Molecular marker linked with major quantitative trait locus (QTL) of hundred-kernel weight of arachis hypogaea and application of molecular marker
A technique based on molecular markers and all kinds of kernels, which is applied in the fields of molecular genetics and crop molecular breeding, can solve the problems of general effect, low phenotypic contribution rate of molecular markers, and large QTL intervals
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
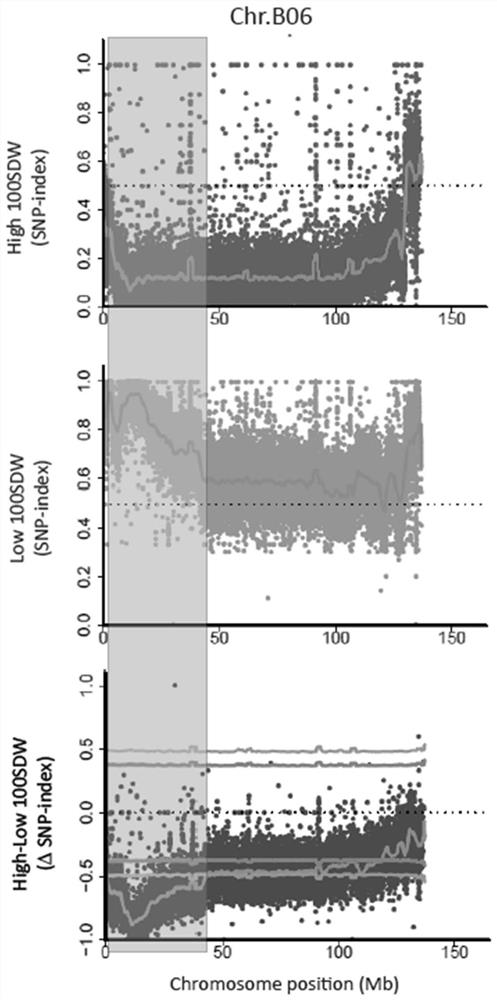
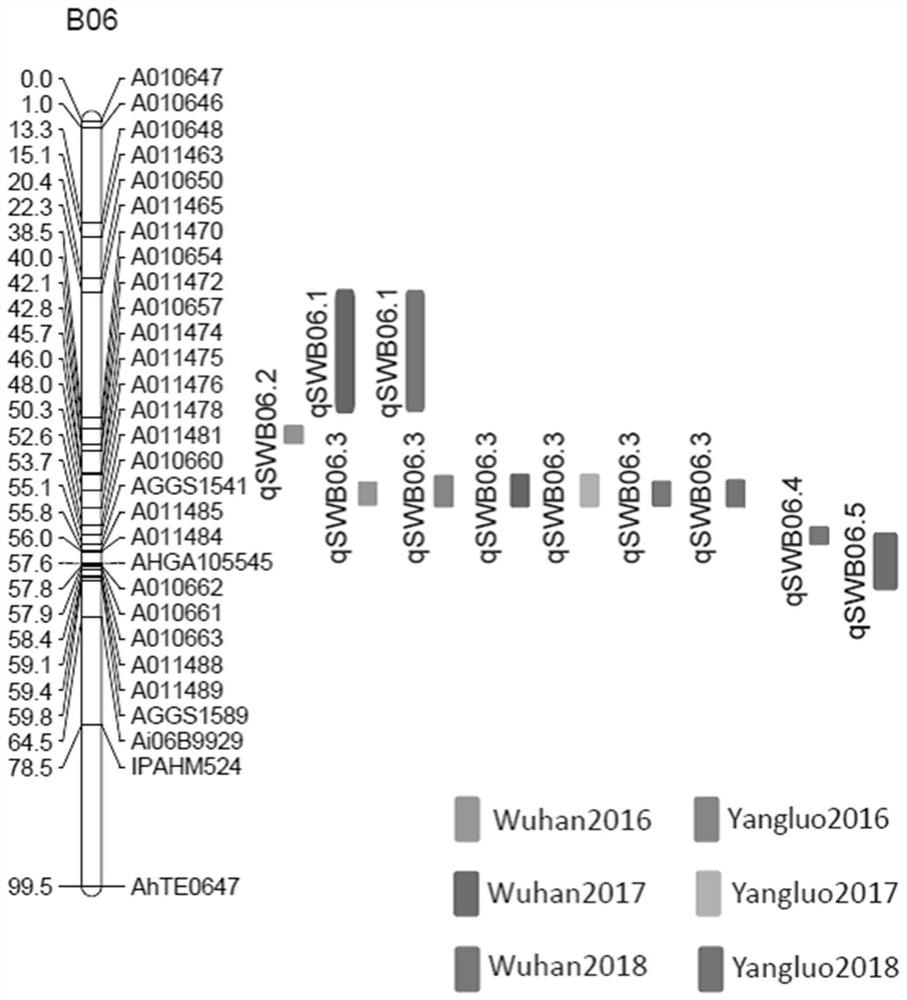
[0043] This embodiment provides a method for screening molecular markers linked to the main effect QTL locus of peanut weight, which specifically includes the following steps:
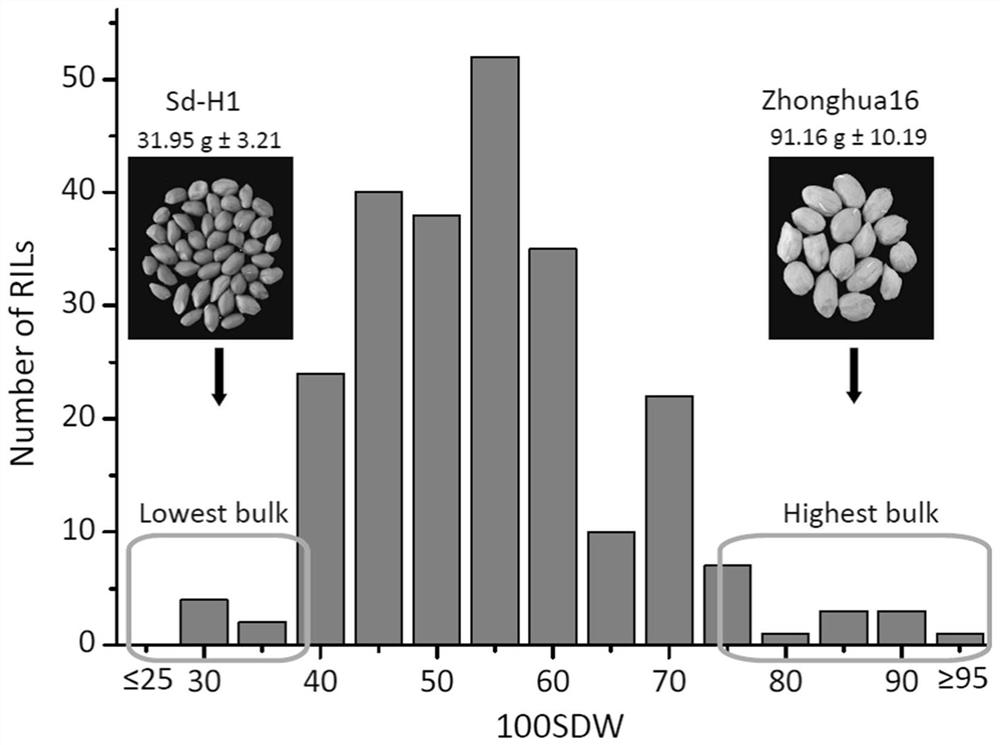
[0044] 1. Construct a recombinant inbred line (RIL) population with Zhonghua 16 and sd-H1 as parents. The female parent, Zhonghua 16, is a high-yield, high-oil peanut variety with pearl bean-shaped large fruits, and the male parent, sd-H1, is a multi-grained small-fruited peanut variety. For low-yielding peanut varieties, the two parents have large differences in agronomic traits such as grain weight (100-kernel weight), such as figure 1 shown.
[0045]2. Zhonghua 16, sd-H1 and F6 (2016), F7 (2017), F8 (2018) generation RIL population 242 strains were sown in two experimental bases in Wuhan and Yangluo three times in completely randomized blocks After the material matures naturally, it is harvested and dried in the sun. According to the standard in the "Peanut Germplasm Resources Description Specifica...
Embodiment 2
[0050] In this embodiment, the linkage between the two molecular markers related to the weight of 100 kernels and the traits in peanut varieties obtained in Example 1 is verified, as follows:
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com