
[0060] (2) DNA sample extraction and detection
[0061] DNA from each rice sample was extracted according to the CTAB method (Murray & Thompson 1980). DNA purity and integrity of each sample were analyzed by agarose gel electrophoresis; 260 / 280 The ratio is between 1.8 and 2.0); Qubit accurately quantifies the sample DNA concentration (sample concentration > 20ng / μL).
[0062] (3) PCR amplification
[0063] PCR amplification was performed according to the method of Panaud et al. (1996) with slight modifications. The 20 μL reaction system includes 0.15 μM primer, 200 μM dNTP, 1× PCR reaction buffer (50 mM KCl, 10 mM Tris-HCl pH8.3, 1.5 mM MgCl 2 , 0.01% gelatin), 50-100ng of DNA template, 1UTaq enzyme...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com