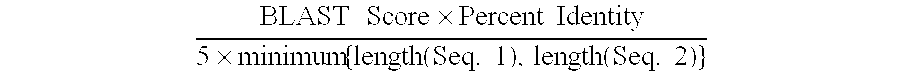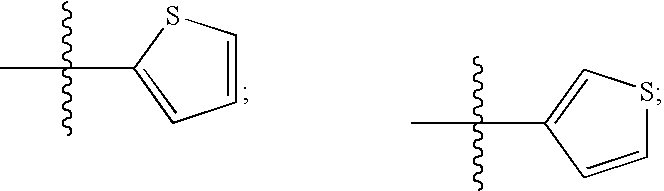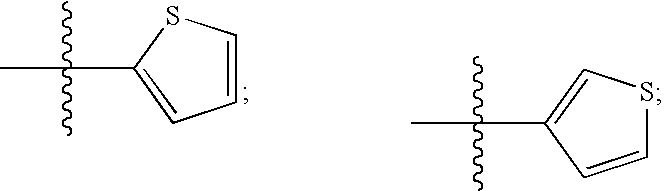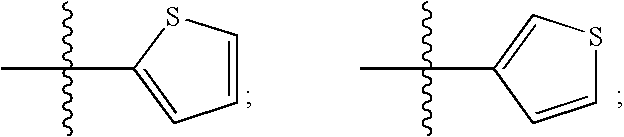Patents
Literature
389 results about "Human proteins" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Proteins are essential nutrients for the human body. They are one of the building blocks of body tissue, and can also serve as a fuel source. As a fuel, proteins provide as much energy density as carbohydrates: 4 kcal (17 kJ) per gram; in contrast, lipids provide 9 kcal (37 kJ) per gram.
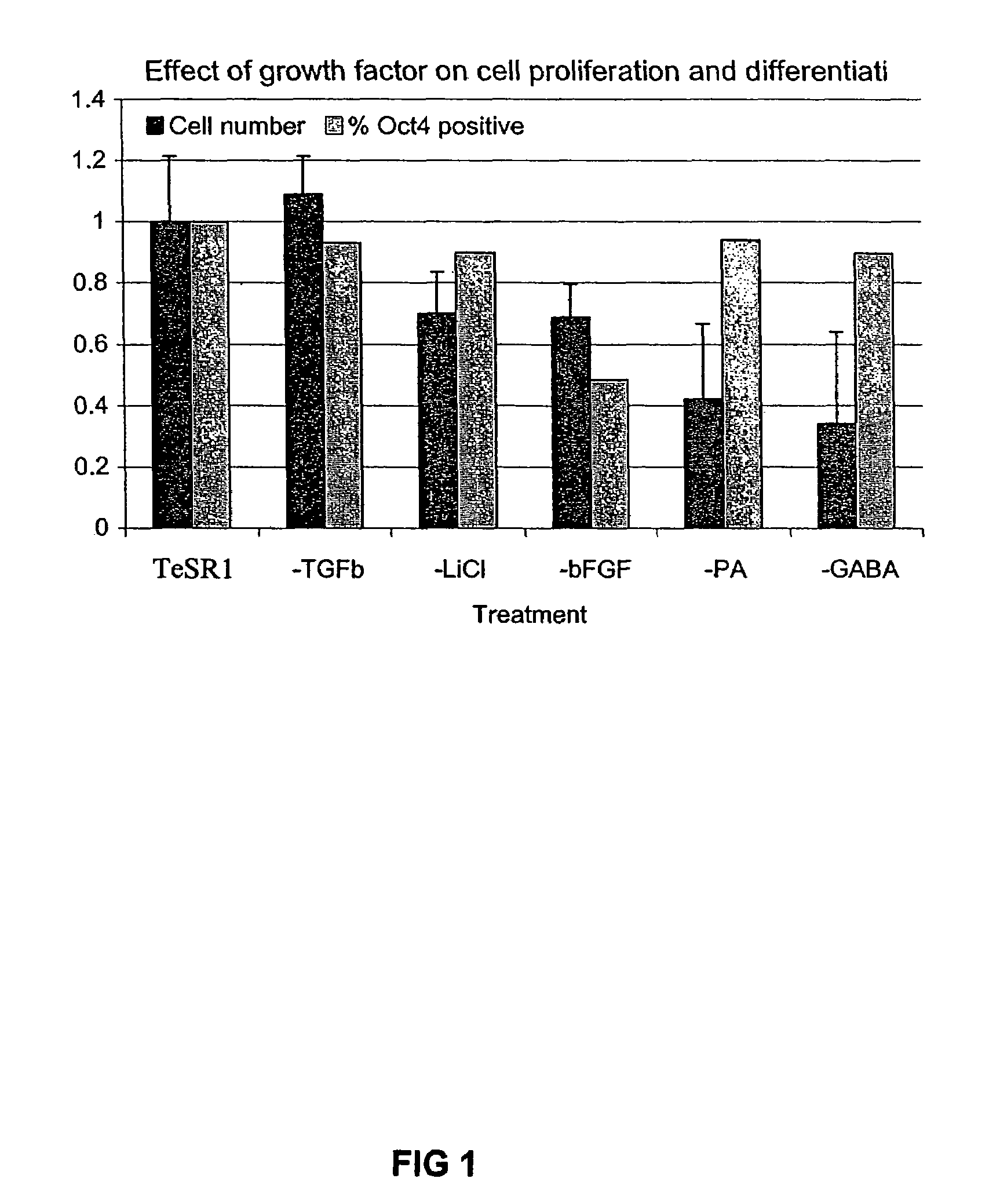
Culturing human embryonic stem cells in medium containing pipecholic acid and gamma amino butyric acid
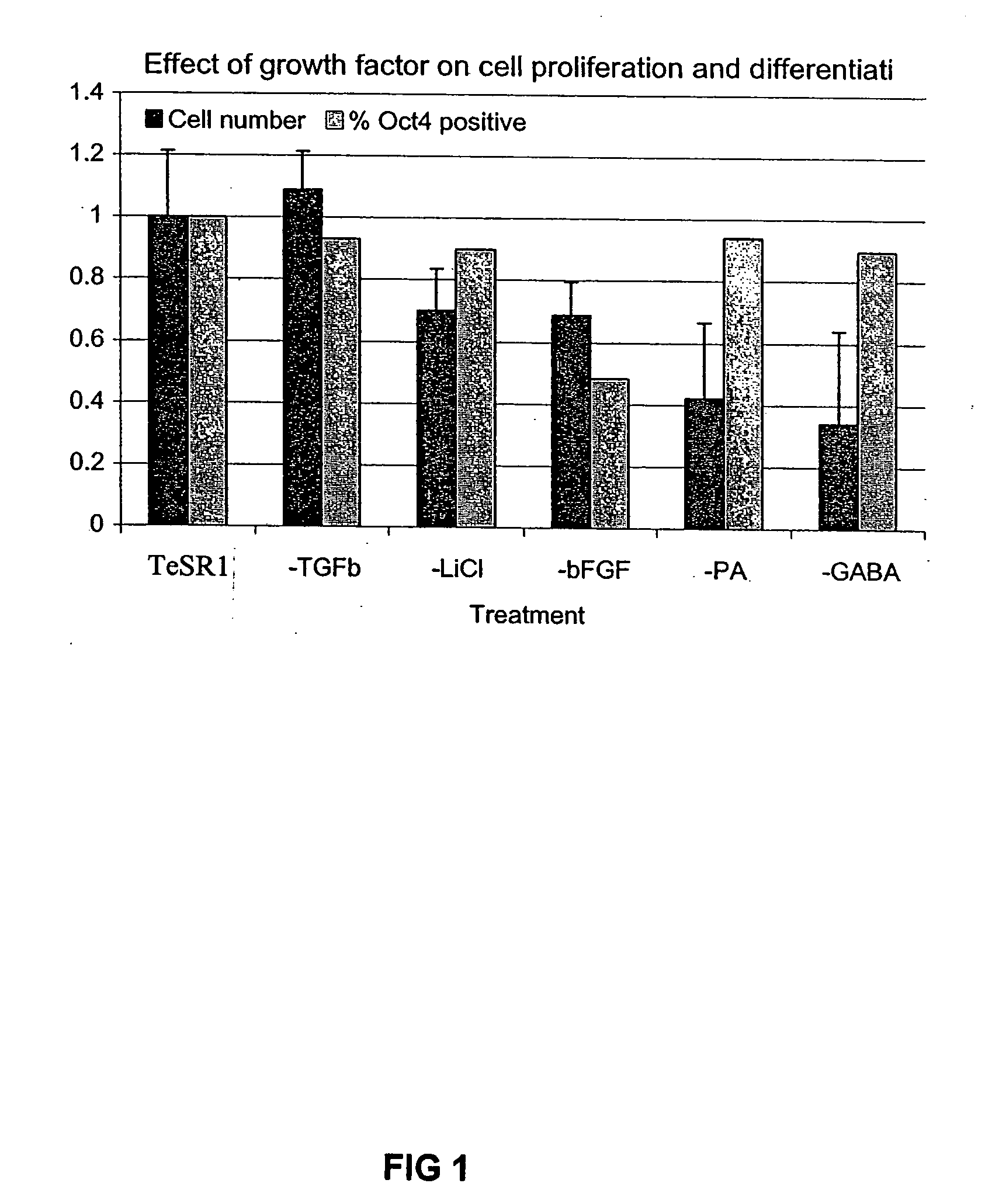
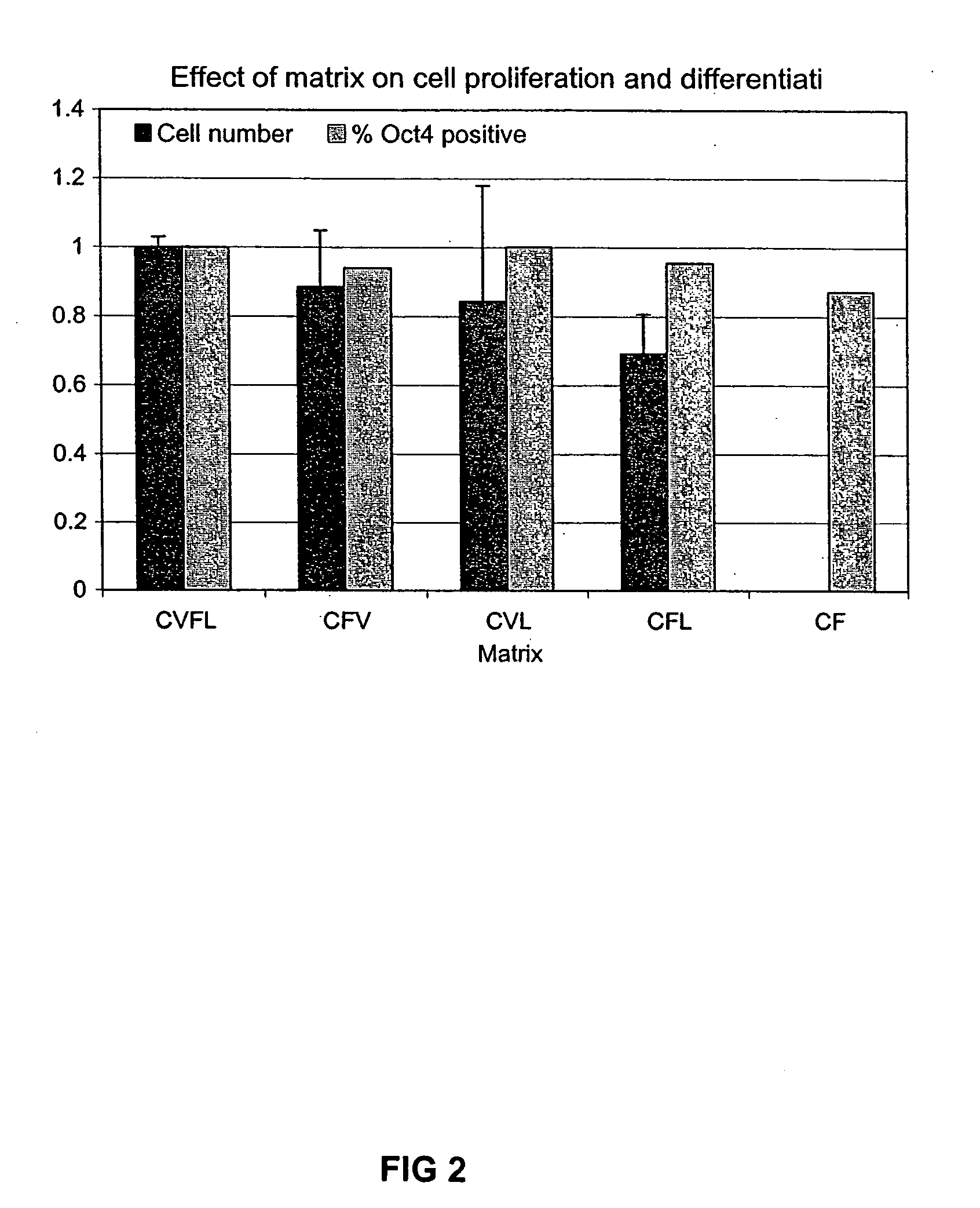
Previous methods for culturing human embryonic stem cells have required either fibroblast feeder cells or a medium which has been exposed to fibroblast feeder cells in order to maintain the stem cells in an undifferentiated state. It has now been found that if high levels of fibroblast growth factor are used in a medium with gamma amino butyric acid, pipecholic acid, lithium and lipids, the stem cells will remain undifferentiated indefinitely through multiple passages, even without feeder cells or conditioned medium. A humanized matrix of human proteins can be used as a basement matrix to culture the cells. New lines of human embryonic stem cells made using these culture conditions, the medium and the matrix, will never have been exposed to animal cells, animal products, feeder cells or conditioned medium.
Owner:WISCONSIN ALUMNI RES FOUND
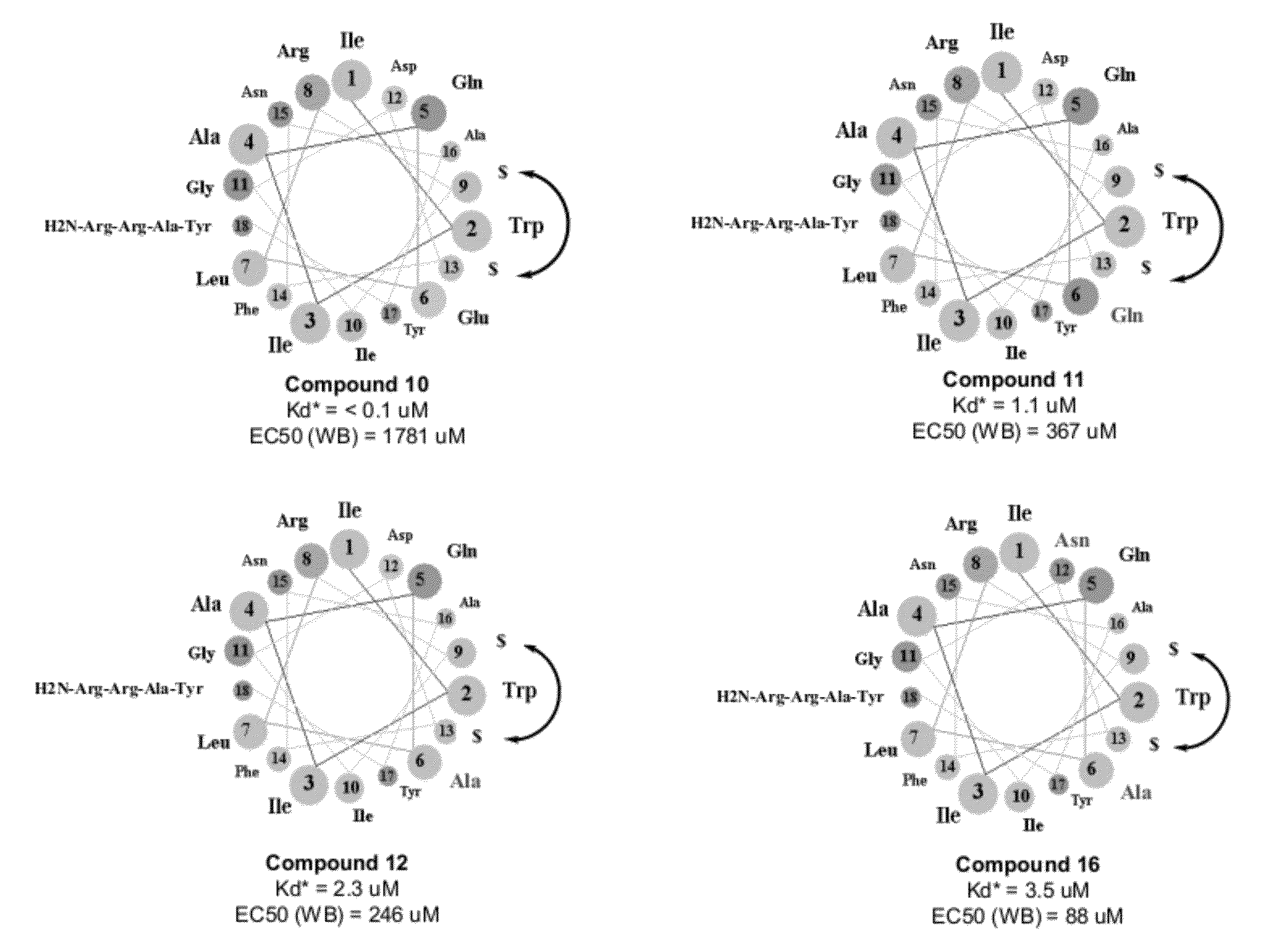
Peptidomimetic macrocycles
InactiveUS20100216688A1Low affinityEasy to transportPeptide/protein ingredientsAntipyreticHuman proteinsCell penetration
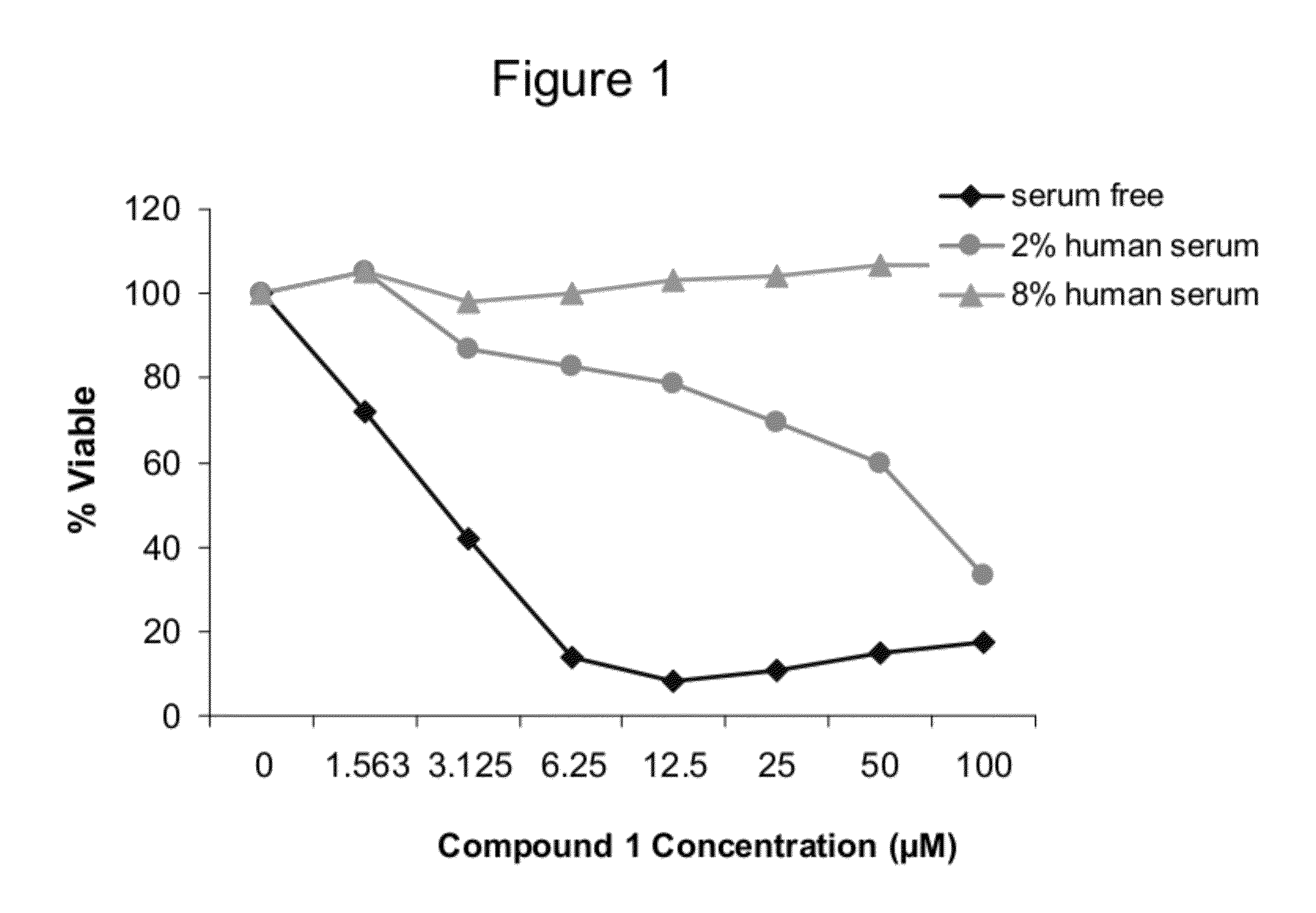
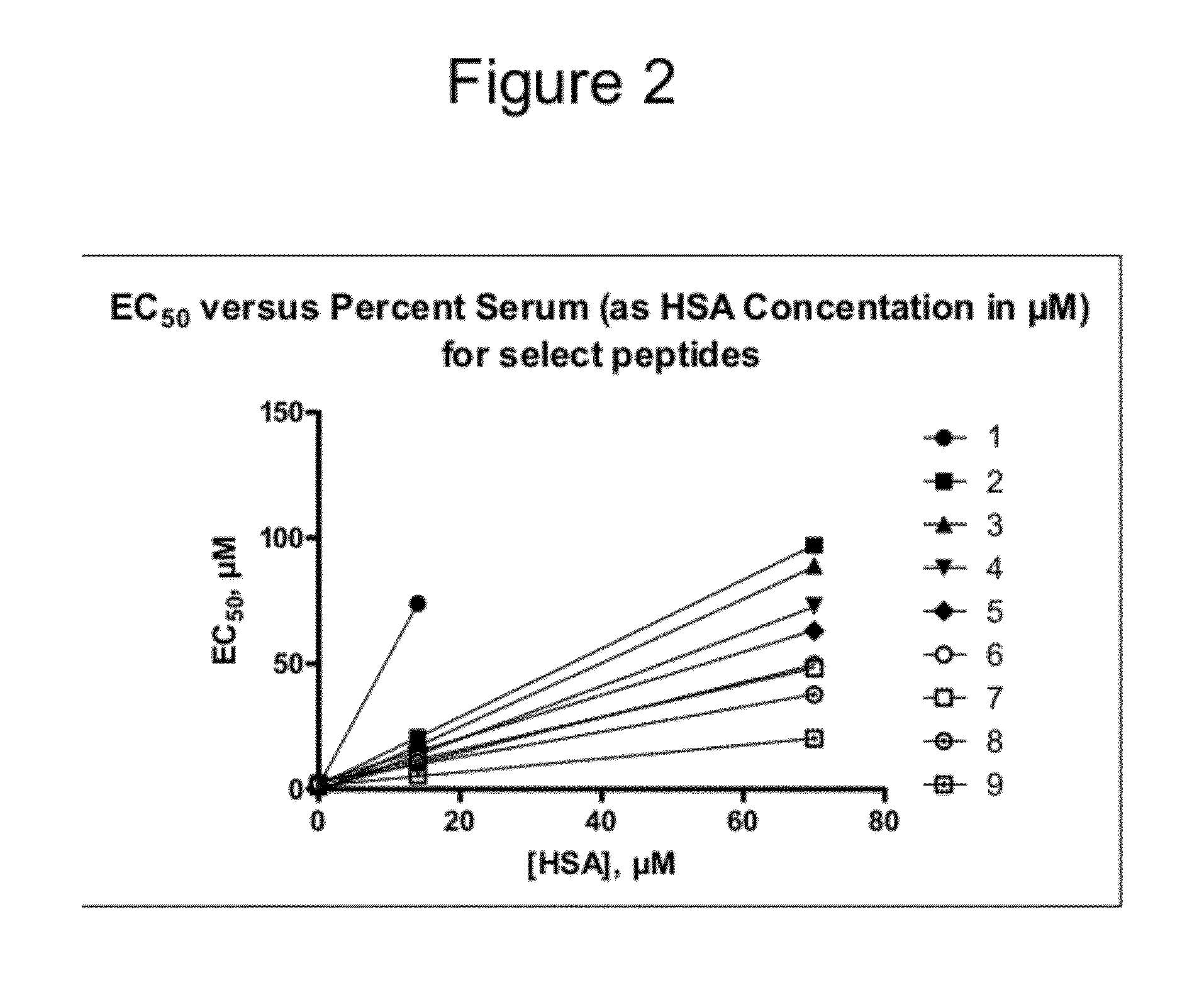
The present invention provides biologically active crosslinked polypeptides with improved properties relative to their corresponding precursor polypeptides, having good cell penetration properties and reduced binding to human proteins. The invention additionally provides methods of identifying and making such improved polypeptides.
Owner:AILERON THERAPEUTICS INC
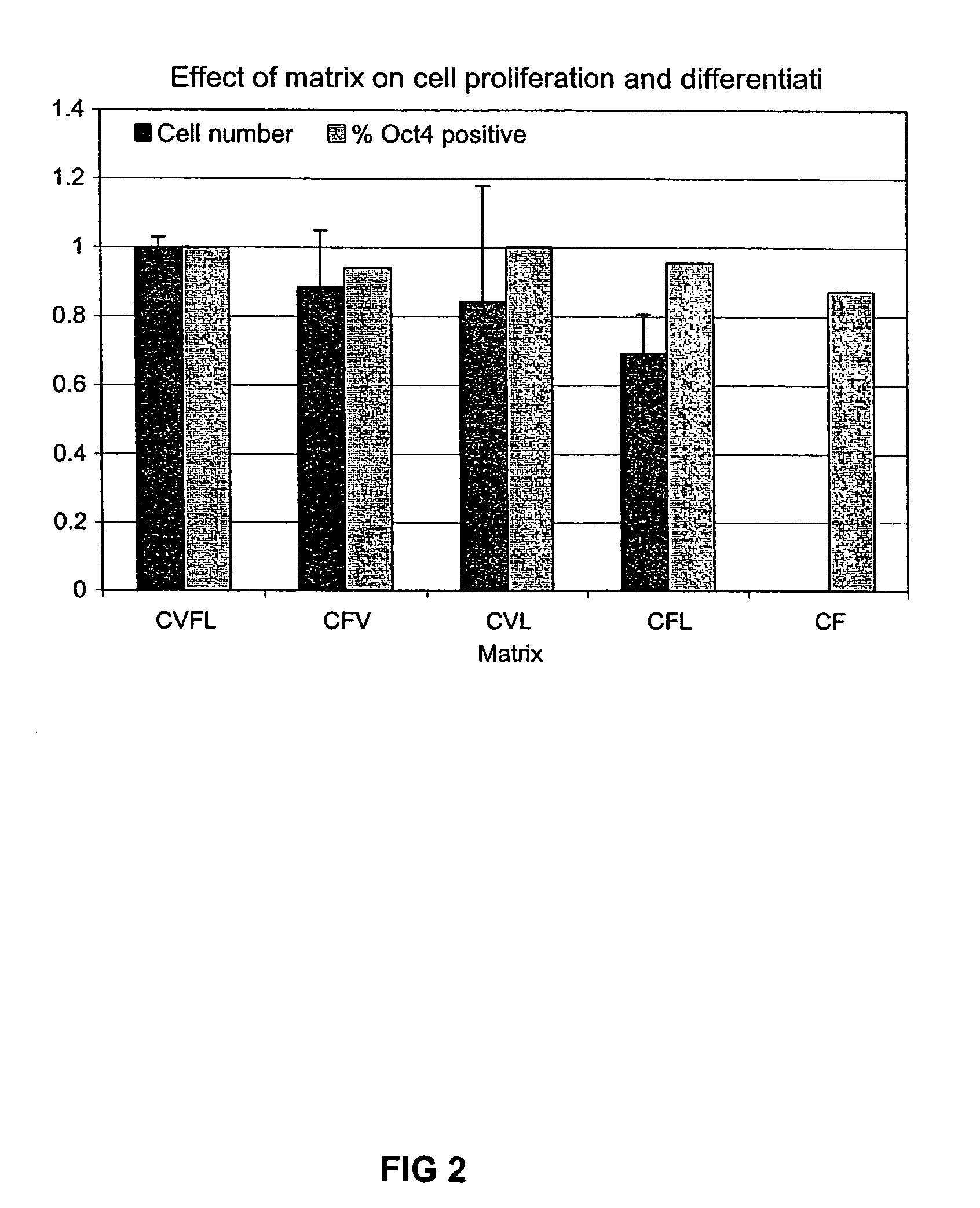
Culturing human embryonic stem cells
Previous methods for culturing human embryonic stem cells have required either fibroblast feeder cells or a medium which has been exposed to fibroblast feeder cells in order to maintain the stem cells in an undifferentiated state. It has now been found that if high levels of fibroblast growth factor are used in a medium with gamma amino butyric acid, pipecholic acid, lithium and lipids, the stem cells will remain undifferentiated indefinitely through multiple passages, even without feeder cells or conditioned medium. A humanized matrix of human proteins can be used as a basement matrix to culture the cells. New lines of human embryonic stem cells made using these culture conditions, the medium and the matrix, will never have been exposed to animal cells, animal products, feeder cells or conditioned medium.
Owner:WISCONSIN ALUMNI RES FOUND
Protein modification and maintenance molecules
The invention provides human protein modification and maintenance molecules (PMMM) and polynucleotides which identify and encode PMMM. The invention also provides expression vectors, host cells, antibodies, agonists, and antagonists. The invention also provides methods for diagnosing, treating, or preventing disorders associated with aberrant expression of PMMM.
Owner:INCYTE
Crystalline frap complex
InactiveUS6532437B1Reduce the numberHigh resolutionTransferasesDigital computer detailsCrystallographyTernary complex
The invention relates to the human protein FRAP, and in particular to the FKBP12-rapamycin binding domain thereof and to the ternary complex formed by the FRB domain, rapamycin and FKBP12. A new crystalline composition comprising the ternary complex, coordinates defining its three dimensional structure in atomic detail, and uses thereof are disclosed.
Owner:CORNELL RES FOUNDATION INC
Peptidomimetic macrocycles
The present invention provides biologically active crosslinked polypeptides with improved properties relative to their corresponding precursor polypeptides, having good cell penetration properties and reduced binding to human proteins. The invention additionally provides methods of identifying and making such improved polypeptides.
Owner:AILERON THERAPEUTICS INC
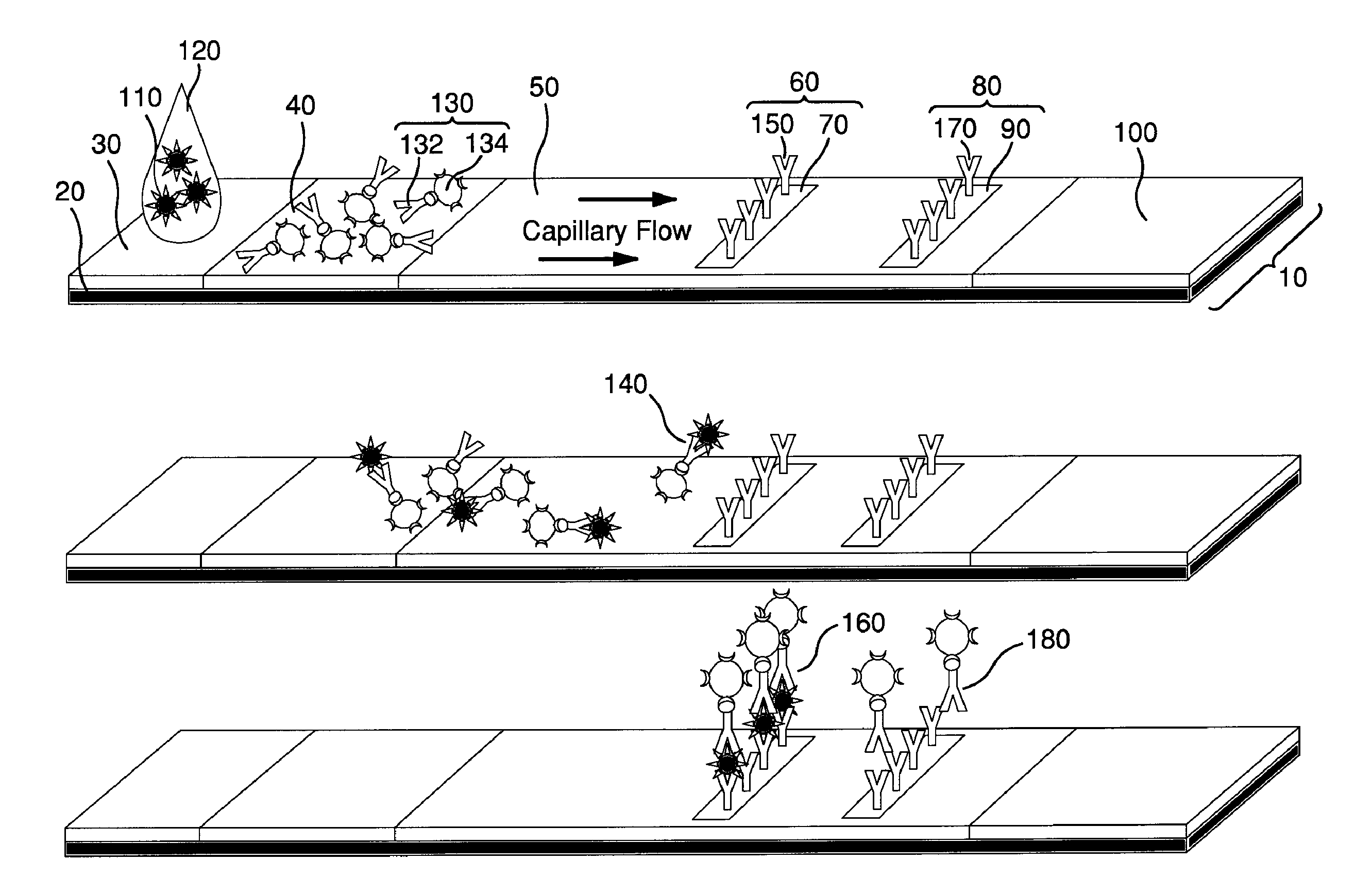
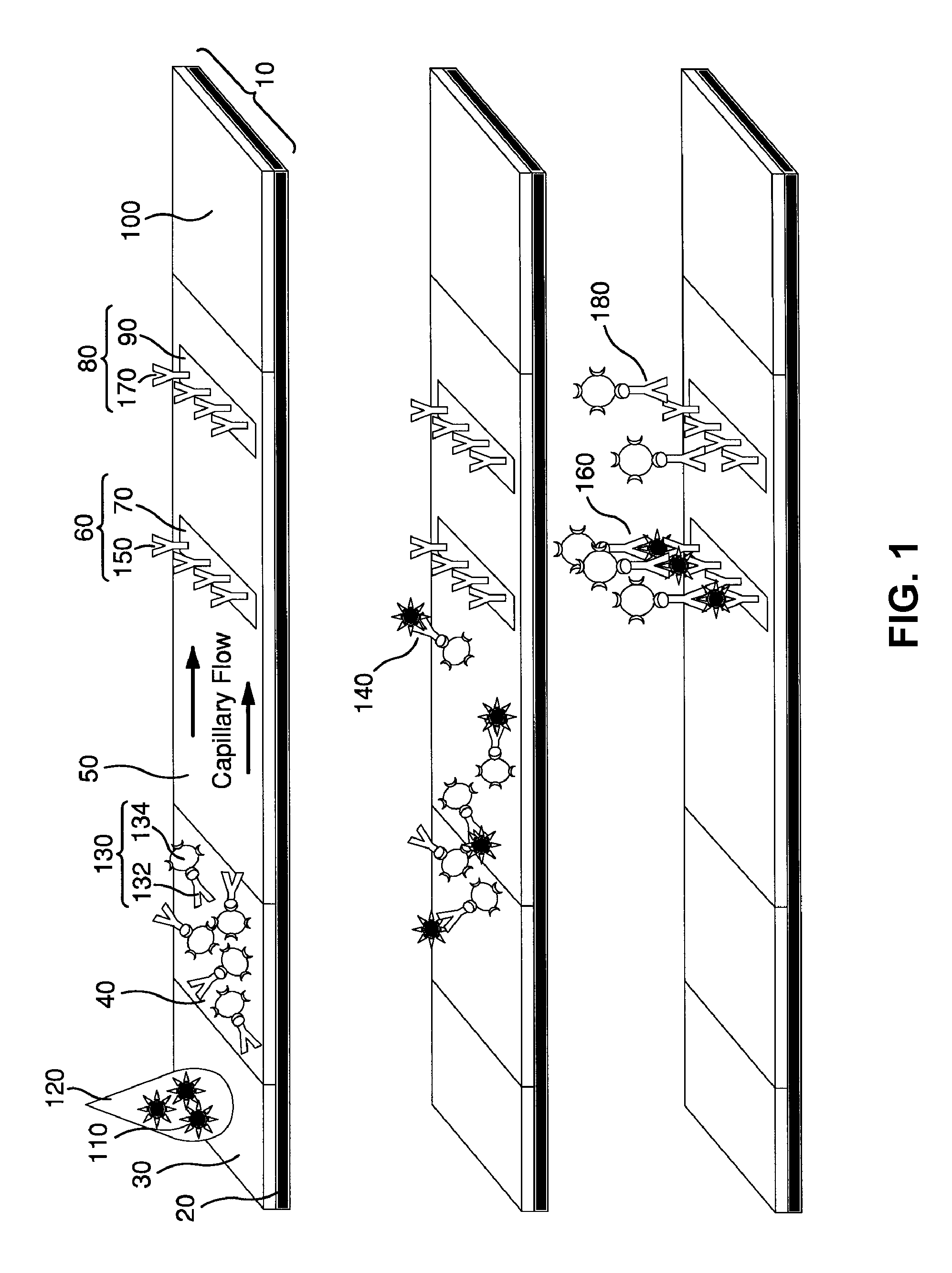
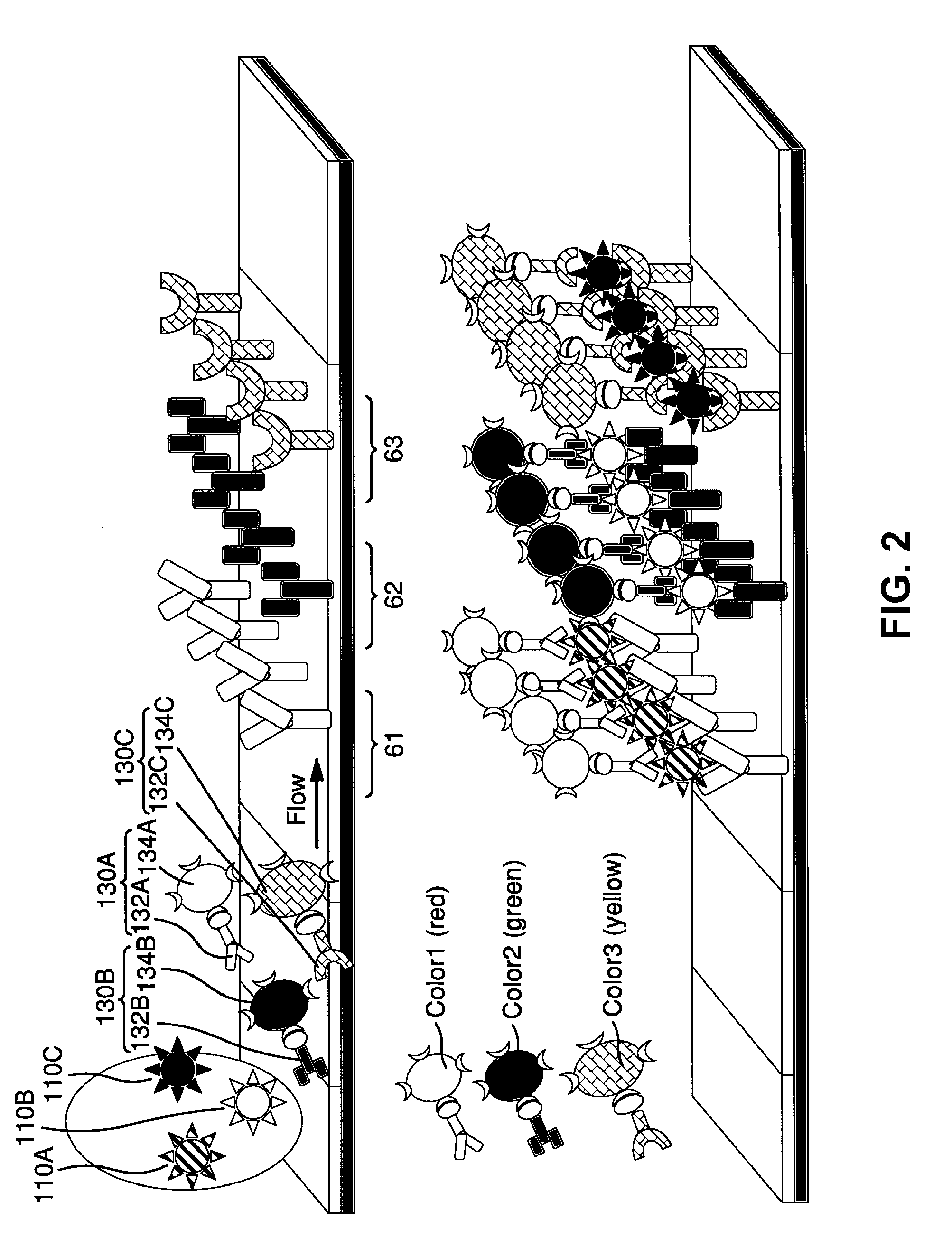
Surface Enhanced Raman Scattering and Multiplexed Diagnostic Assays
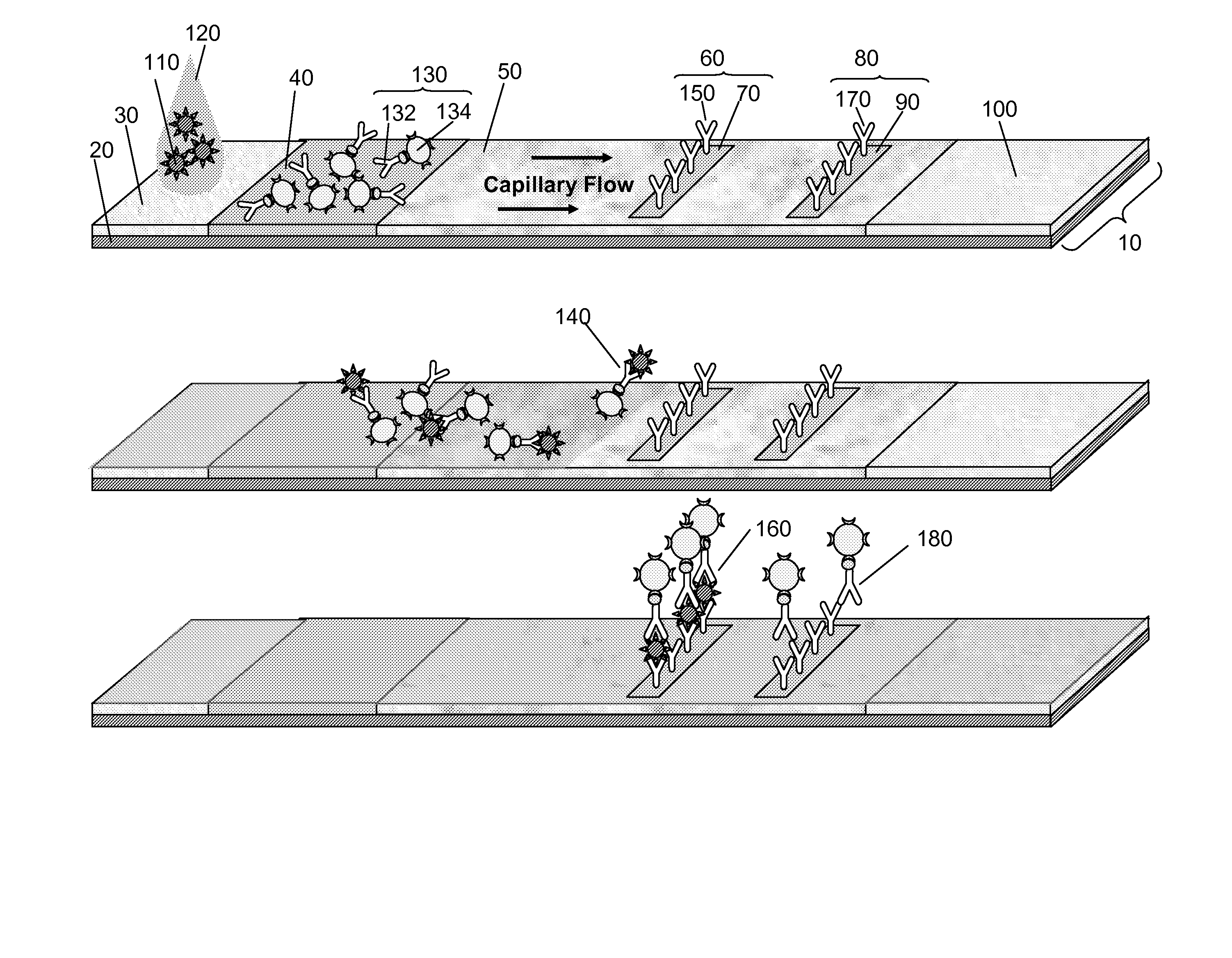
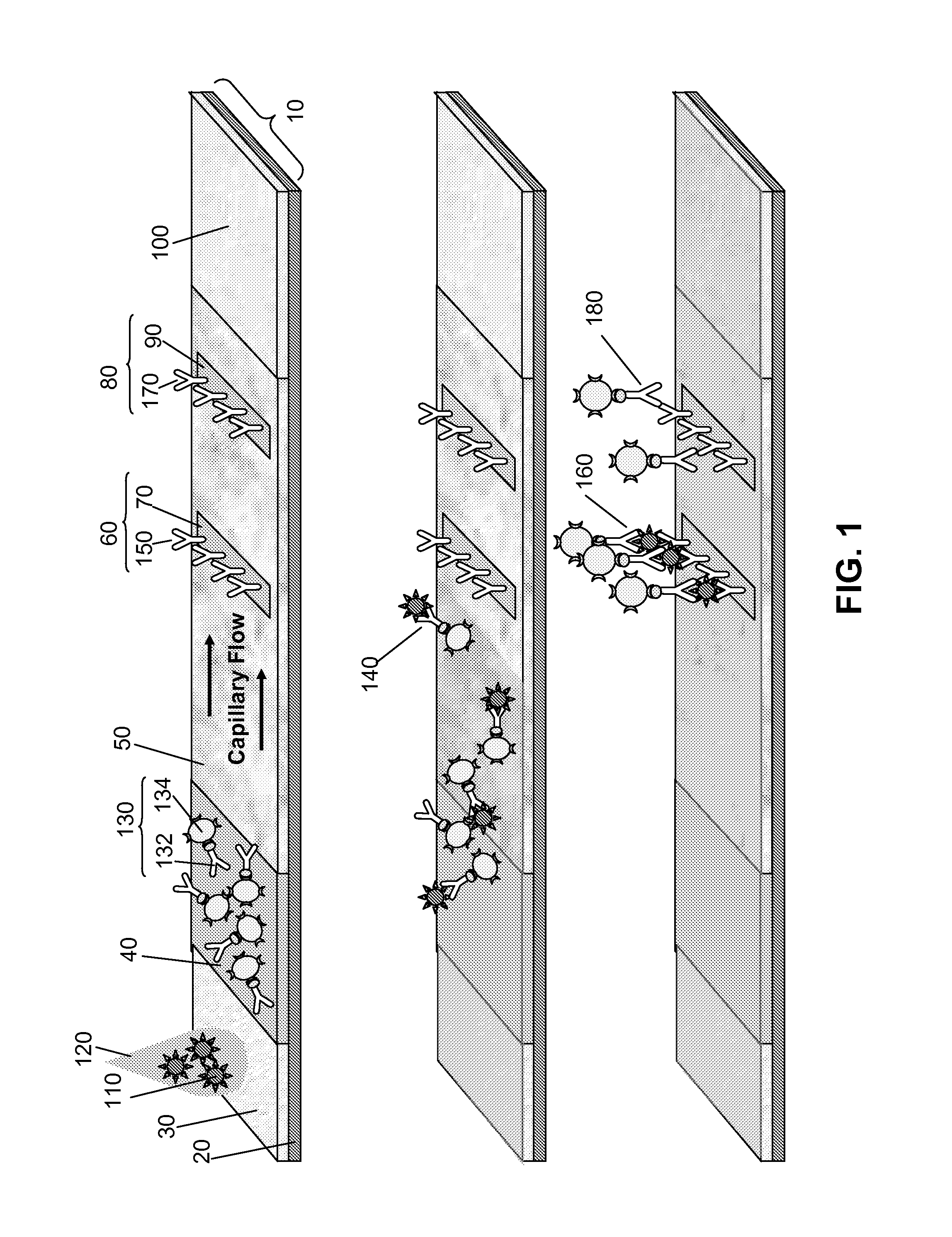
Multiplexed lateral flow assays, related methods, and devices are disclosed which are capable of simultaneously detecting multiple analytes. The assays are preferably immunoassays and can be multiplexed spatially, spectrally, and both spatially and spectrally. Multiplexed assays are disclosed employing quantum dots for applications including the detection of human proteins and the monitoring of microorganisms relevant to water contamination. The multiplexed assays can employ one or more species of Surface Enhanced Raman Scattering nanoparticles, with one or more species having a unique Raman shift spectrum. The invention is widely adaptable to a variety of analytes such as biowarfare agents, human clinical markers, and other substances.
Owner:CALIFORNIA INST OF TECH
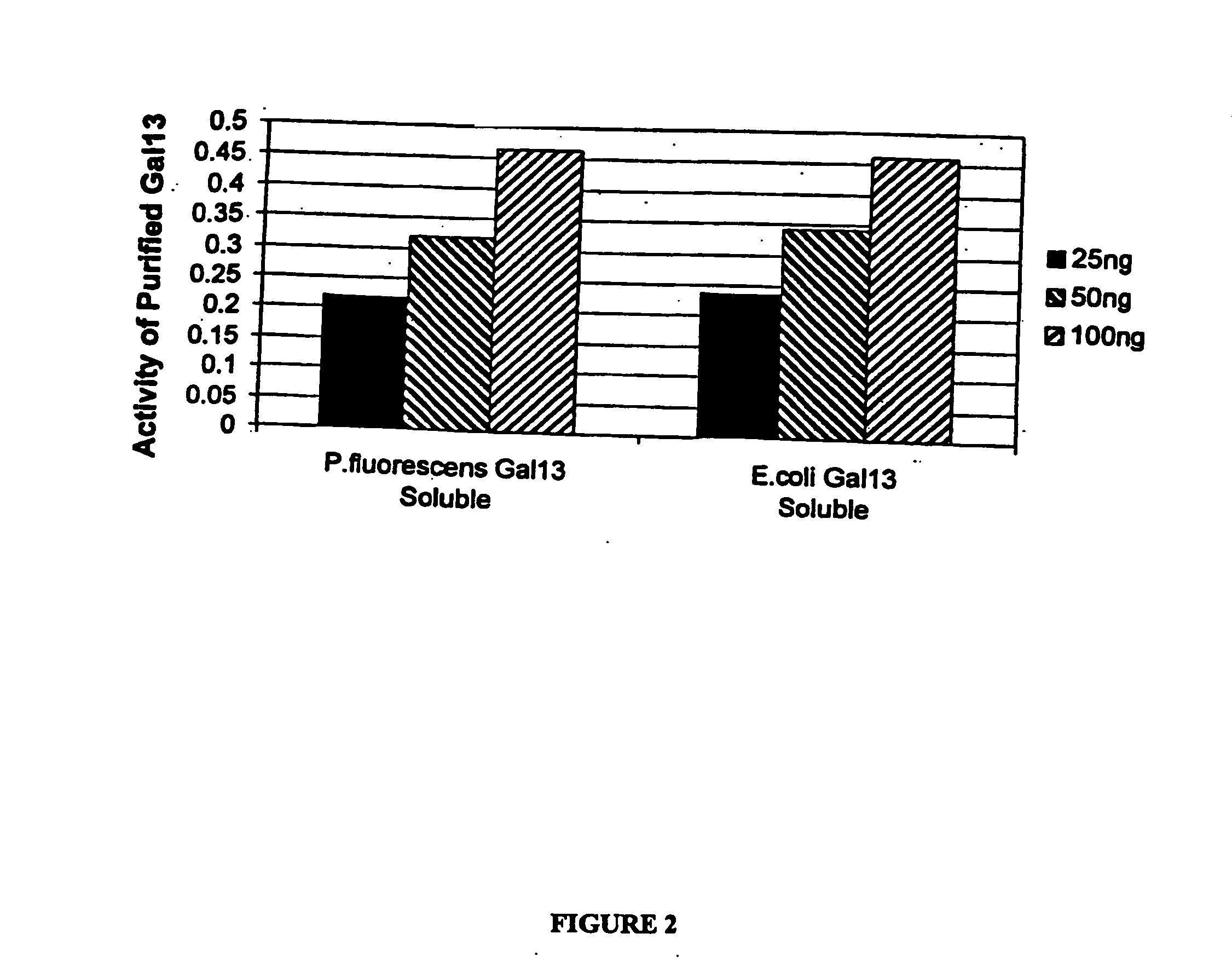
Expression of mammalian proteins in Pseudomonas fluorescens
The invention is a process for improved production of a recombinant mammalian protein by expression in a Pseudomonad, particularly in a Pseudomonas fluorescens organism. The process improves production of mammalian proteins, particularly human or human-derived proteins, over known expression systems such as E. coli in comparable circumstances Processes for improved production of isolated mammalian, particularly human, proteins are provided.
Owner:PELICAN TECH HLDG INC
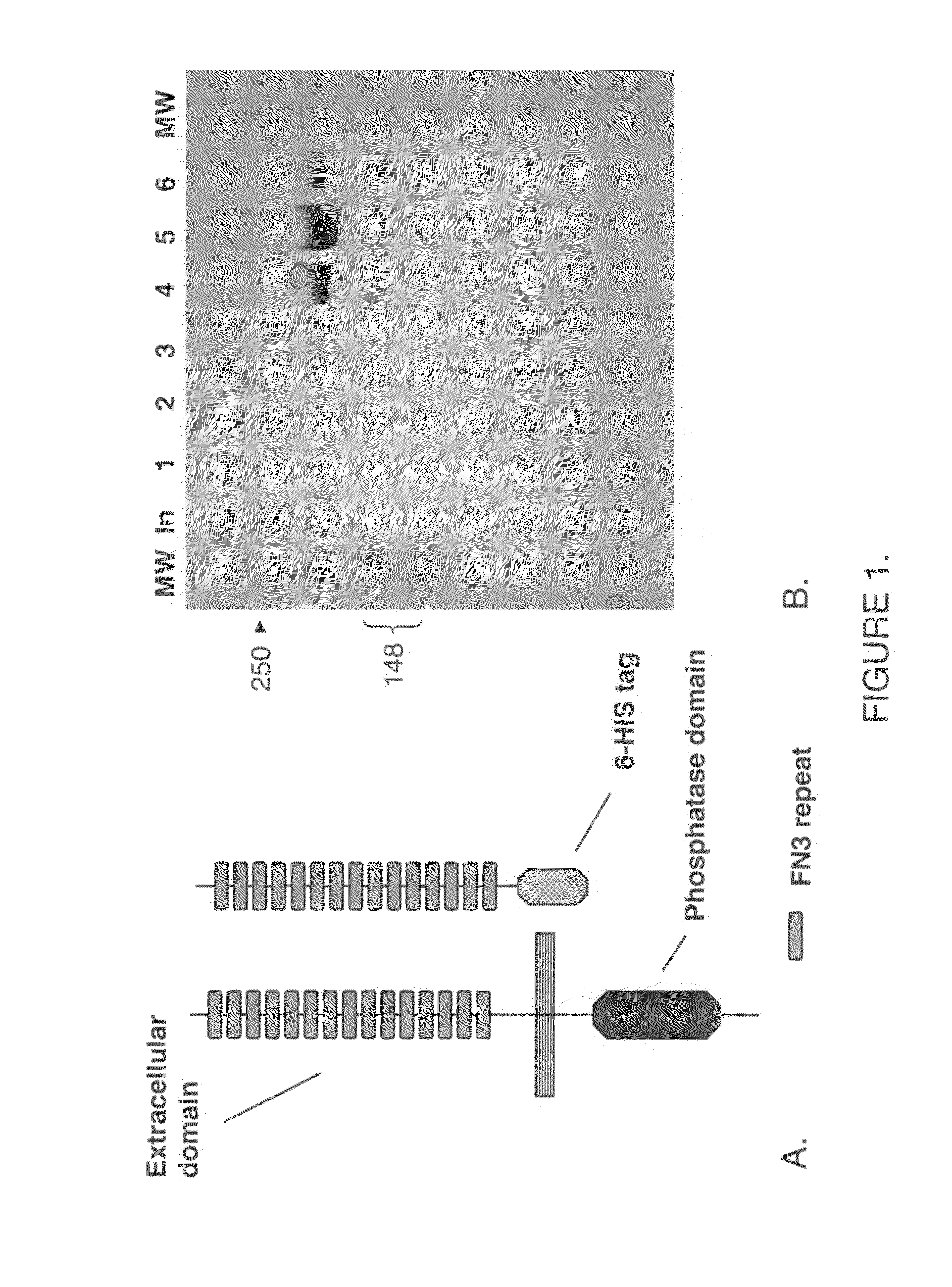
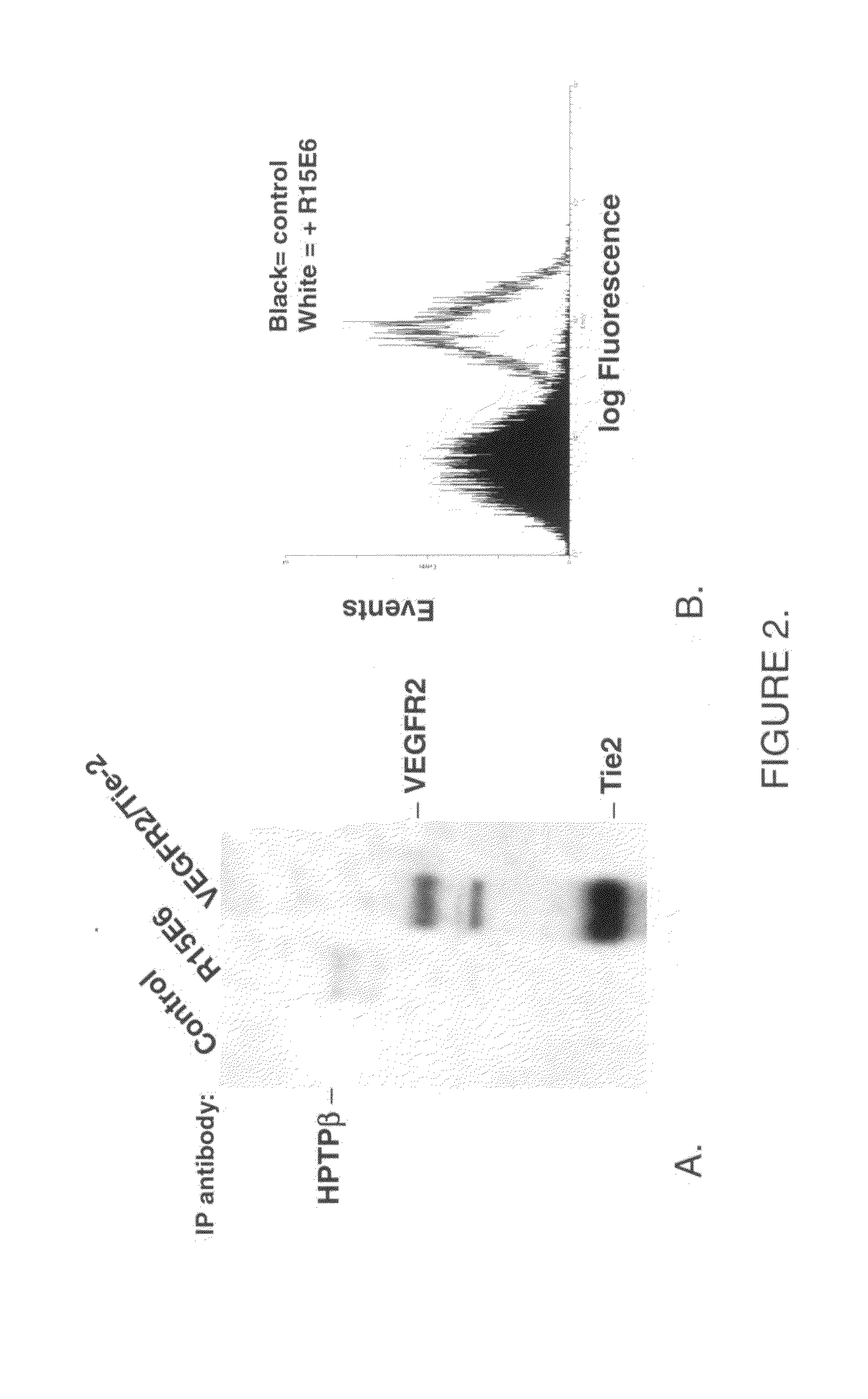
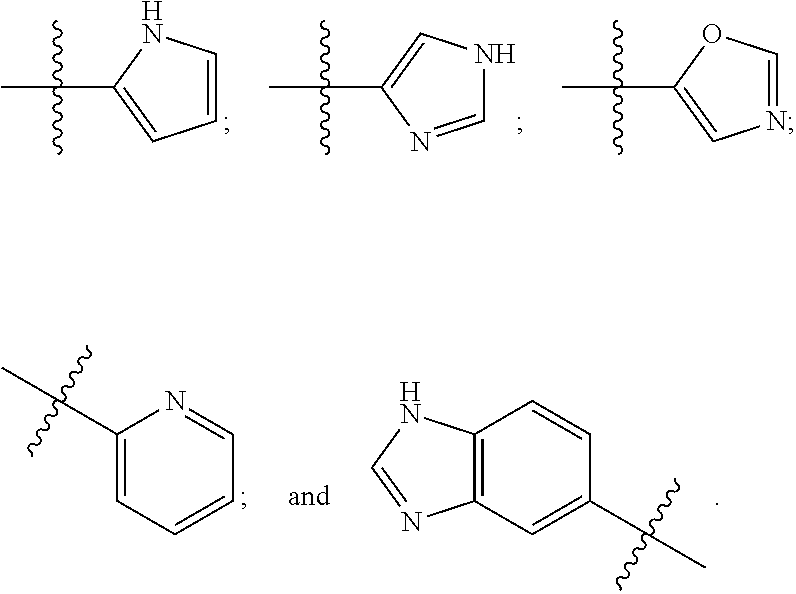
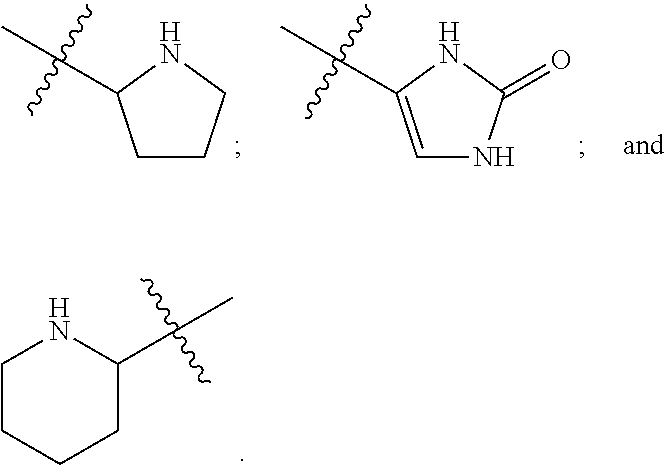
Human protein tyrosine phosphatase inhibitors an methods of use
ActiveUS20080108631A1Regulating angiogenesisAntibacterial agentsBiocidePhosphatase inhibitorTyrosine
The present disclosure relates to compounds effective as human protein tyrosine phosphatase beta (HPTP-β) inhibitors thereby regulating angiogenesis. The present disclosure further relates to compositions comprising said human protein tyrosine phosphatase beta (HPTP-β) inhibitors, and to methods for regulating angiogenesis.
Owner:EYEPOINT PHARMA INC
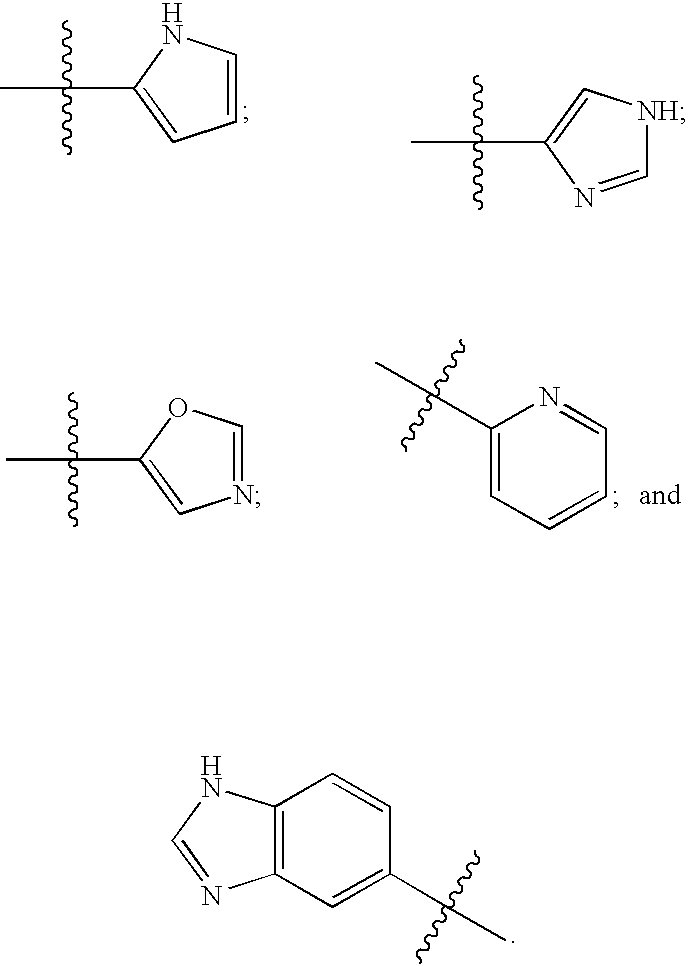
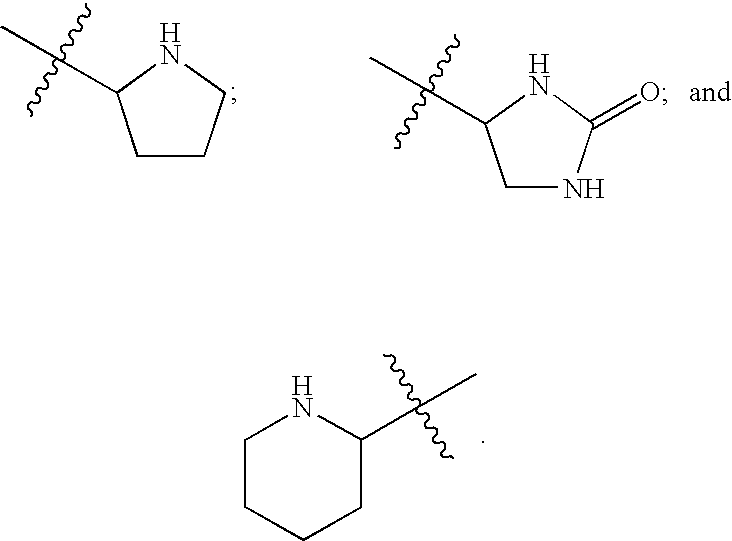
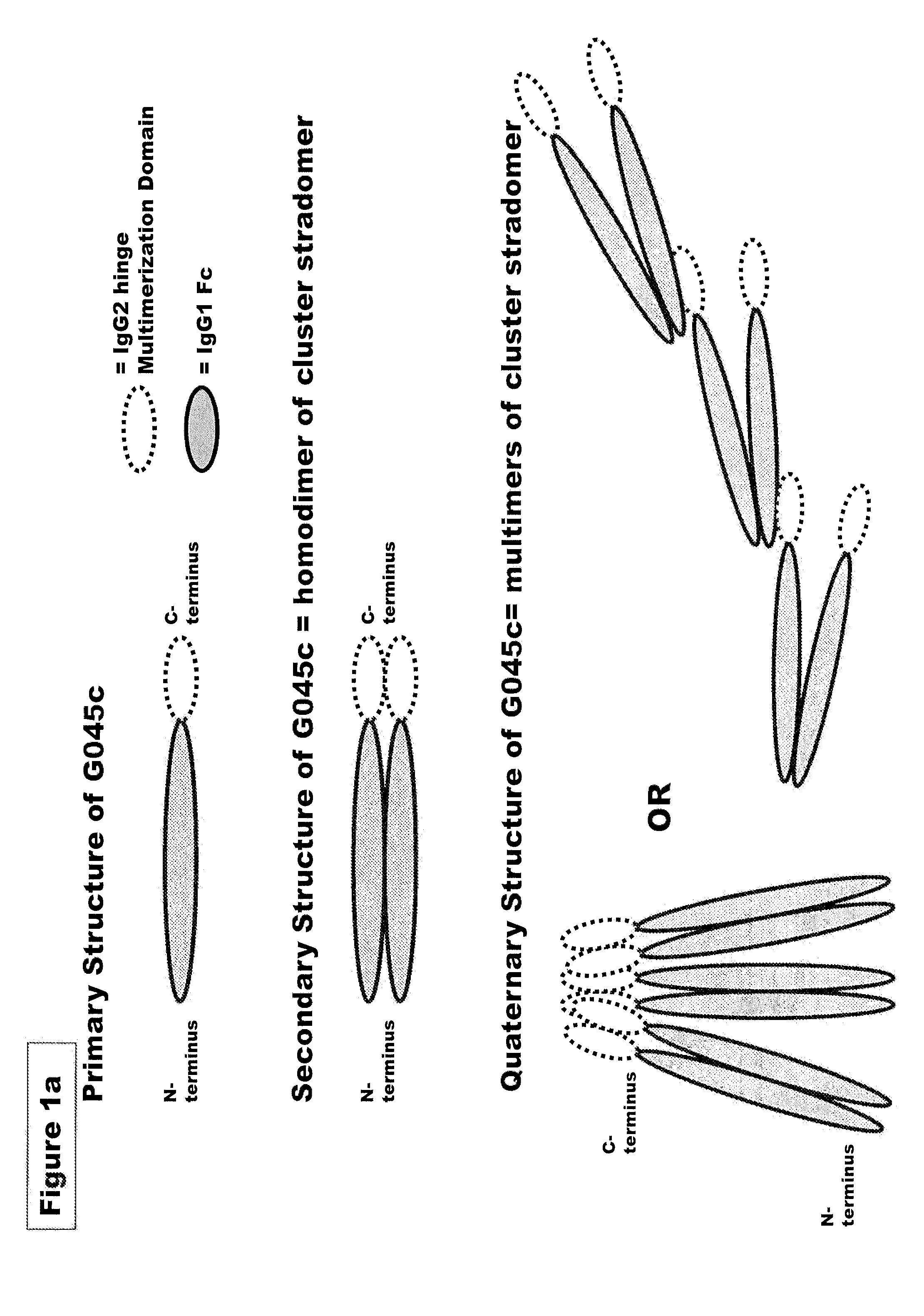
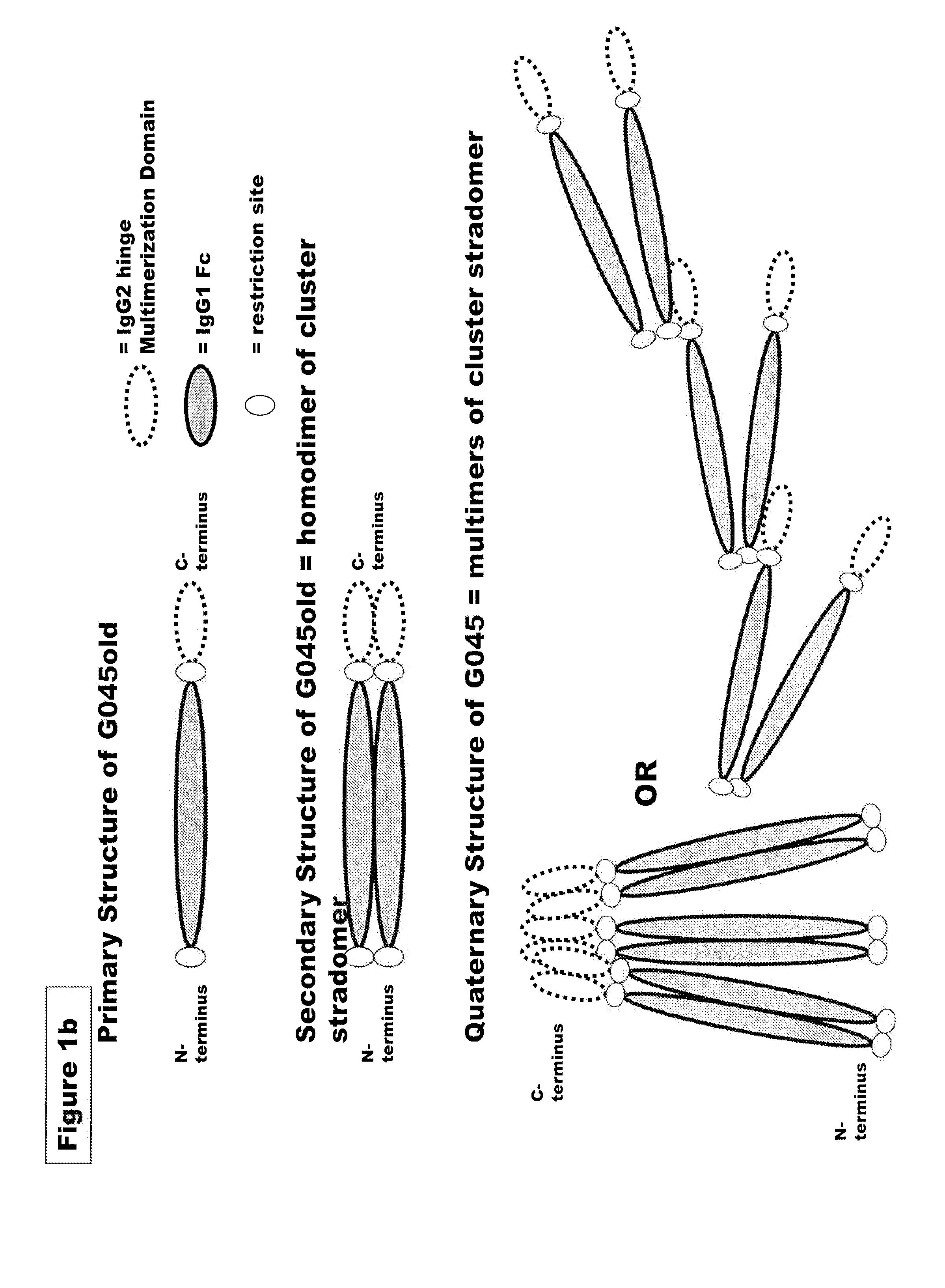
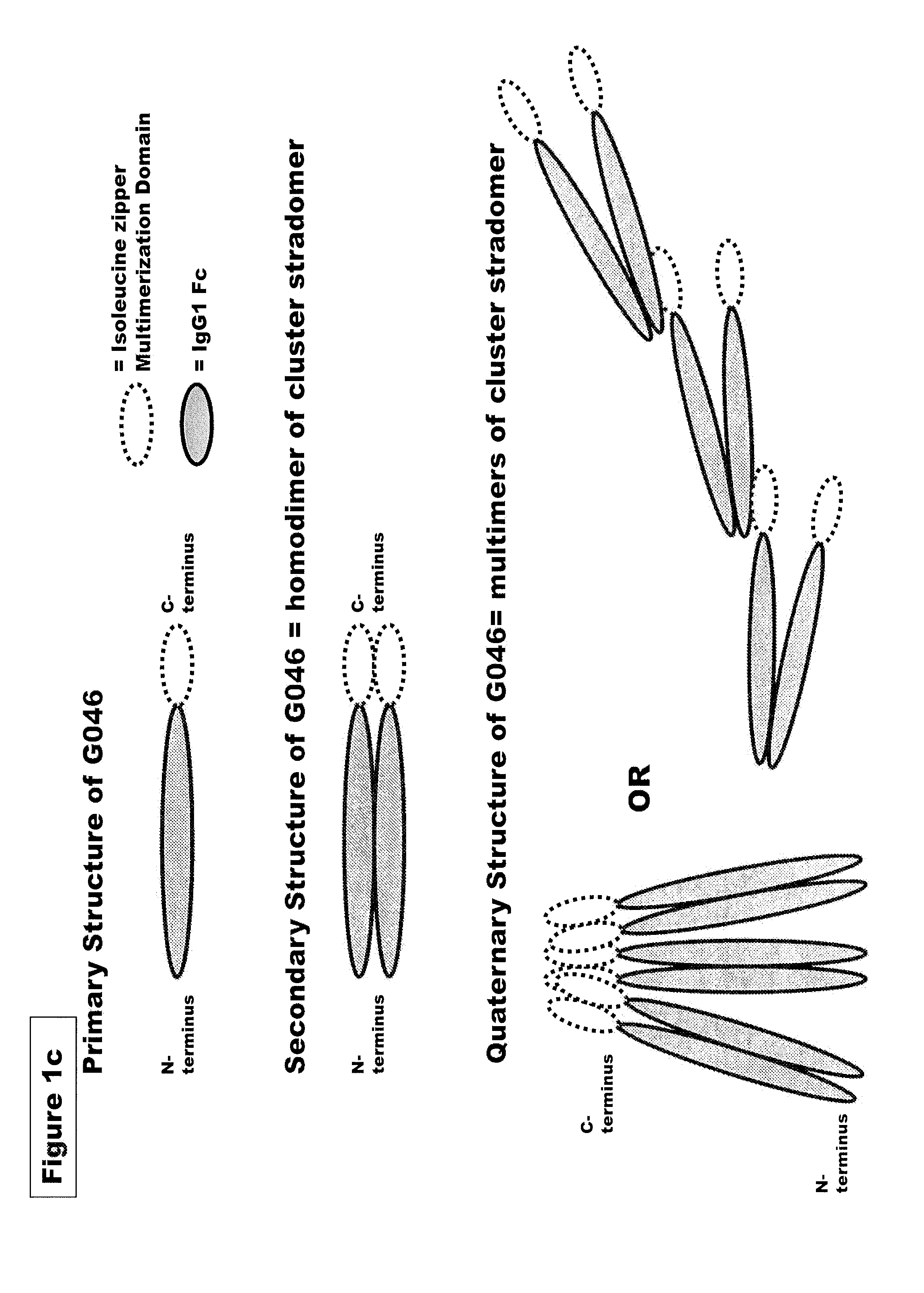
FUSION PROTEINS OF NATURAL HUMAN PROTEIN FRAGMENTS TO CREATE ORDERLY MULTIMERIZED IMMUNOGLOBULIN Fc COMPOSITIONS
InactiveUS20130156765A1High protein loadGood curative effectOrganic active ingredientsAntibody mimetics/scaffoldsDiseaseHuman proteins
The current invention involves a series of fully recombinant multimerized forms of immunoglobulin Fc which thereby present polyvalent immunoglobulin Fc to immune cell receptors. The fusion proteins exist as both homodimeric and highly ordered multimeric fractions, termed stradomers. In comparison to the homodimeric fraction, purified multimeric stradomers have higher affinity and avidity for Fc Rs with slower dissociation and are useful in the treatment and prevention of disease. The current invention demonstrates that directly linking IgG1 Fc regions to multimerization domains leads to enhanced multimerization and biological activity.
Owner:GLIKNIK
Human protein tyrosine phosphatase inhibitors and methods of use
ActiveUS7622593B2Antibacterial agentsOrganic active ingredientsPhosphatase inhibitorAngiogenesis growth factor
The present disclosure relates to compounds effective as human protein tyrosine phosphatase beta (HPTP-β) inhibitors thereby regulating angiogenesis. The present disclosure further relates to compositions comprising said human protein tyrosine phosphatase beta (HPTP-β) inhibitors, and to methods for regulating angiogenesis.
Owner:EYEPOINT PHARMA INC
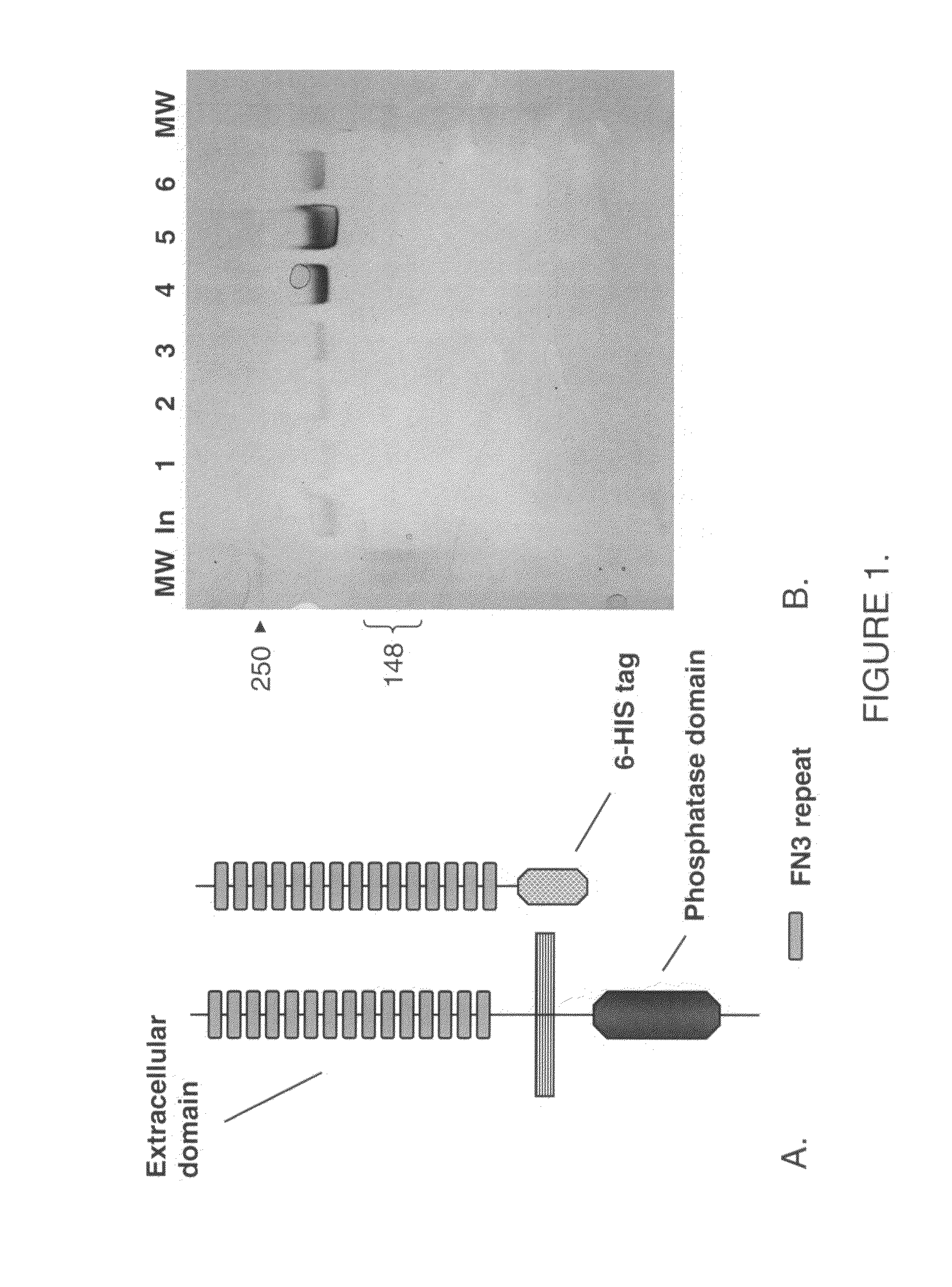
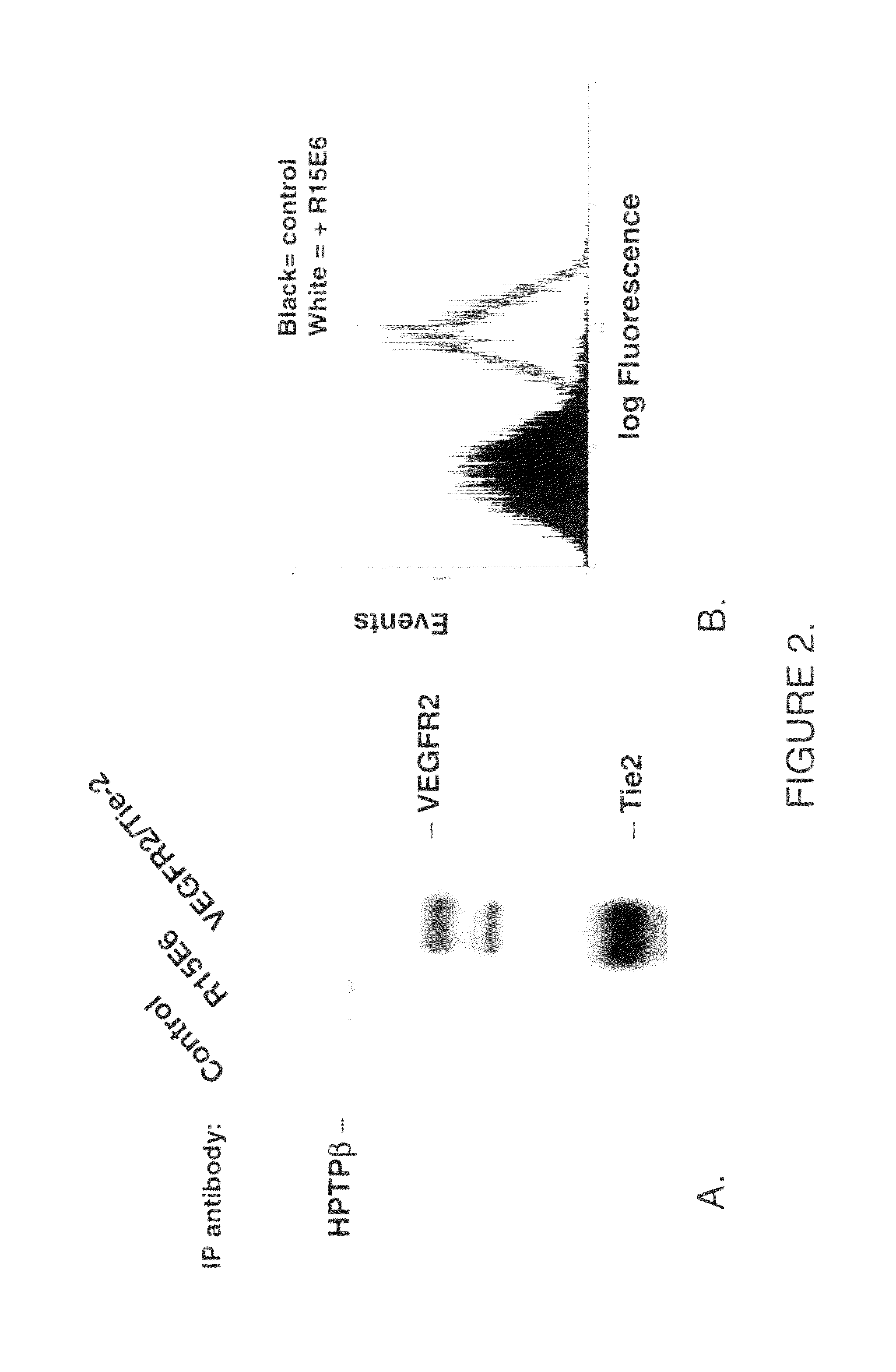
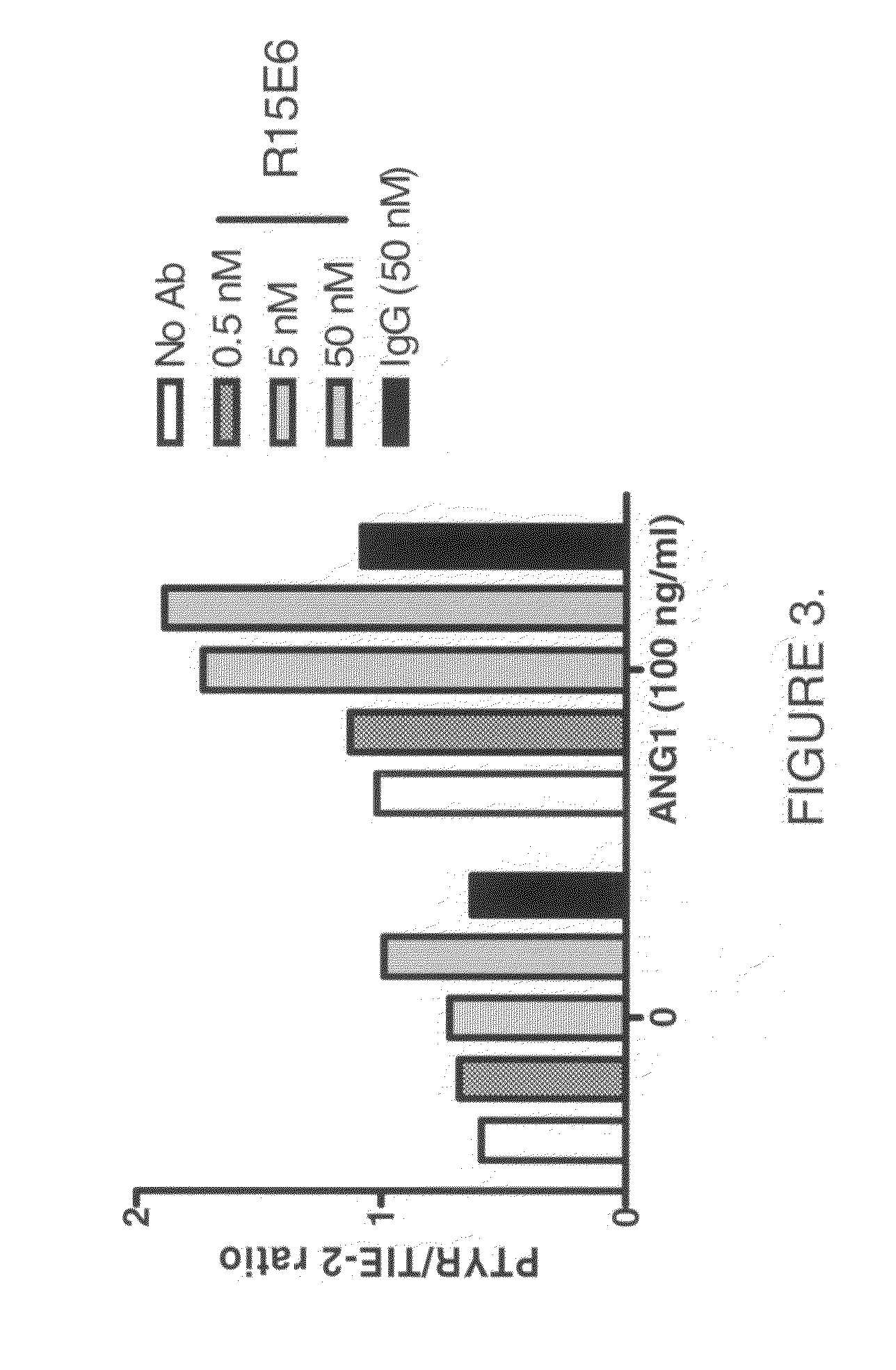
Antibodies that bind human protein tyrosine phosphatase beta (HPTPβ)
Owner:EYEPOINT PHARMA INC
Human protein tyrosine phosphatase inhibitors and methods of use
The present disclosure relates to compounds effective as human protein tyrosine phosphatase beta (HPTP-β) inhibitors thereby regulating angiogenesis. The present disclosure further relates to compositions comprising one or more human protein tyrosine phosphatase beta (HPTP-β) inhibitors, and to methods for regulating angiogenesis.
Owner:EYEPOINT PHARMA INC
Thrombomodulin Derivatives and Conjugates
InactiveUS20080051562A1Extended half-lifeImprove stabilityPeptide/protein ingredientsPeptide sourcesHuman proteinsIn vivo
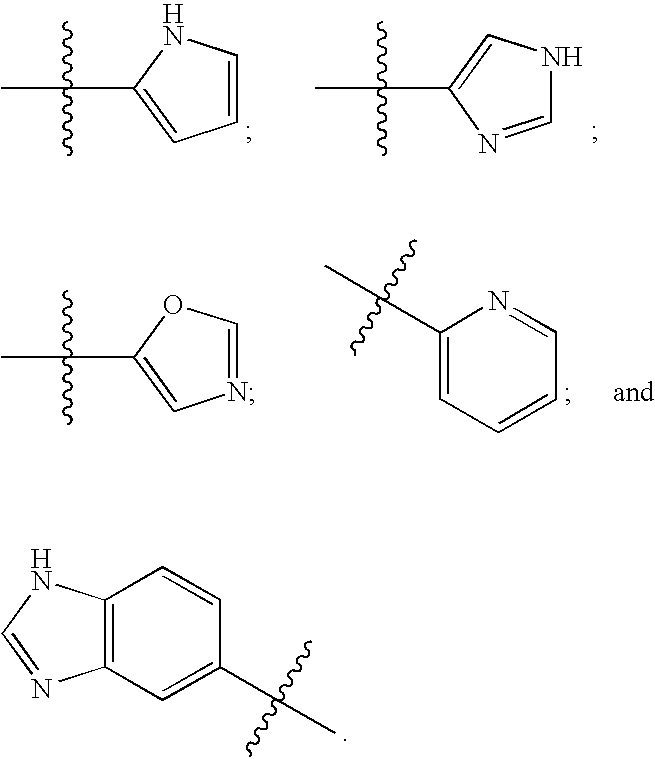
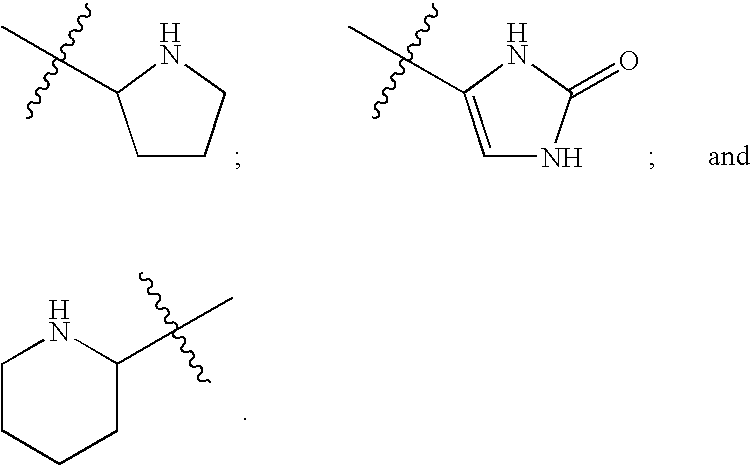
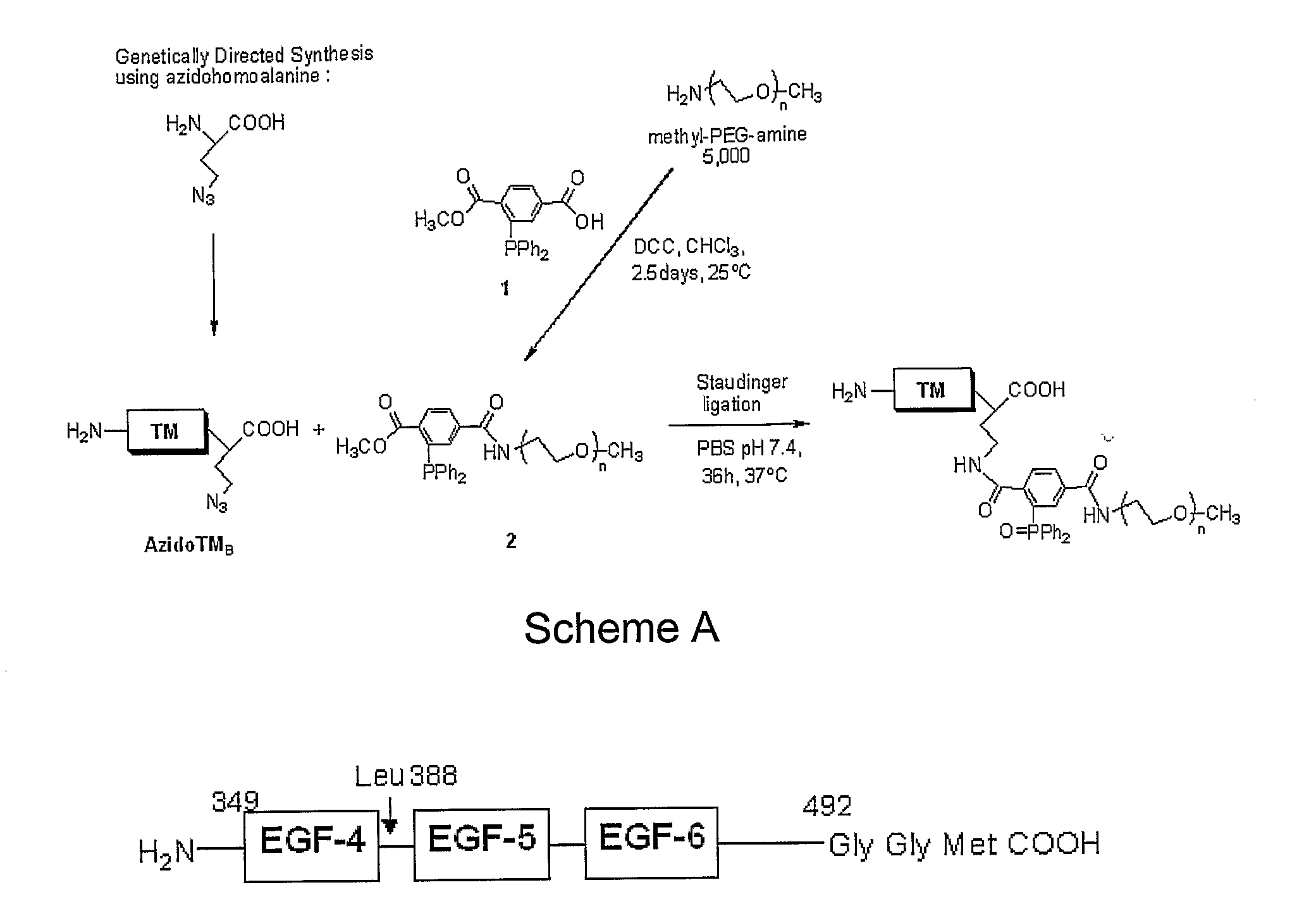
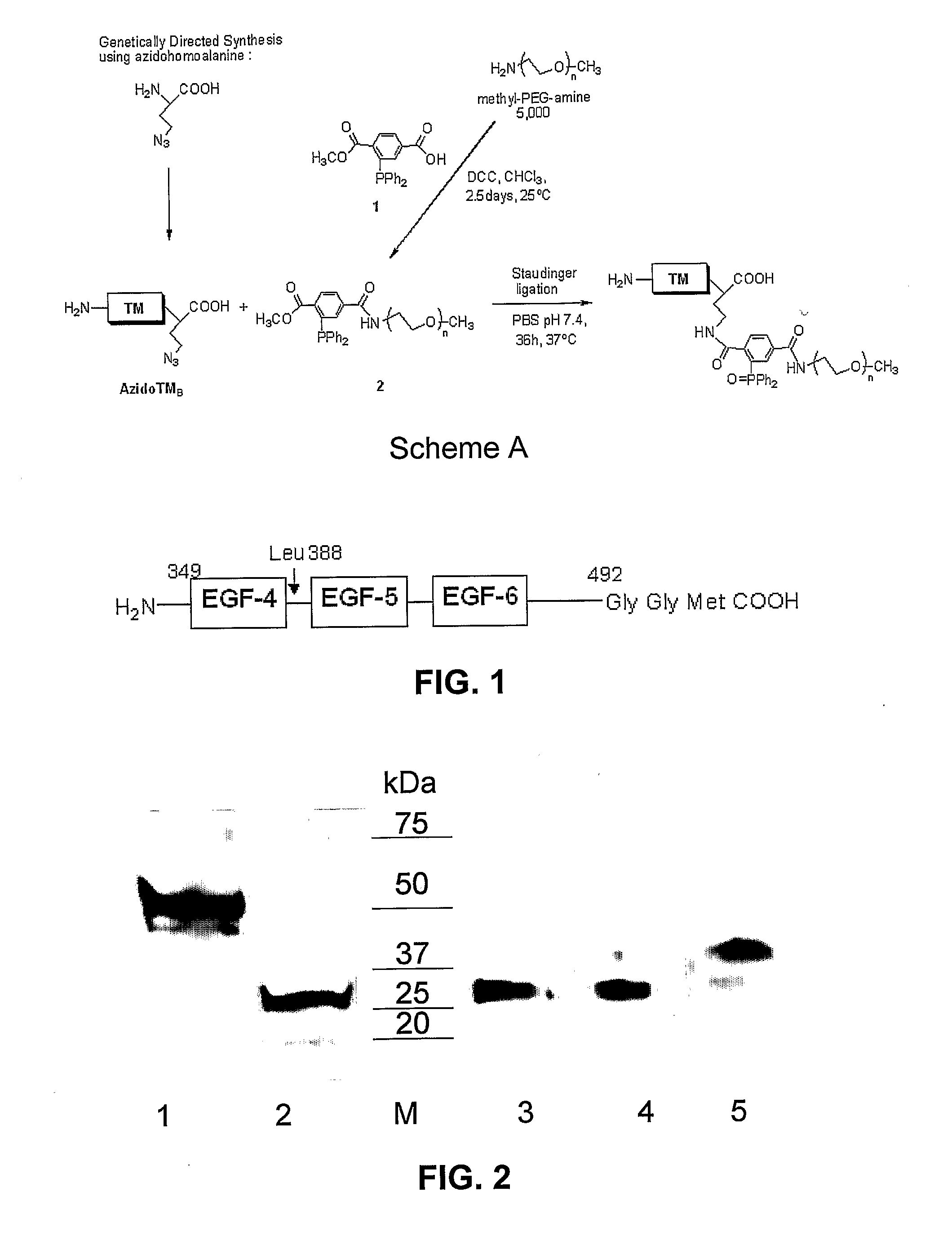
The transmembrane human protein thrombomodulin (TM), as a critical regulator of the protein C pathway, represents the major anticoagulant mechanism that is operative in both normal and injured blood vessels under physiologic conditions in vivo. Compositions and methods are disclosed relating to thrombomodulin derivatives and conjugates, including methods for site-specific pegylation and compositions of a truncated thrombomodulin derivative.
Owner:EMORY UNIVERSITY
Pharmaceutical proteins, human therapeutics, human serum albumin, insulin, native cholera toxic b submitted on transgenic plastids
InactiveUS20030204864A1Eliminate needLarge biomassBiocidePeptide/protein ingredientsEscherichia coliInsulin-like growth factor
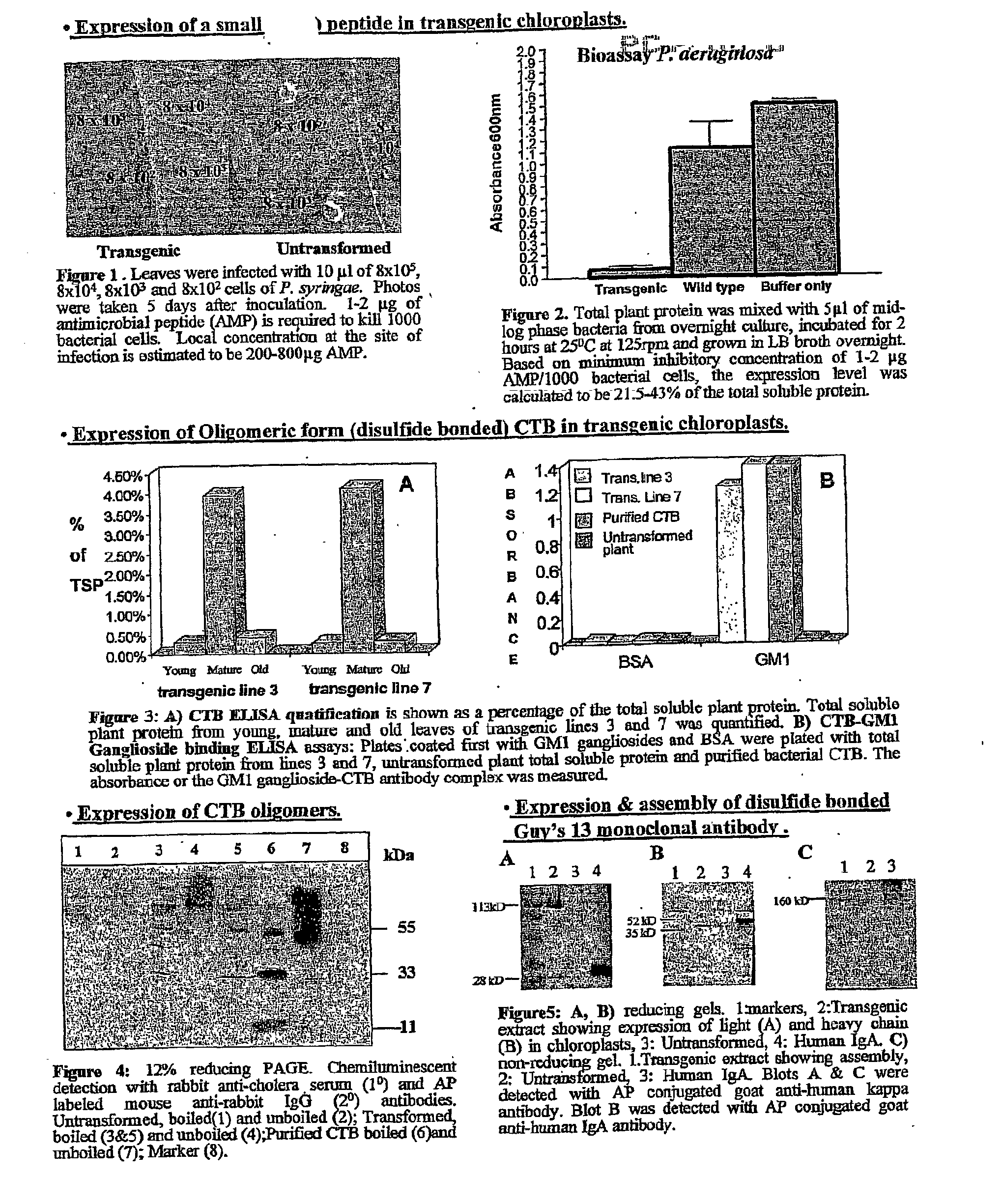
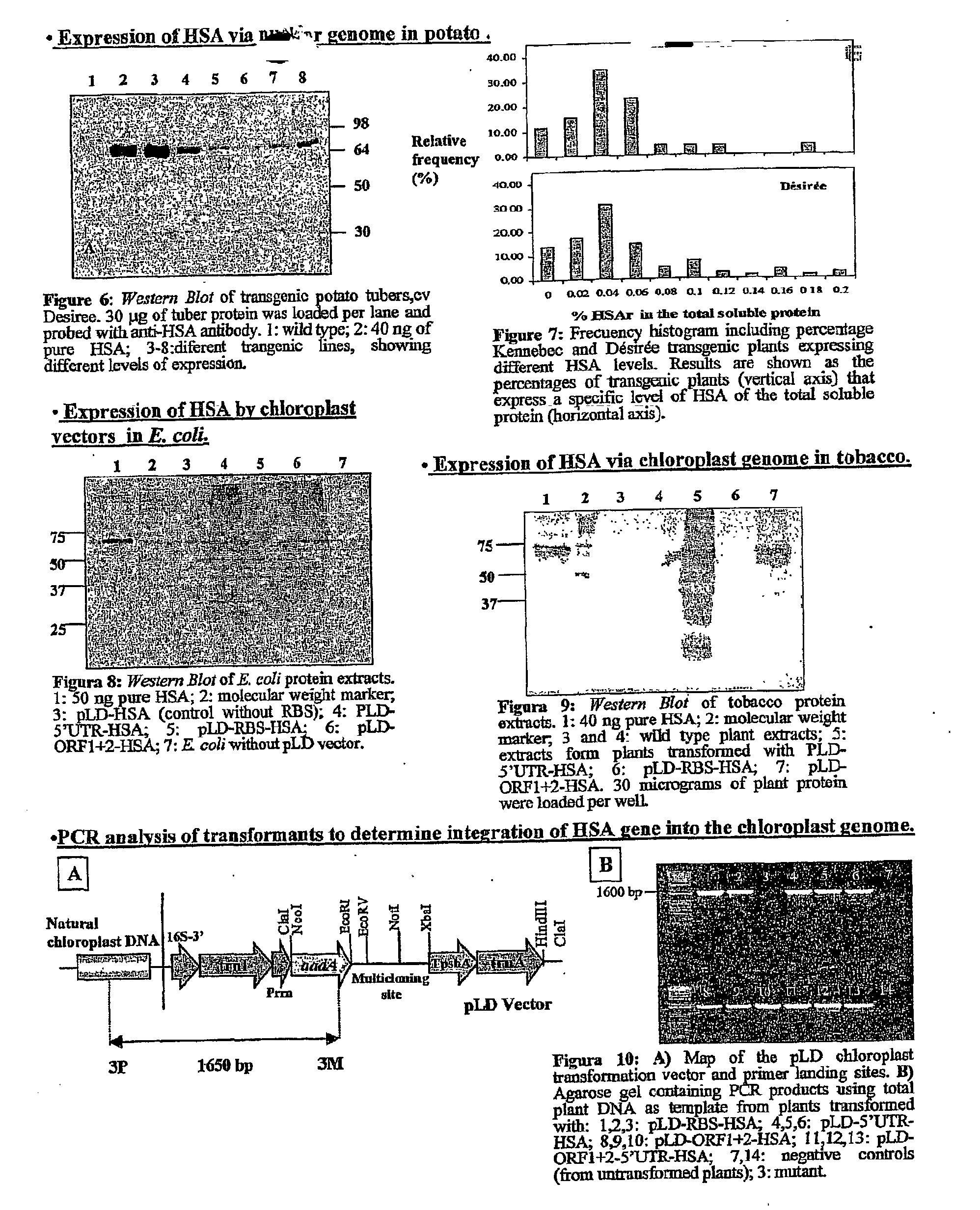
Transgenic chloroplast technology could provide a viable solution to the production of Insulin-like Growth Factor I (IGF-I), Human Serum Albumin (HSA), or interferons (IFN) because of hyper-expression capabilities, ability to fold and process eukaryotic proteins with disulfide bridges (thereby eliminating the need for expensive post-purification processing). Tobacco is an ideal choice because of its large biomass, ease of scale-up (million seeds per plant), genetic manipulation and impending need to explore alternate uses for this hazardous crop. Therefore, all three human proteins will be expressed as follows: a) Develop recombinant DNA vectors for enhanced expression via tobacco chloroplast genomes b) generate transgenic plants c) characterize transgenic expression of proteins or fusion proteins using molecular and biochemical methods d) large scale purification of therapeutic proteins from transgenic tobacco and comparison of current purification / processing methods in E. coli or yeast e) Characterization and comparison of therapeutic proteins (yield, purity, functionality) produced in yeast or E. coli with transgenic tobacco f) animal testing and pre-clinical trials for effectiveness of the therapeutic proteins. Mass production of affordable vaccines can be achieved by genetically engineering plants to produce recombinant proteins that are candidate vaccine antigens. The B subunits of Enteroxigenic E. coli (LTB) and cholera toxin of Vibrio cholerae (CTB) are examples of such antigens. When the native LTB gene was expressed via the tobacco nuclear genome, LTB accumulated at levels less than 0.01% of the total soluble leaf protein. Production of effective levels of LTB in plants, required extensive codon modification. Amplification of an unmodified CTB coding sequence in chloroplasts, up to 10,000 copies per cell, resulted in the accumulation of up to 4.1% of total soluble tobacco leaf protein as oligomers (about 410 fold higher expression levels than that of the unmodified LTB gene). PCR and Southern blot analyses confirmed stable integration of the CTB gene into the chloroplast genome. Western blot analysis showed that chloroplast synthesized CTB assembled into oligomers and was antigenically identical to purified native CTB. Also, GM1,-ganglioside binding assays confirmed that chloroplast synthesized CTB binds to the intestinal membrane receptor of cholera toxin, indicating correct folding and disulfide bond formation within the chloroplast. In contrast to stunted nuclear transgenic plants, chloroplast transgenic plants were morphologically indistinguishable from untransformed plants, when CTB was constitutively expressed. The introduced gene was stably inherited in the subsequent generation as confirmed by PCR and Southern blot analyses. Incrased production of an efficient transmucosal carrier molecule and delivery system, like CTB, in transgenic chloroplasts makes plant based oral vaccines and fusion proteins with CTB needing oral administration a much more practical approach.
Owner:AUBURN UNIV +1
Peptides derived from STEAP1
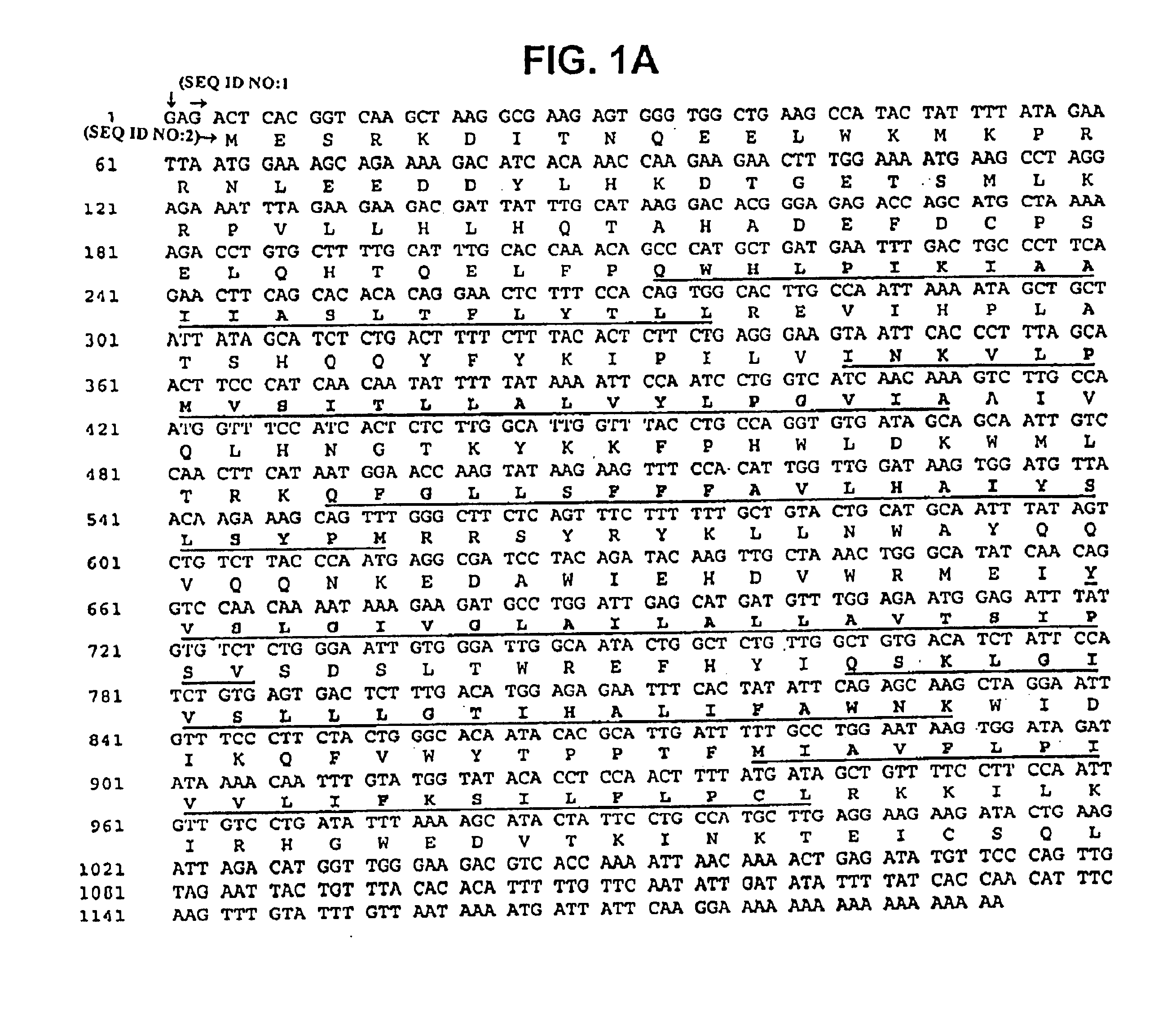
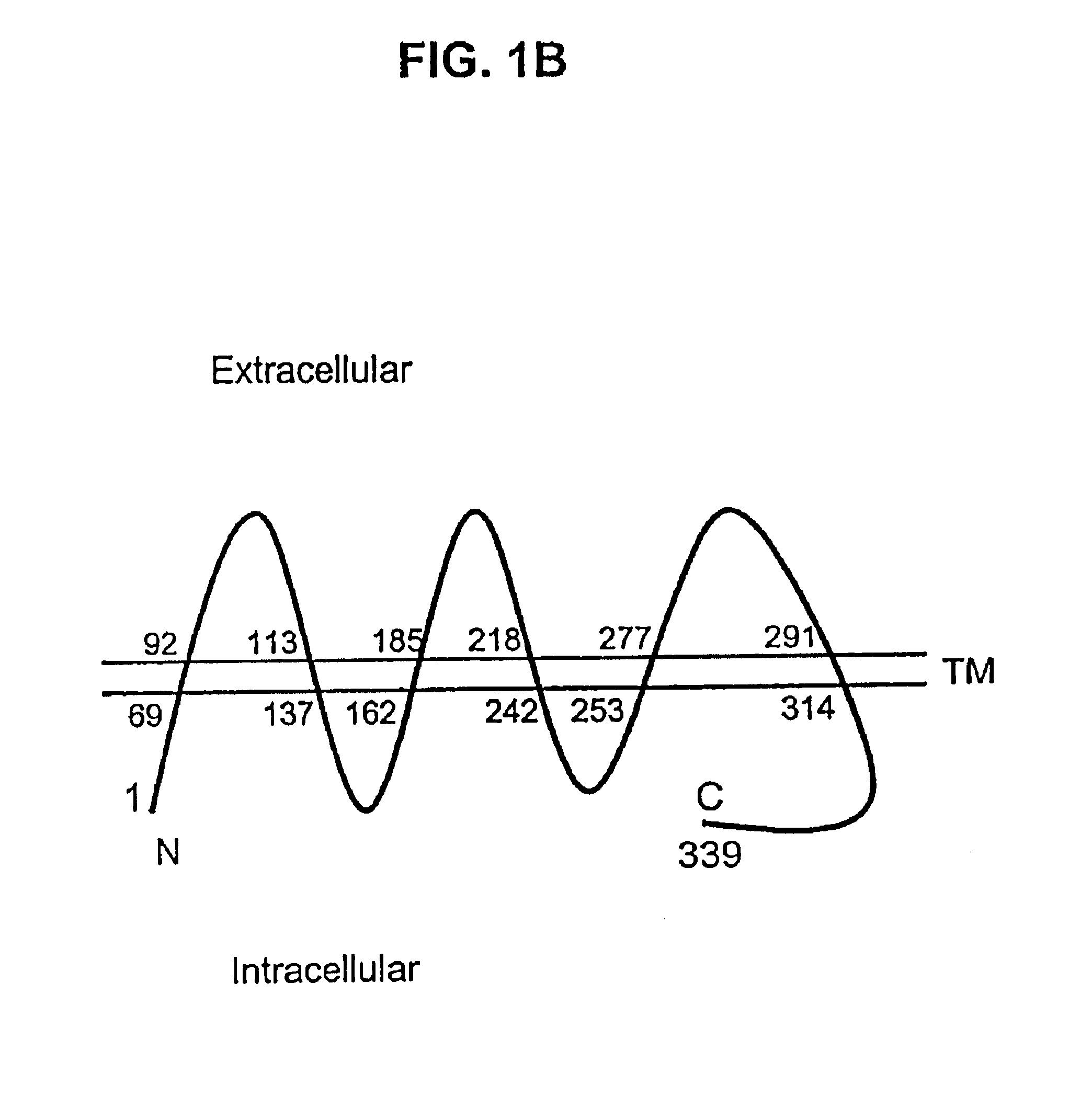
Described is a novel family of cell surface serpentine transmembrane antigens. Two of the proteins in this family are exclusively or predominantly expressed in the prostate, as well as in prostate cancer, and thus members of this family have been termed “STEAP” (serpentine transmembrane antigens of the prostate). Four particular human STEAPs are described and characterized herein. The human STEAPs exhibit a high degree of structural conservation among them but show no significant structural homology to any known human proteins. The prototype member of the STEAP family, STEAP-1, appears to be a type IIIa membrane protein expressed predominantly in prostate cells in normal human tissues. Structurally, STEAP-1 is a 339 amino acid protein characterized by a molecular topology of six transmembrane domains and intracellular N- and C-termini, suggesting that it folds in a “serpentine” manner into three extracellular and two intracellular loops. STEAP-1 protein expression is maintained at high levels across various stages of prostate cancer. Moreover, STEAP-1 is highly over-expressed in certain other human cancers.
Owner:AGENSYS
Expression of mammalian proteins in Pseudomonas fluorescens
The invention is a process for improved production of a recombinant mammalian protein by expression in a Pseudomonad, particularly in a Pseudomonas fluorescens organism. The process improves production of mammalian proteins, particularly human or human-derived proteins, over known expression systems such as E. coli in comparable circumstances Processes for improved production of isolated mammalian, particularly human, proteins are provided.
Owner:PELICAN TECH HLDG INC
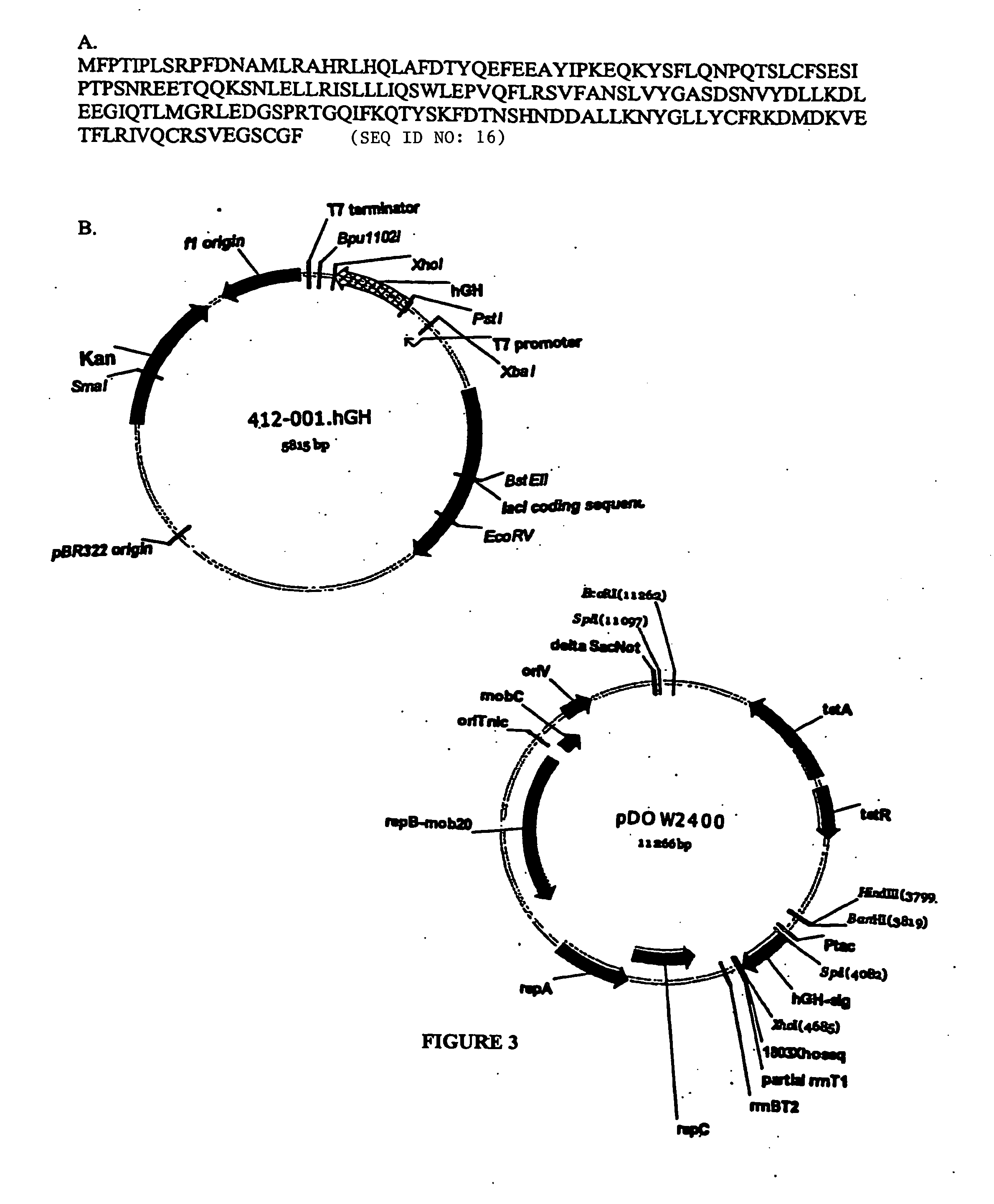
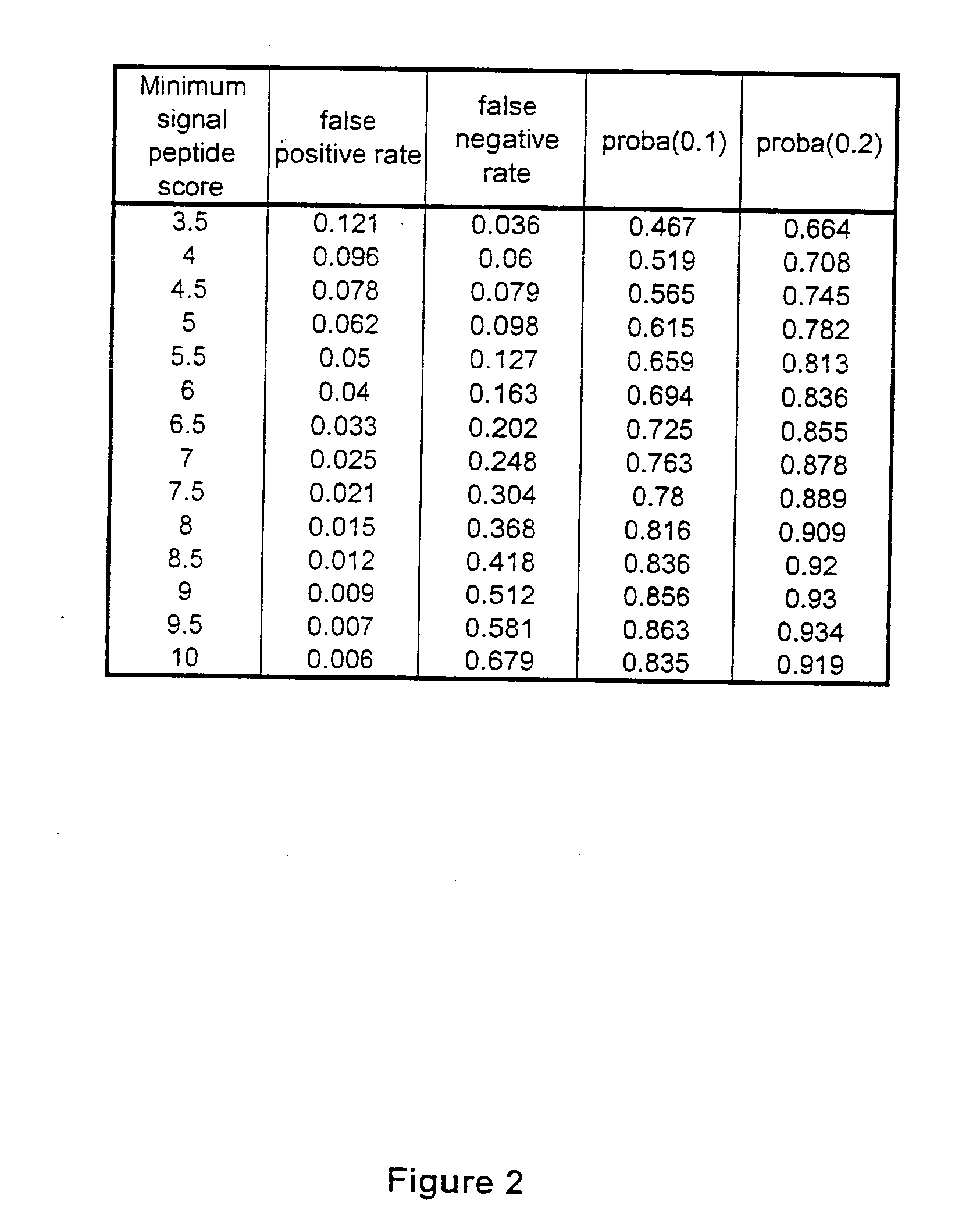
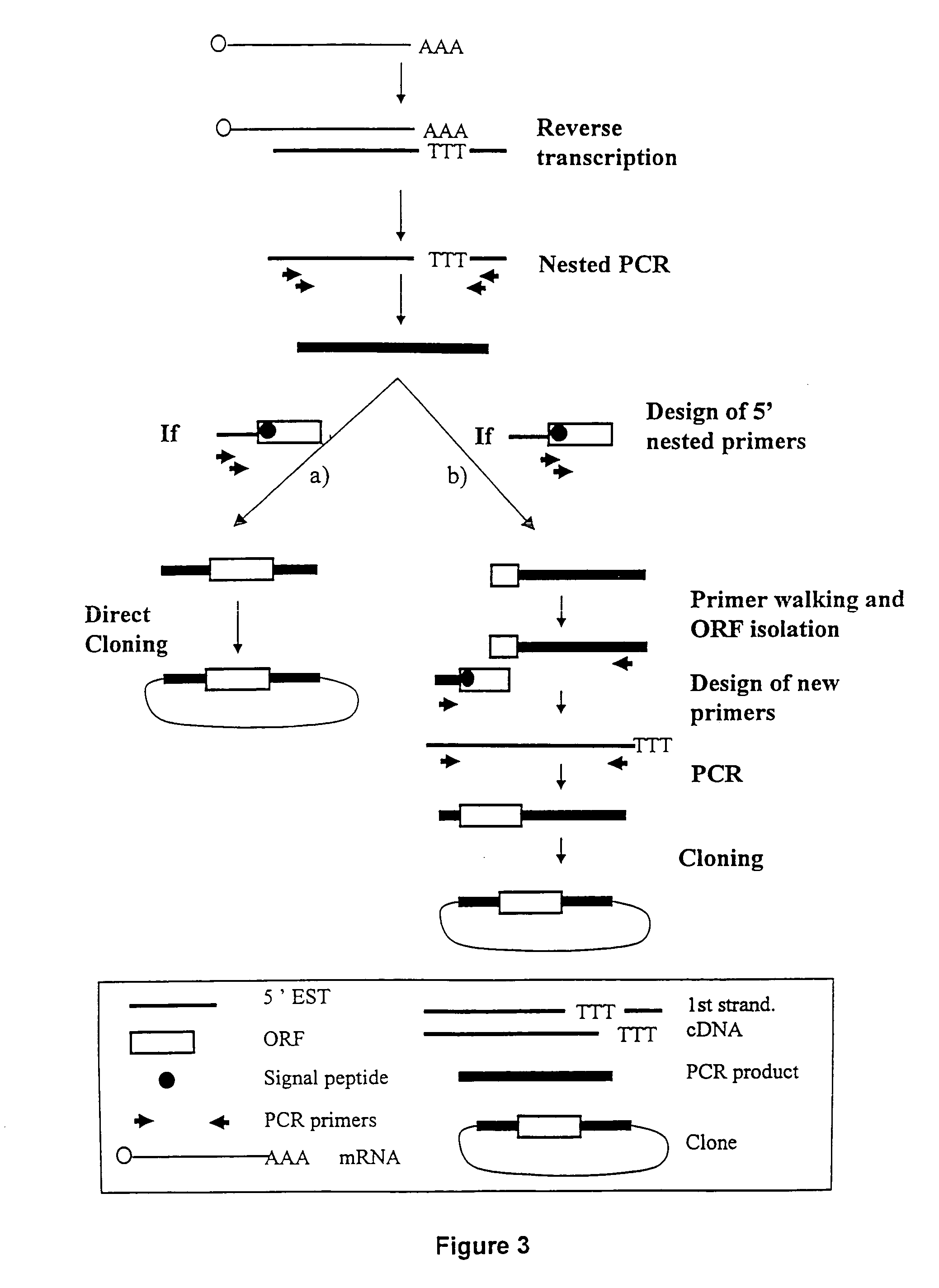
ESTs and encoded human proteins
The sequences of 5′ ESTs and consensus contigated 5′ESTs derived from mRNAs encoding secreted proteins are disclosed. The 5′ ESTs and consensus contigated 5′ESTs may be to obtain cDNAs and genomic DNAs corresponding to the 5′ ESTs and consensus contigated 5′ESTs. The 5′ ESTs and consensus contigated 5′ESTs may also be used in diagnostic, forensic, gene therapy, and chromosome mapping procedures. Upstream regulatory sequences may also be obtained using the 5′ ESTs and consensus contigated 5′ESTs. The 5′ ESTs and consensus contigated 5′ESTs may also be used to design expression vectors and secretion vectors.
Owner:SERONO GENETICS INST SA
Surface Enhanced Raman Scattering and Multiplexed Diagnostic Assays
InactiveUS20100055721A1Bioreactor/fermenter combinationsBiological substance pretreatmentsMicroorganismAssay
Multiplexed lateral flow assays, related methods, and devices are disclosed which are capable of simultaneously detecting multiple analytes. The assays are preferably immunoassays and can be multiplexed spatially, spectrally, and both spatially and spectrally. Multiplexed assays are disclosed employing quantum dots for applications including the detection of human proteins and the monitoring of microorganisms relevant to water contamination. The multiplexed assays can employ one or more species of Surface Enhanced Raman Scattering nanoparticles, with one or more species having a unique Raman shift spectrum. The invention is widely adaptable to a variety of analytes such as biowarfare agents, human clinical markers, and other substances.
Owner:CALIFORNIA INST OF TECH
Human protein tyrosine phosphatase inhibitors and method of use
ActiveUS8329916B2Antibacterial agentsOrganic active ingredientsPhosphatase inhibitorAngiogenesis growth factor
The present disclosure relates to compounds effective as human protein tyrosine phosphatase beta (HPTP-β) inhibitors thereby regulating angiogenesis. The present disclosure further relates to compositions comprising said human protein tyrosine phosphatase beta (HPTP-β) inhibitors, and to methods for regulating angiogenesis.
Owner:EYEPOINT PHARMA INC
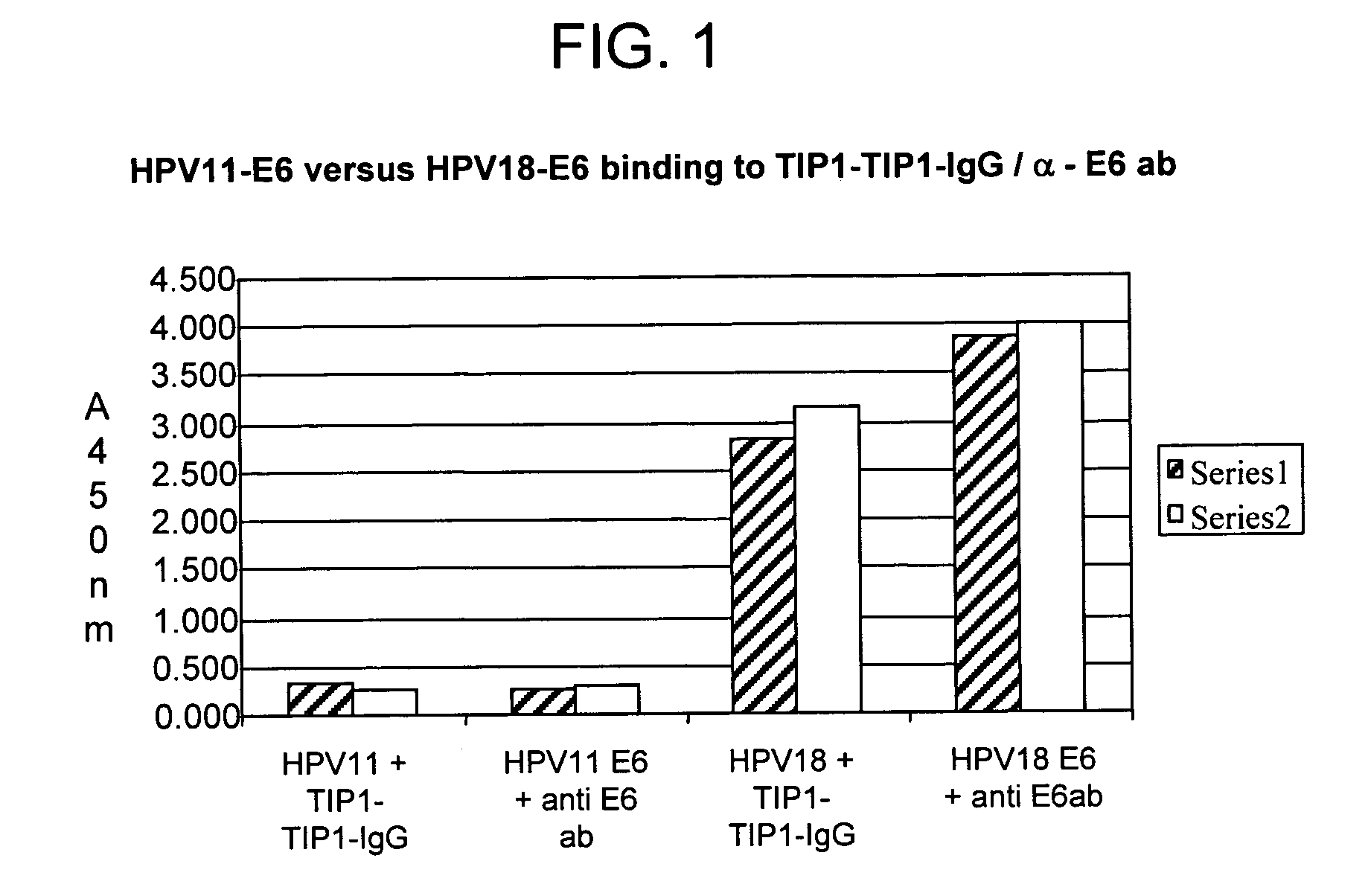
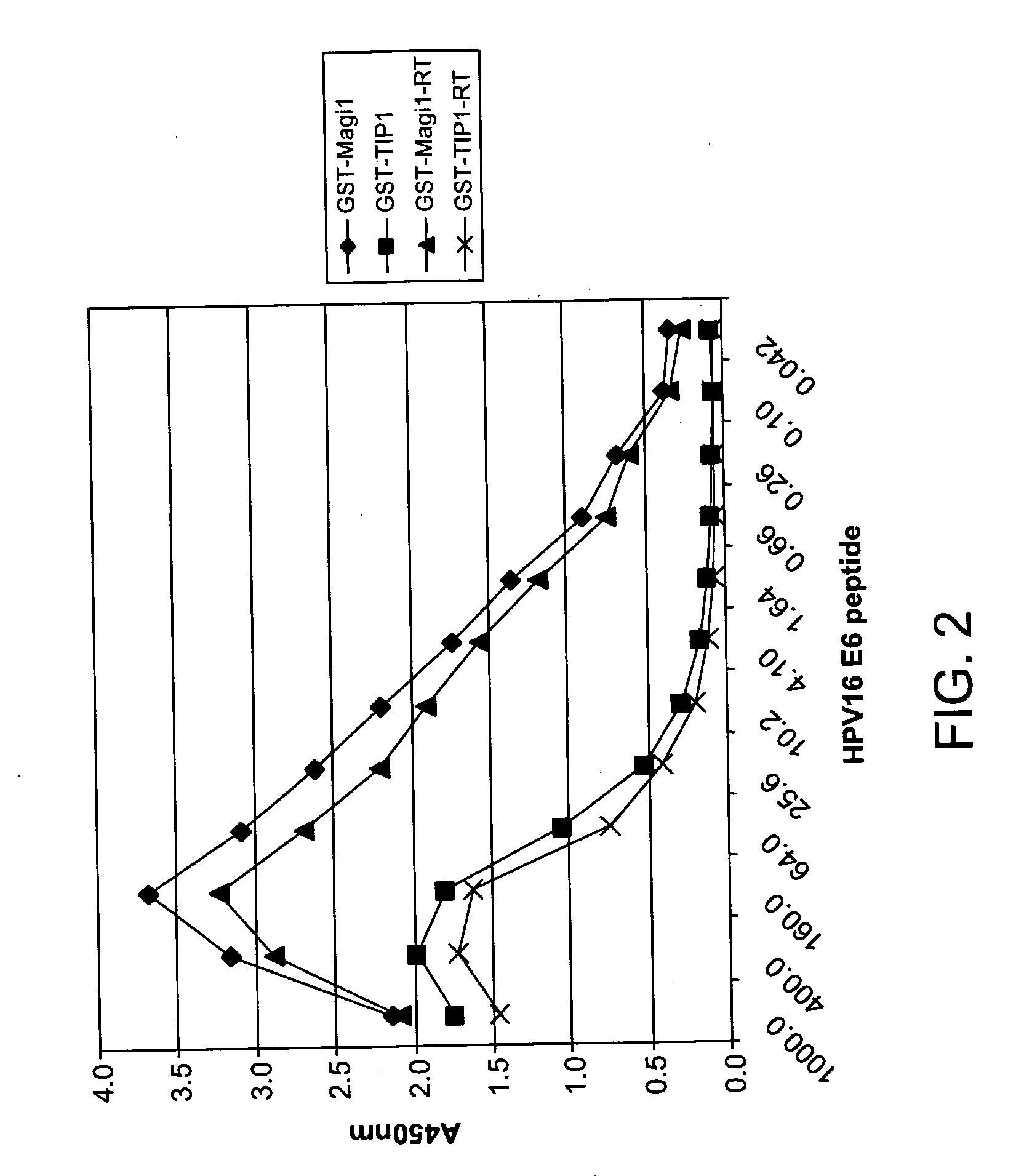
Methods of diagnosing cervical cancer
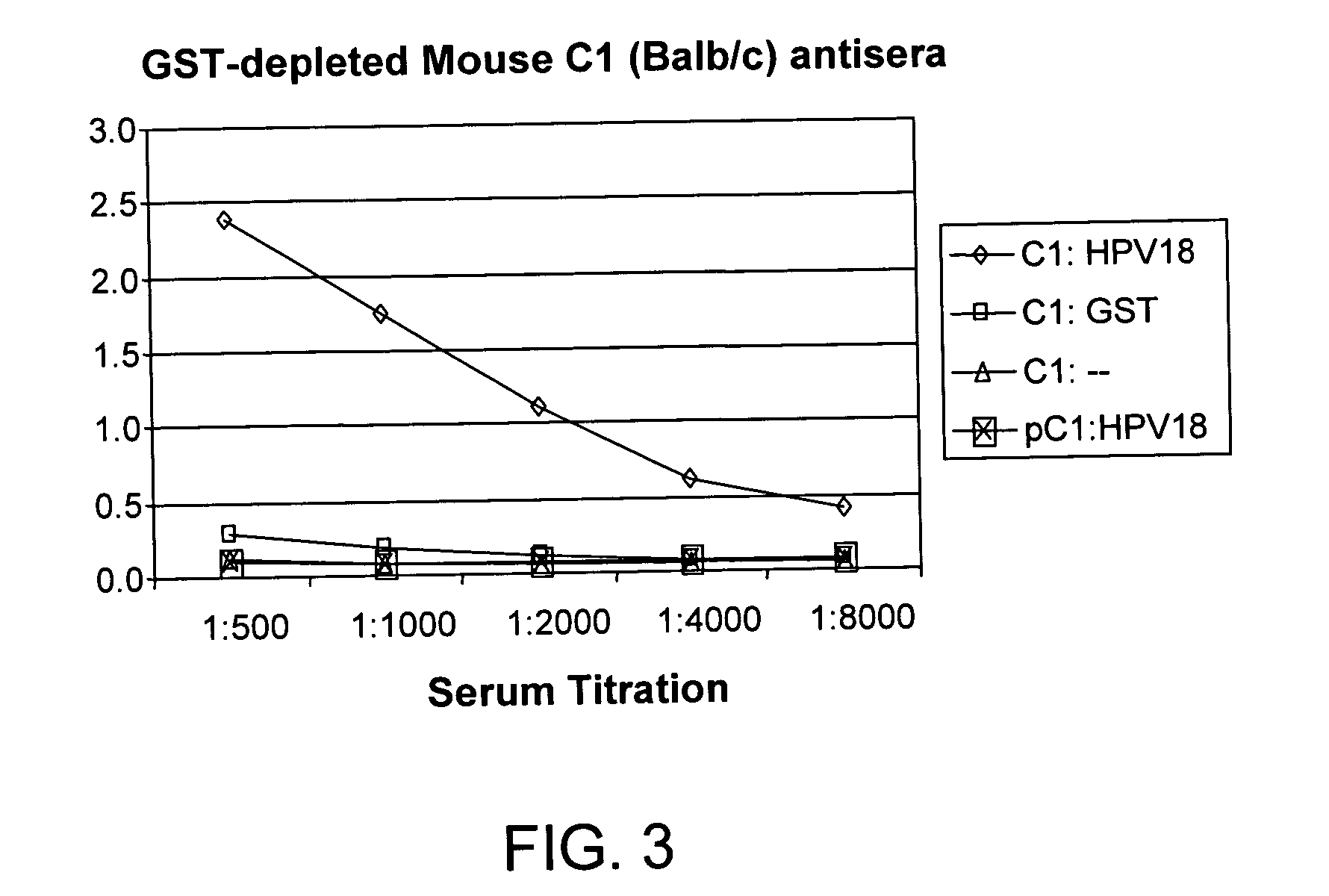
The invention provides reagents and methods for detecting pathogen infections in human samples. This detection utilizes specific proteins to detect the presence of pathogen proteins or abnormal expression of human proteins resulting from pathogen infections. Specific methods, compositions and kits are disclosed herein for the detection of oncogenic Human papillomavirus E6 proteins in clinical samples.
Owner:ARBOR VITA CORP
Human protein tyrosine phosphatase inhibitors and methods of use
The present disclosure relates to compounds effective as human protein tyrosine phosphatase beta (HPTP-β) inhibitors thereby regulating angiogenesis. The present disclosure further relates to compositions comprising said human protein tyrosine phosphatase beta (HPTP-β) inhibitors, and to methods for regulating angiogenesis.
Owner:EYEPOINT PHARMA INC
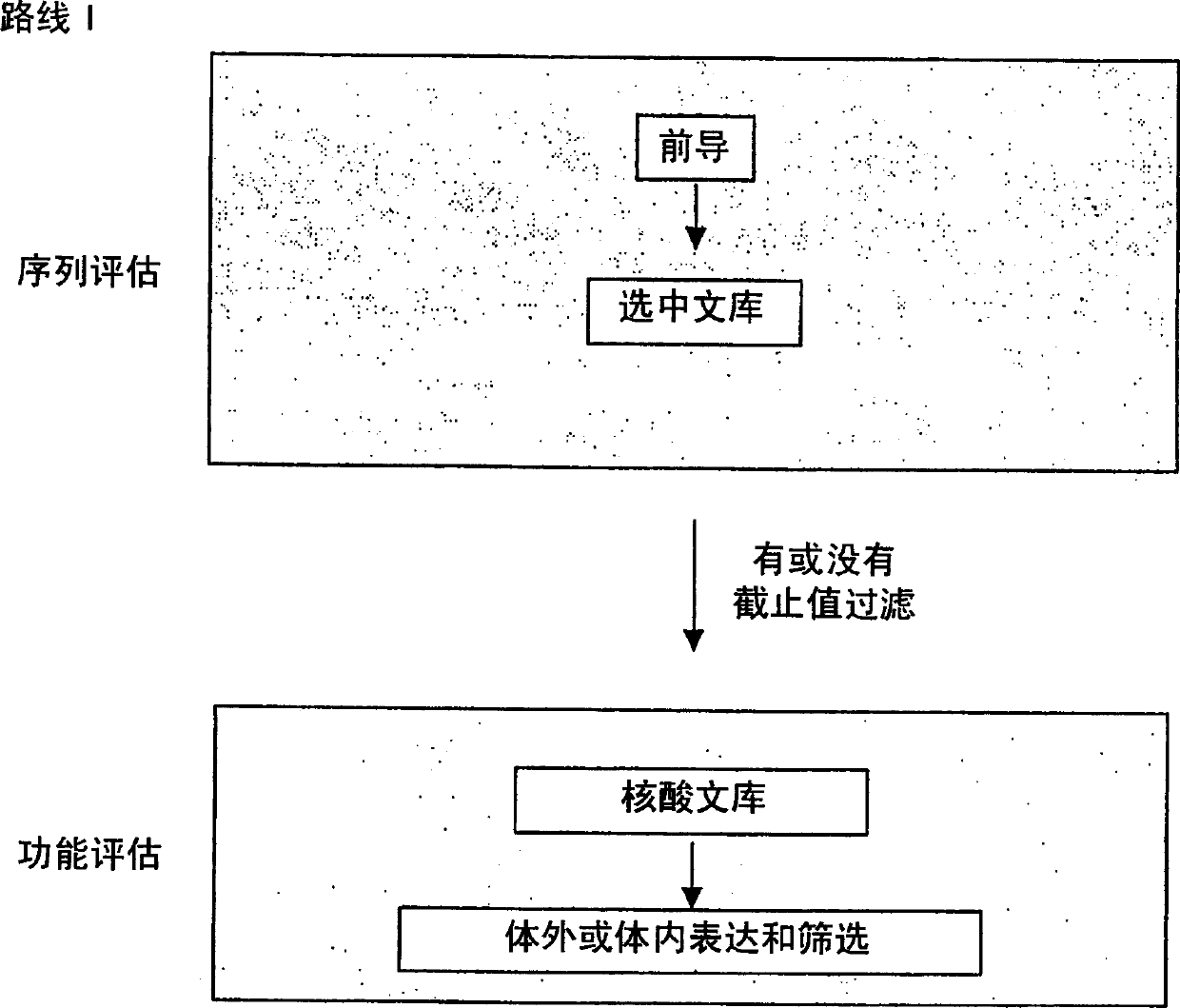
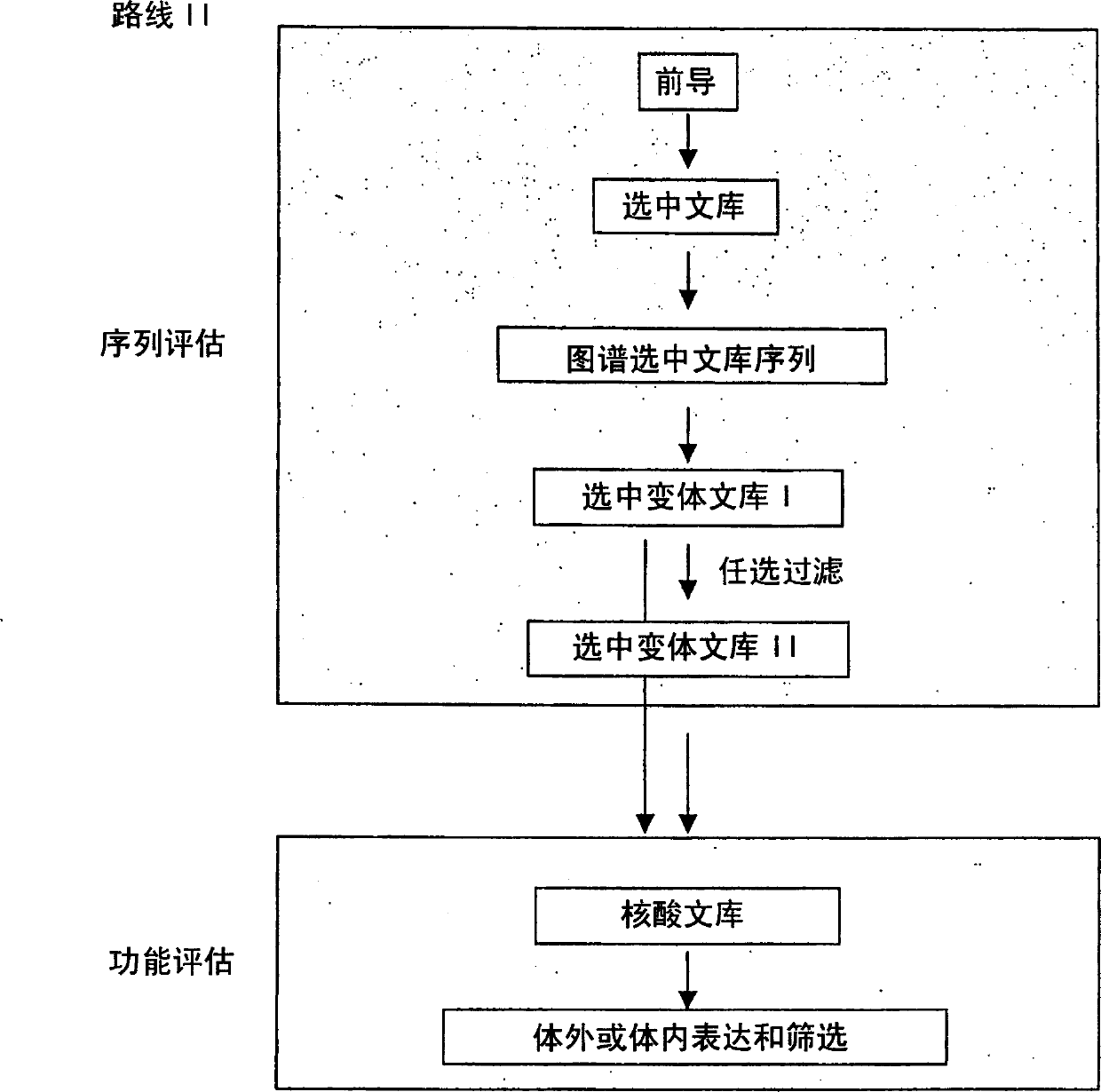
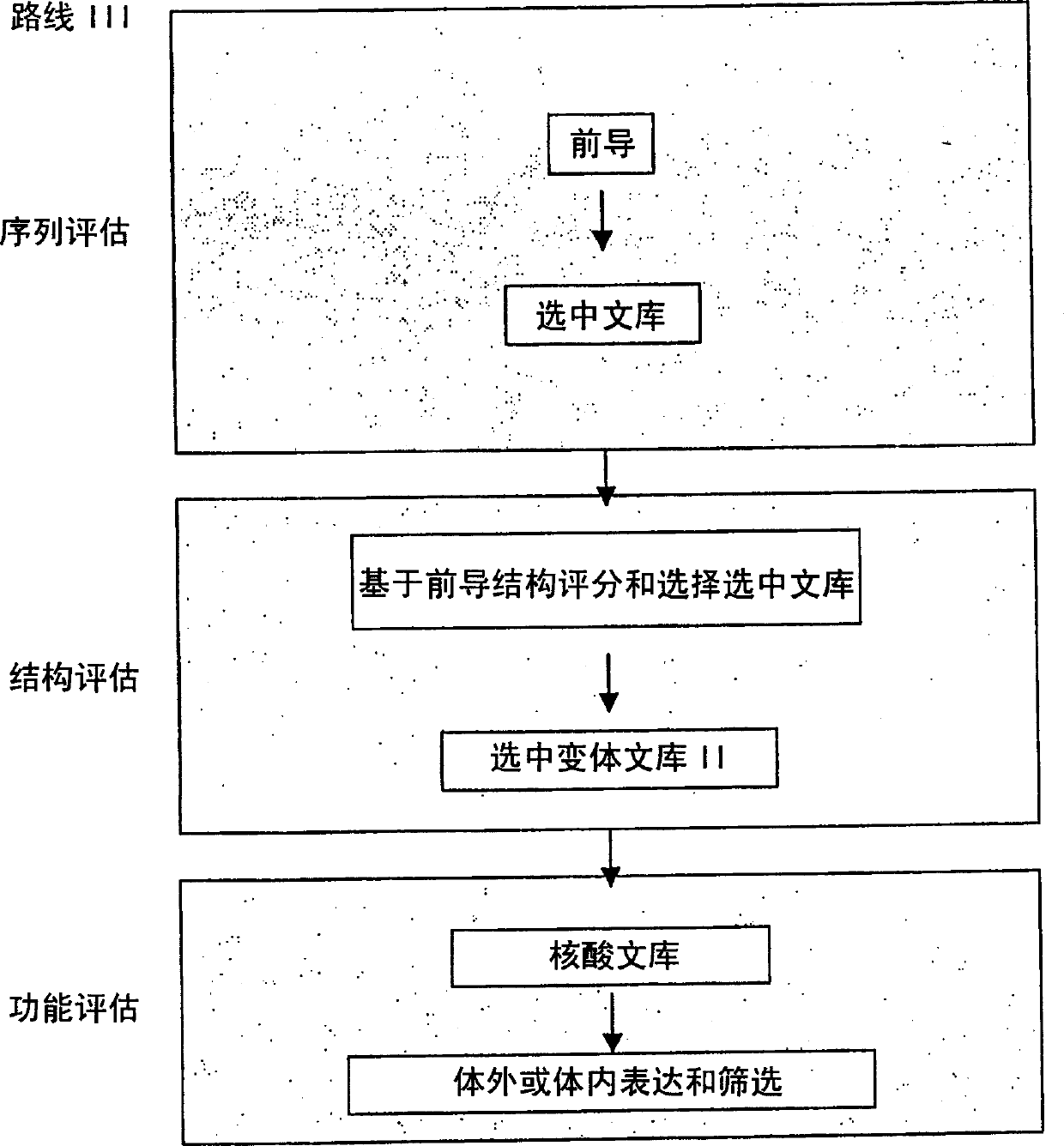
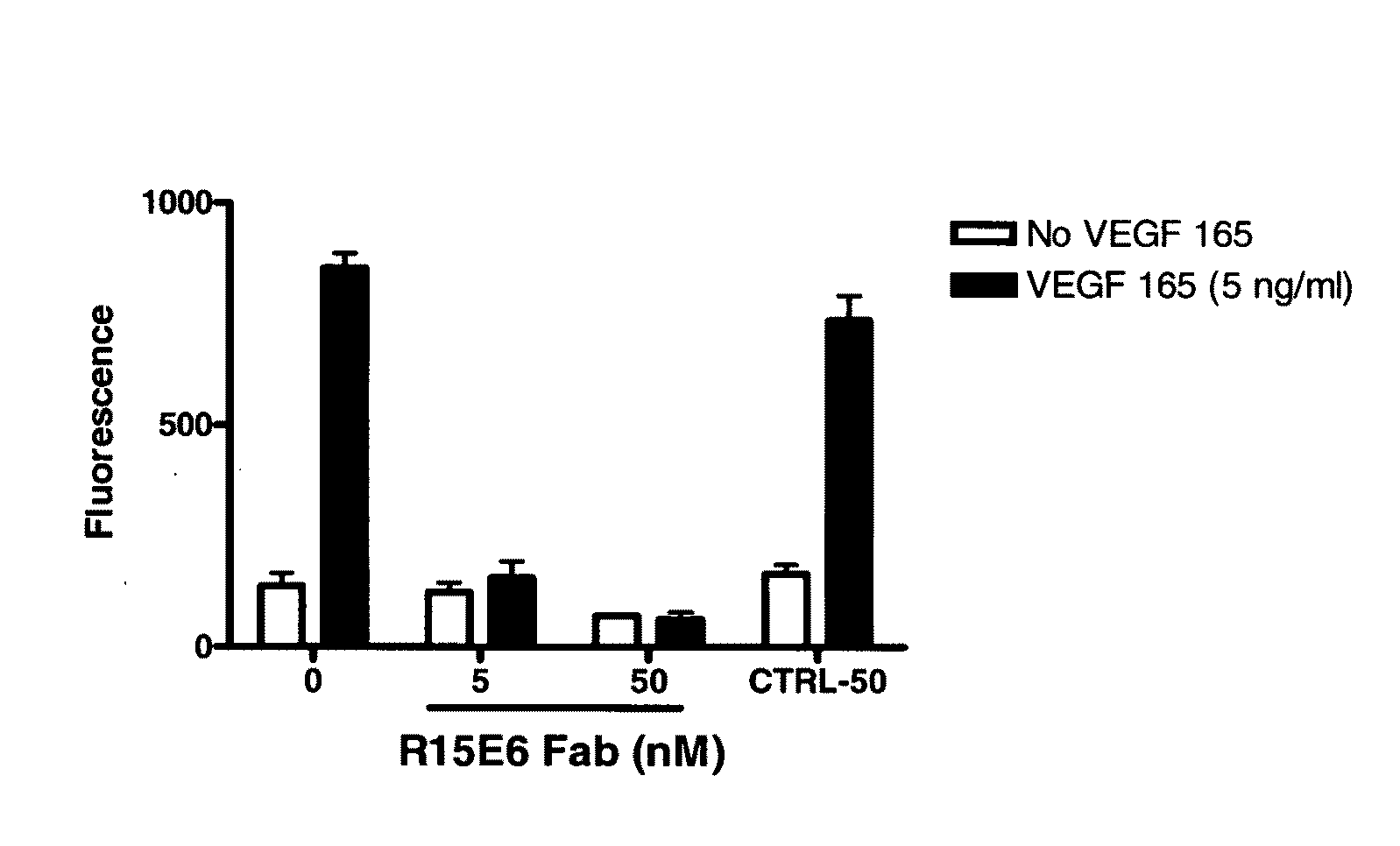
Generation and selection of protein library in silico
The present invention provides methods for efficiently generating and screening protein libraries for optimal proteins with desired biological functions, such as improved binding affinity for biologically and / or therapeutically important target molecules. The method is performed in silico in a high-throughput fashion by mining the ever-expanding database of protein sequences in all living things, especially humans. In one embodiment, a method of constructing a designer protein library comprises the steps of: providing an amino acid sequence derived from a lead protein, referred to as a lead sequence; comparing the lead sequence with a plurality of test protein sequences; Select at least two peptide fragments having at least 15% sequence identity with the leader sequence from each test protein sequence, and the selected peptide fragments form a selection library; a library of designed proteins is formed by replacing the leader sequence with the selection library. Libraries of designed proteins can be expressed in vitro or in vivo to generate libraries of recombinant proteins that can be screened for new or improved functions relative to lead proteins, such as antibodies against a therapeutically important target.
Owner:埃博马可西斯公司
Antibodies that bind human protein tyrosine phosphatase beta (HPTPbeta) and uses thereof
Antibodies and antigen binding fragments thereof that bind to human protein tyrosine phosphatase beta (HPTPβ), and uses thereof.
Owner:EYEPOINT PHARMA INC
Human protein tyrosine phosphatase inhibitors and methods of use
ActiveUS20130023543A1Inhibit angiogenesisOrganic active ingredientsBiocidePhosphatase inhibitorAngiogenesis growth factor
The present disclosure relates to compounds effective as human protein tyrosine phosphatase beta (HPTP-β) inhibitors thereby regulating angiogenesis. The present disclosure further relates to compositions comprising said human protein tyrosine phosphatase beta (HPTP-β) inhibitors, and to methods for regulating angiogenesis.
Owner:EYEPOINT PHARMA INC
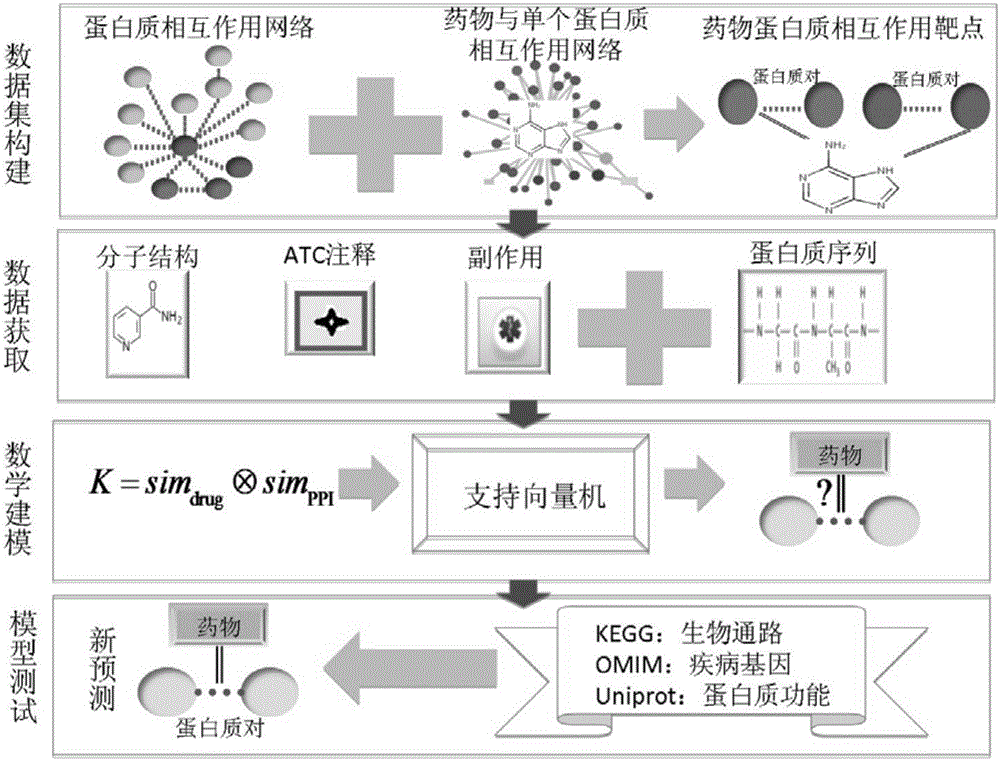
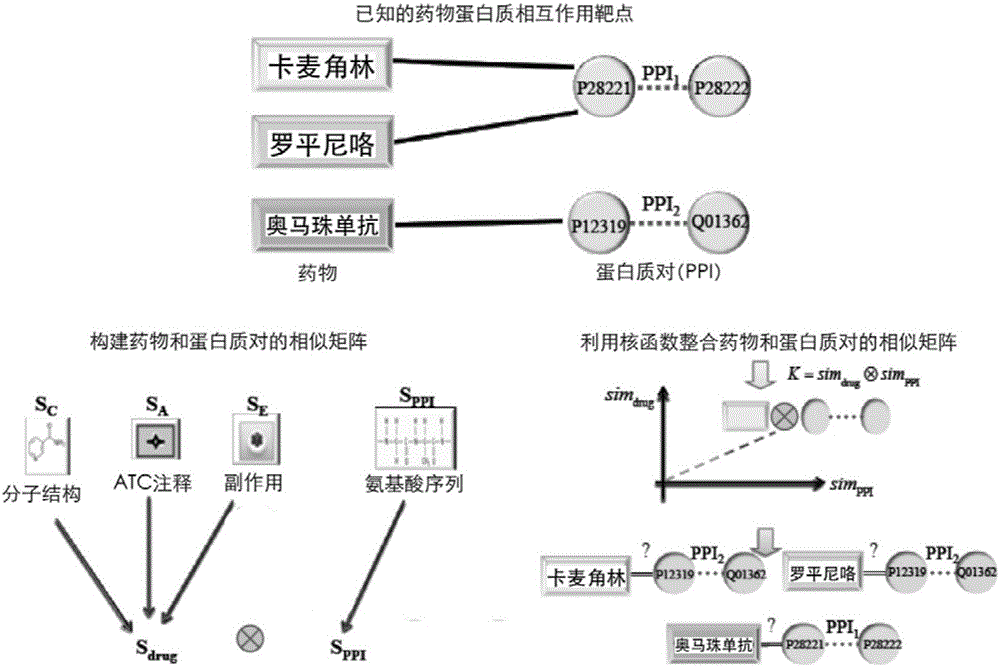
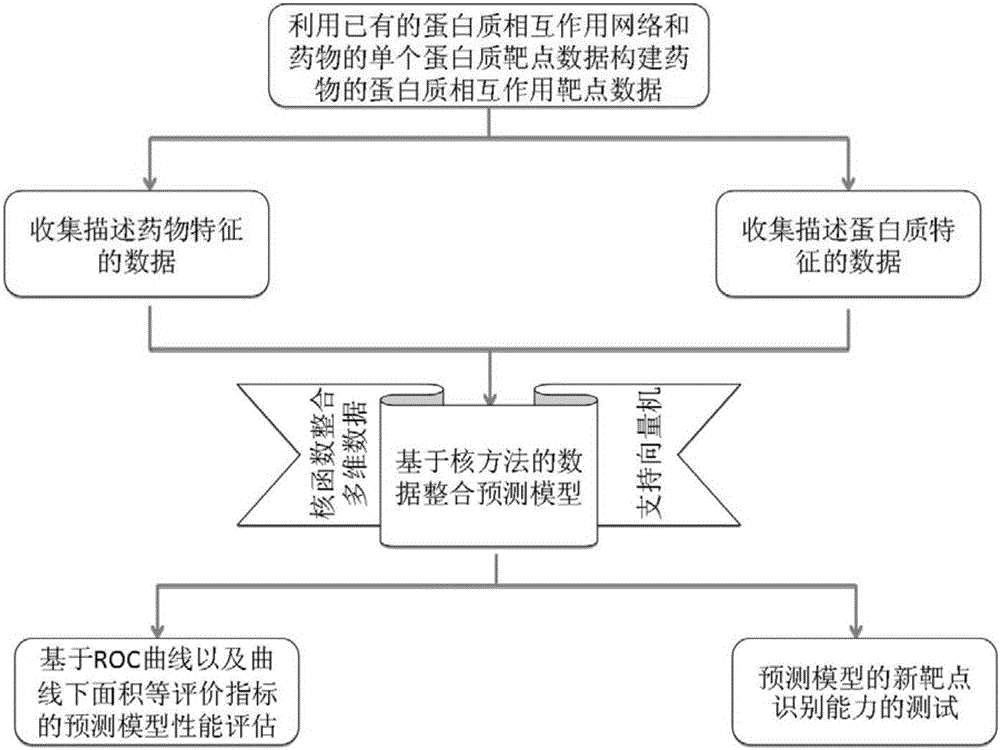
Method and system for predicting protein interaction target point of drug
InactiveCN105160206AWiden the search spaceImprove classification effectSpecial data processing applicationsEvolutionary biologyProtein targetData set
The invention relates to a method and a system for predicting a protein interaction target point of a drug. The method comprises: 1) collecting a human protein interaction network and single protein target point data of the drug, and constructing an interactive protein target point data set of the drug; 2) obtaining description data of the drug and proteins; 3) constructing a bigraph for representing an interactive relationship between the drug and a protein pair, constructing a similar matrix for representing drug similarity and protein pair similarity, establishing a kernel function for correlating the similar matrix of the drug and the protein pair, and establishing a prediction model through a machine learning algorithm; and 4) performing independent set testing by utilizing unknown drug and interactive protein pair, and predicting a possibly existent unknown drug protein interaction target point, and verifying a prediction result through database and document retrieval. According to the method and the system, the search space of the drug target point can be expanded and the more specific drug protein interaction target point with the best classification performance can be obtained.
Owner:ACAD OF MATHEMATICS & SYSTEMS SCIENCE - CHINESE ACAD OF SCI +1
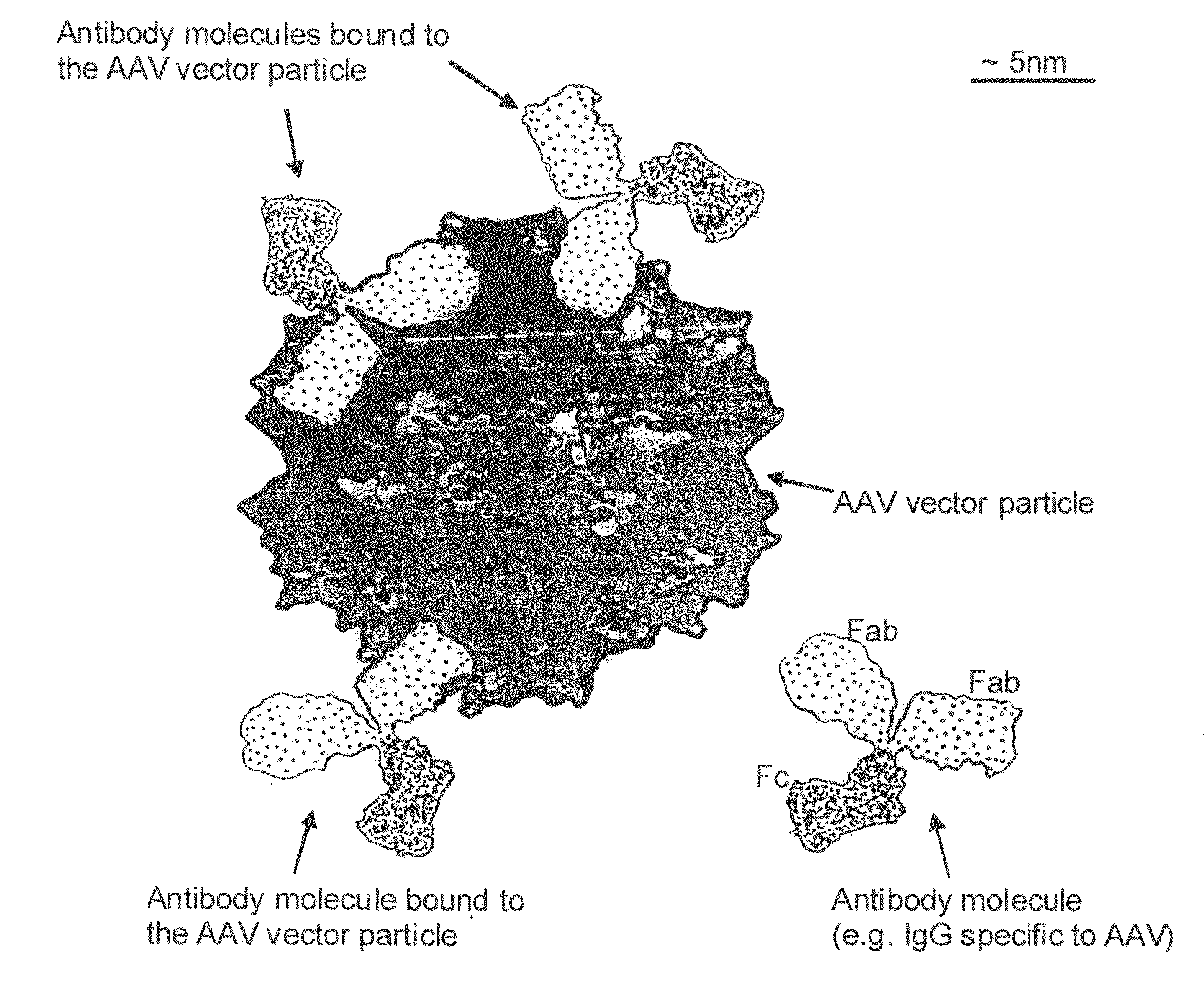
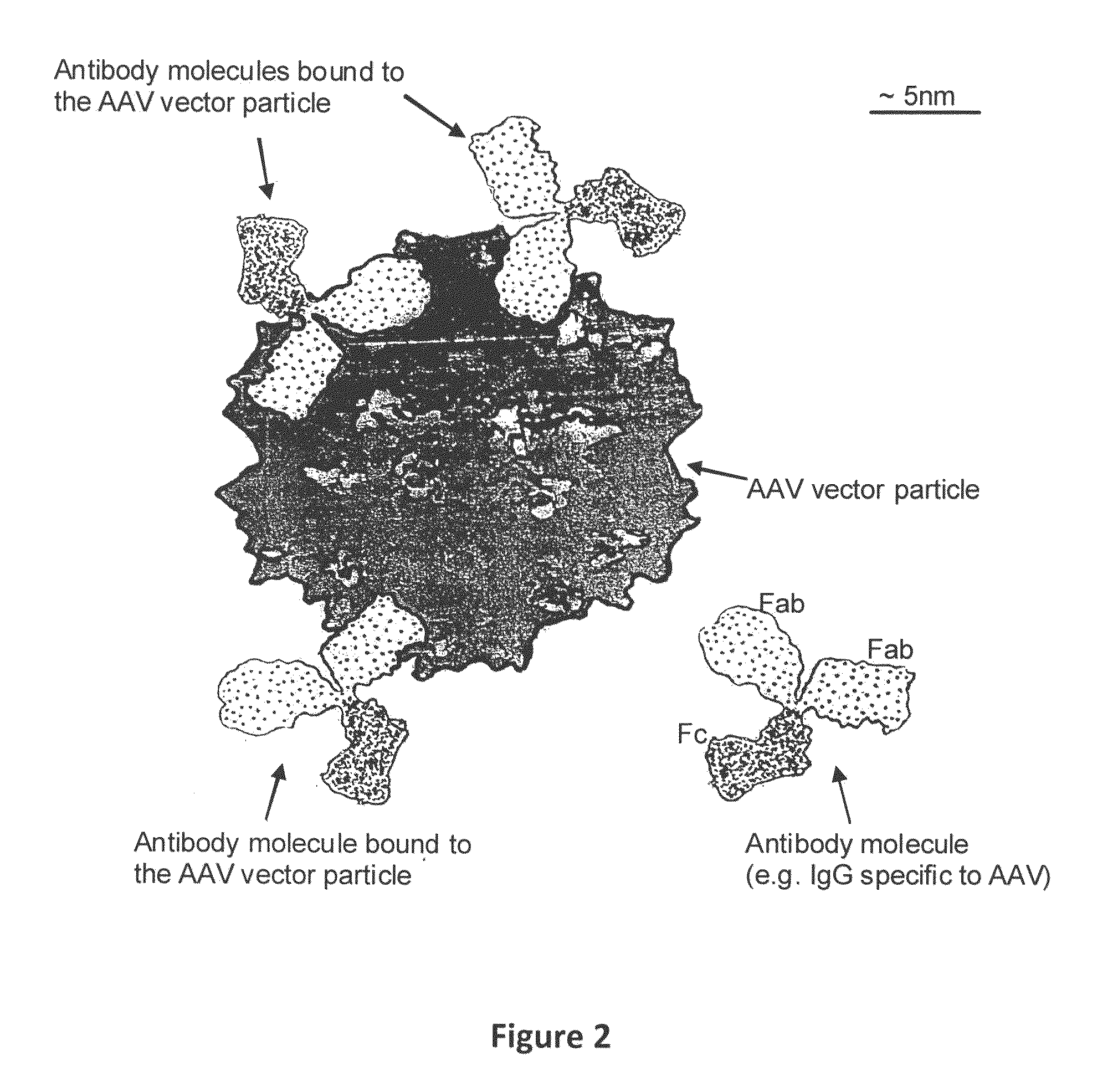
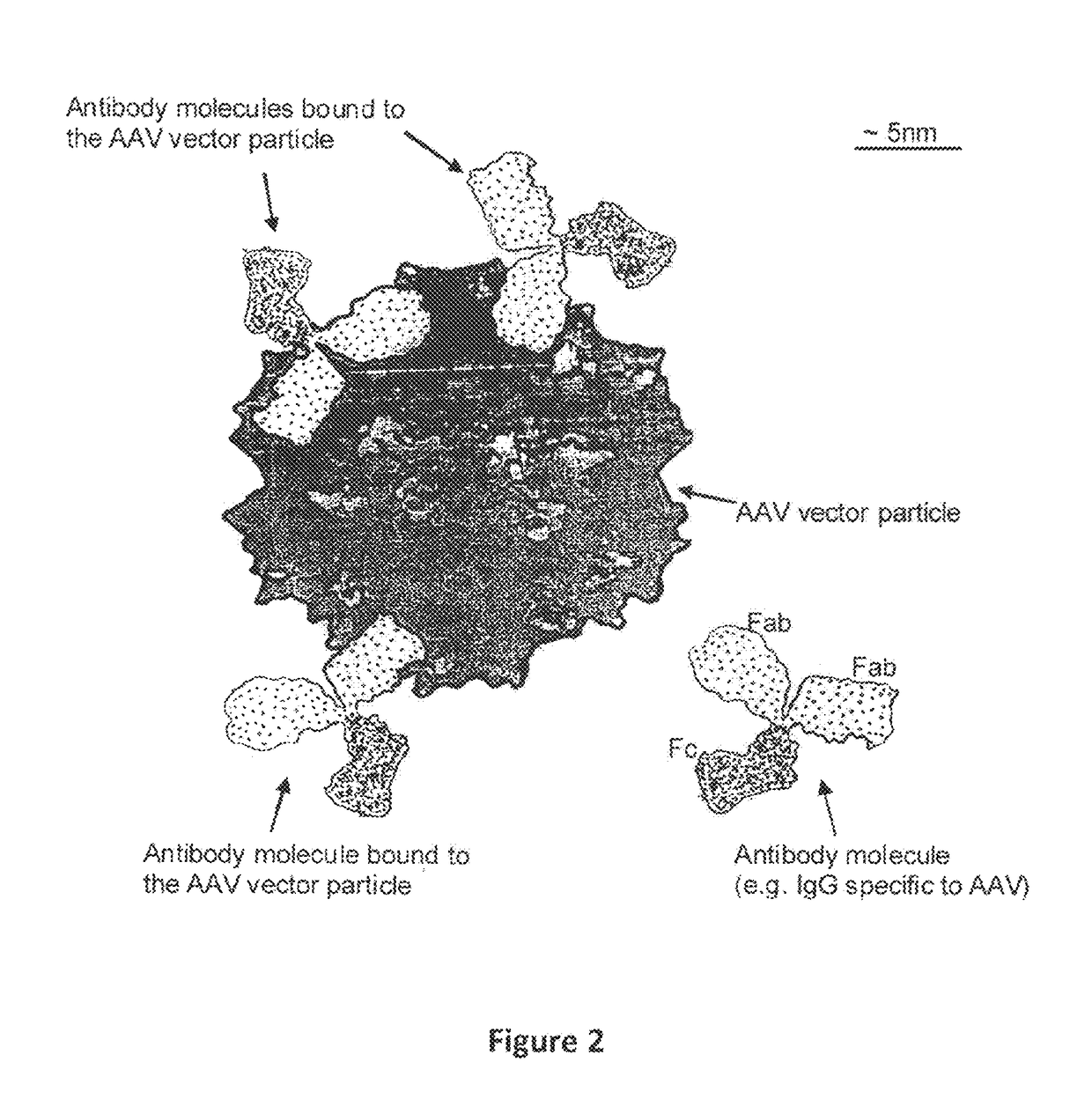
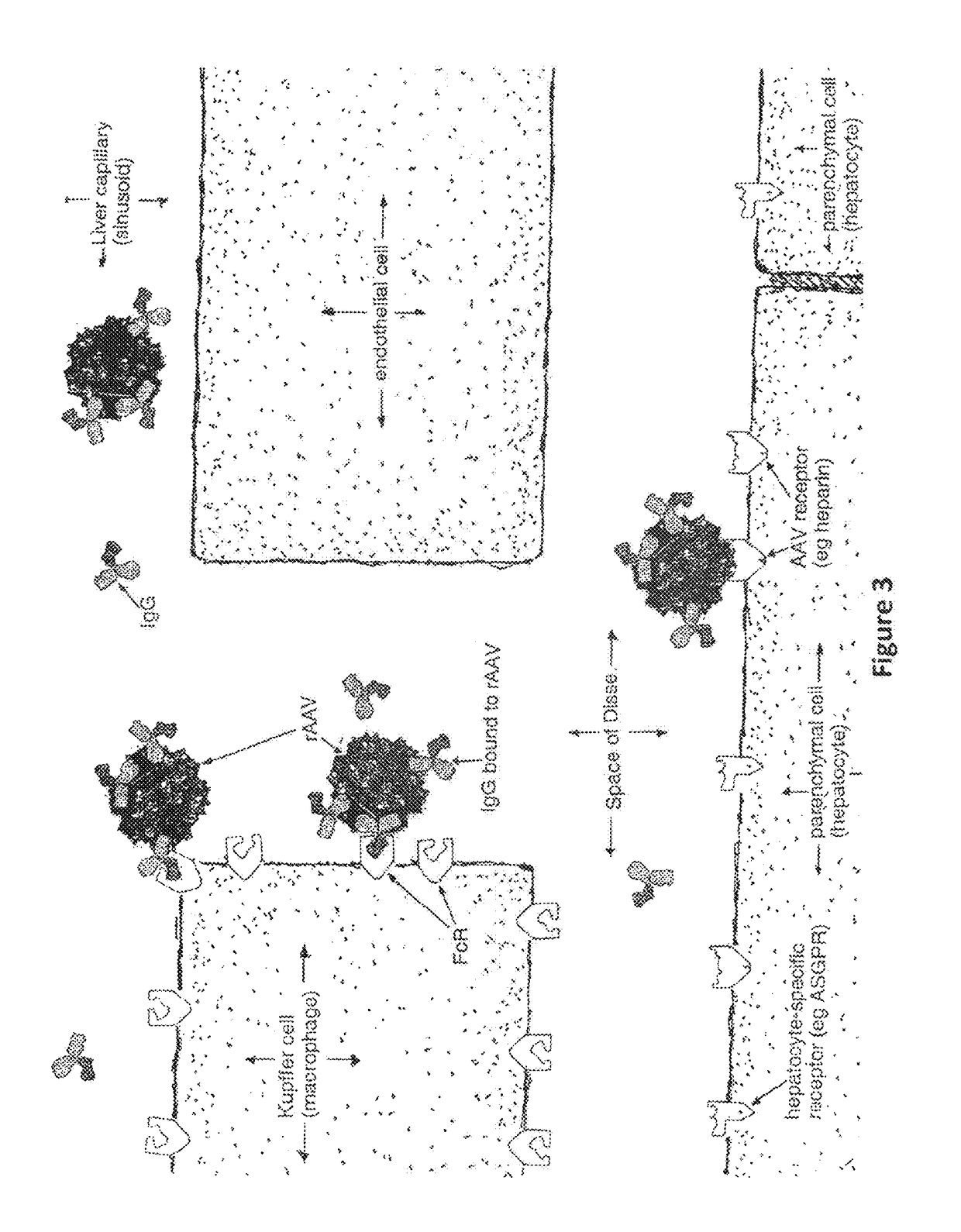
Humanized Viral Vectors and Methods of Use Thereof
InactiveUS20100098666A1Reducing antibody mediated clearanceReducing immune responseBiocidePeptide/protein ingredientsGene transferHuman proteins
The present invention provides humanized viral vectors and methods of use thereof for delivery of transgenes or therapeutic nucleic acids to human subjects. Humanized viral vectors are modified from known viral vectors such as those based on AAV by coating their surface with a human protein such as human serum albumin and optionally a lipid coating or formulation, so that the foreign or non-human nature of the vector is masked. The coating is performed in a manner that reduces or prevents binding of antibodies to the vector surface, thereby reducing or preventing antibody-mediated clearance of vector, but still allowing the vector to transduce target cells and achieve therapeutic gene transfer. Such humanized vectors therefore evade pre-existing immune surveillance, reduce immune responses, and achieve therapeutic gene transfer in the presence of pre-existing antibodies to the viral vector.
Owner:THE CHILDRENS HOSPITAL OF PHILADELPHIA
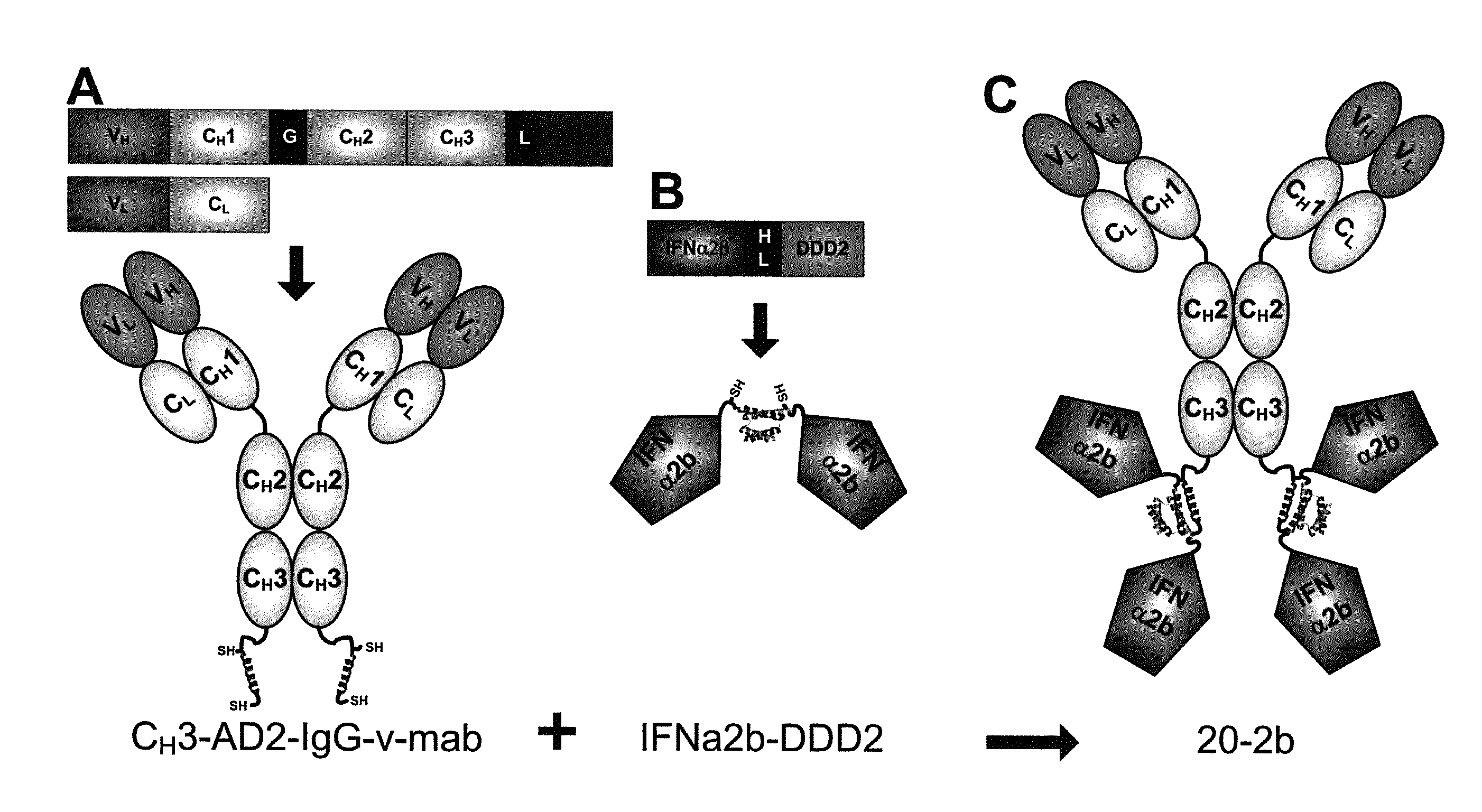
Targeting interferon-lambda with antibodies potently enhances Anti-tumor and Anti-viral activities
ActiveUS20130136718A1Useful effectReduce amyloid plaqueMaterial nanotechnologyPeptide/protein ingredientsAntigen bindingAsthma
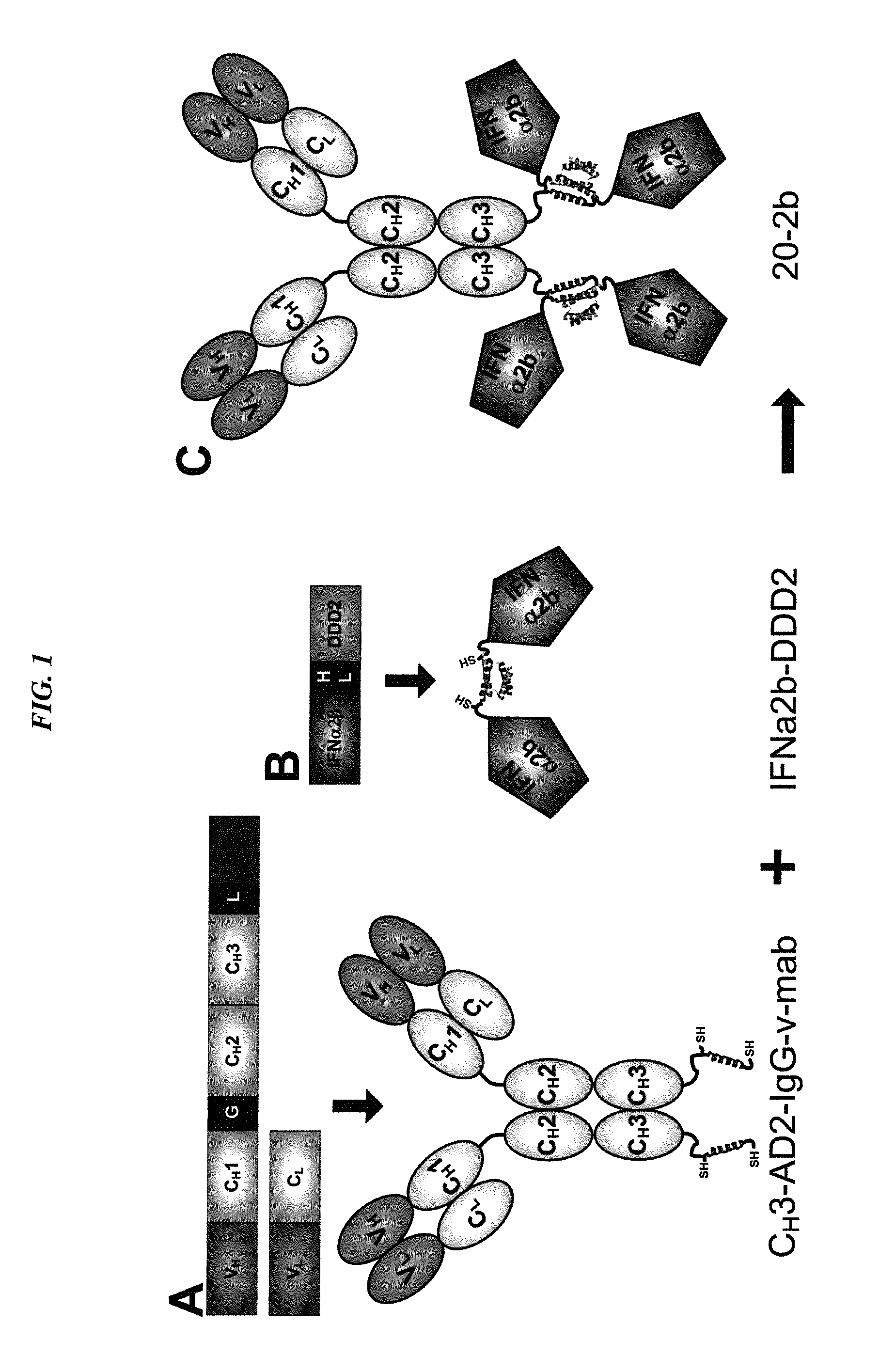
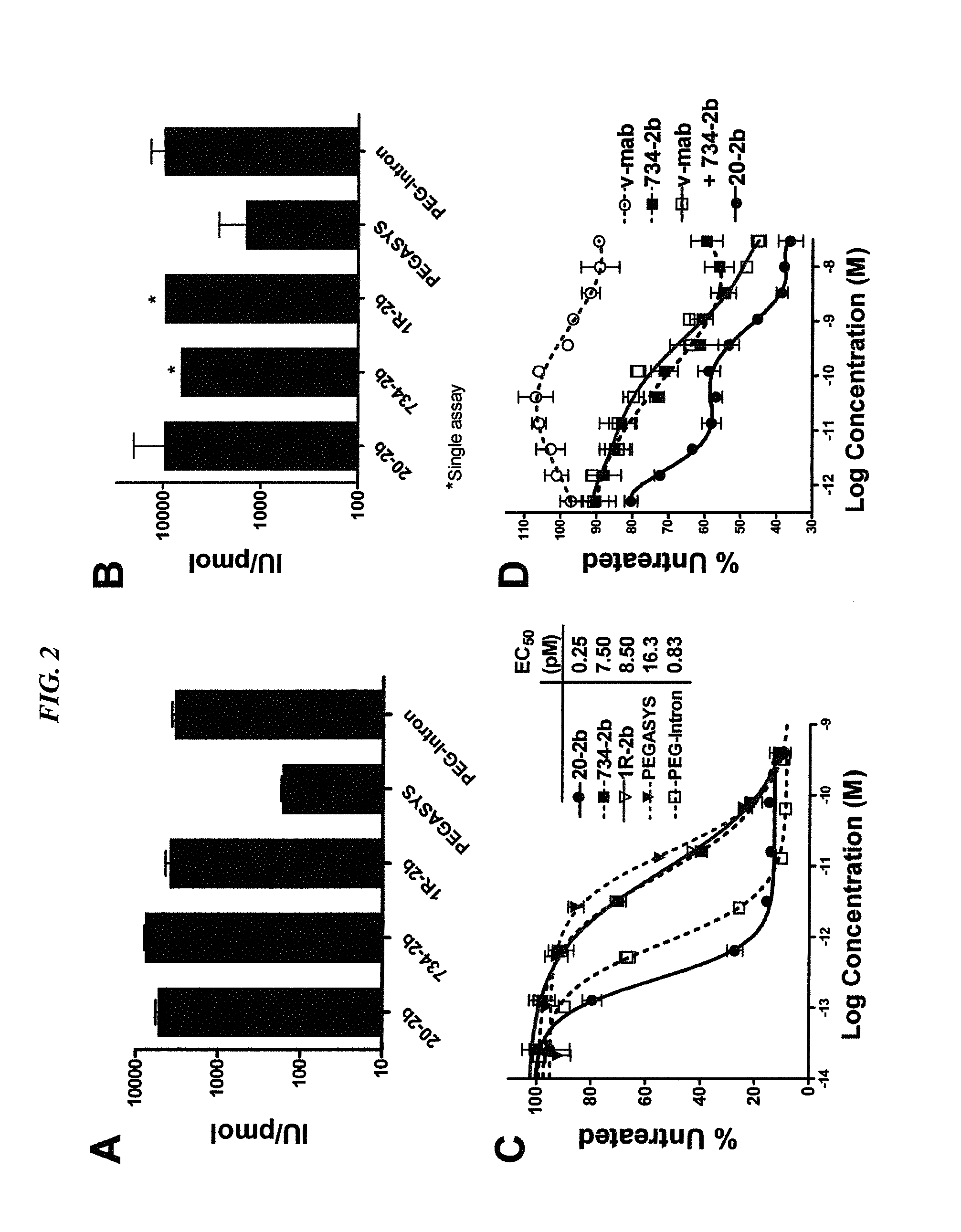
The present invention concerns methods and compositions for forming complexes of interferon-λ with an antibody or antigen-binding antibody fragment. In preferred embodiments, the interferon-λ and the antibody or fragment are fusion proteins, each comprising a dimerization and docking domain (DDD) moiety from human protein kinase A or an anchor domain (AD) moiety from an A-kinase anchoring protein (AKAP). In more preferred embodiments, the interferon-antibody complex is more efficacious for treatment of cancer, asthma, Alzheimer's disease, multiple sclerosis or viral infection than interferon-λ alone, antibody alone, or the combination of unconjugated interferon-λ and antibody.
Owner:IBC PHARMACEUTICALS INC
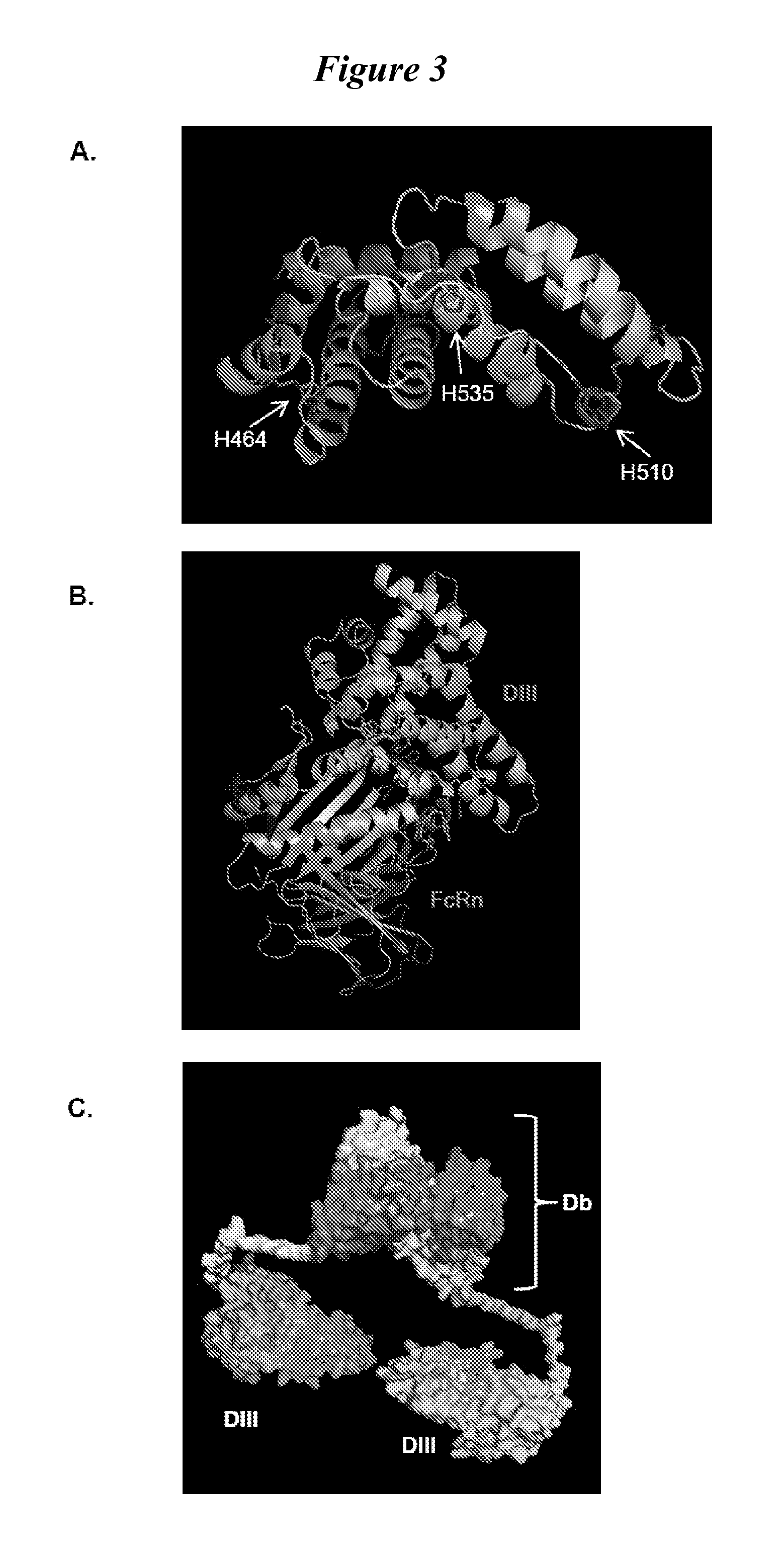
Human protein scaffold with controlled serum pharmacokinetics
InactiveUS20120076728A1Peptide/protein ingredientsAntibody mimetics/scaffoldsHuman proteinsSequence identity
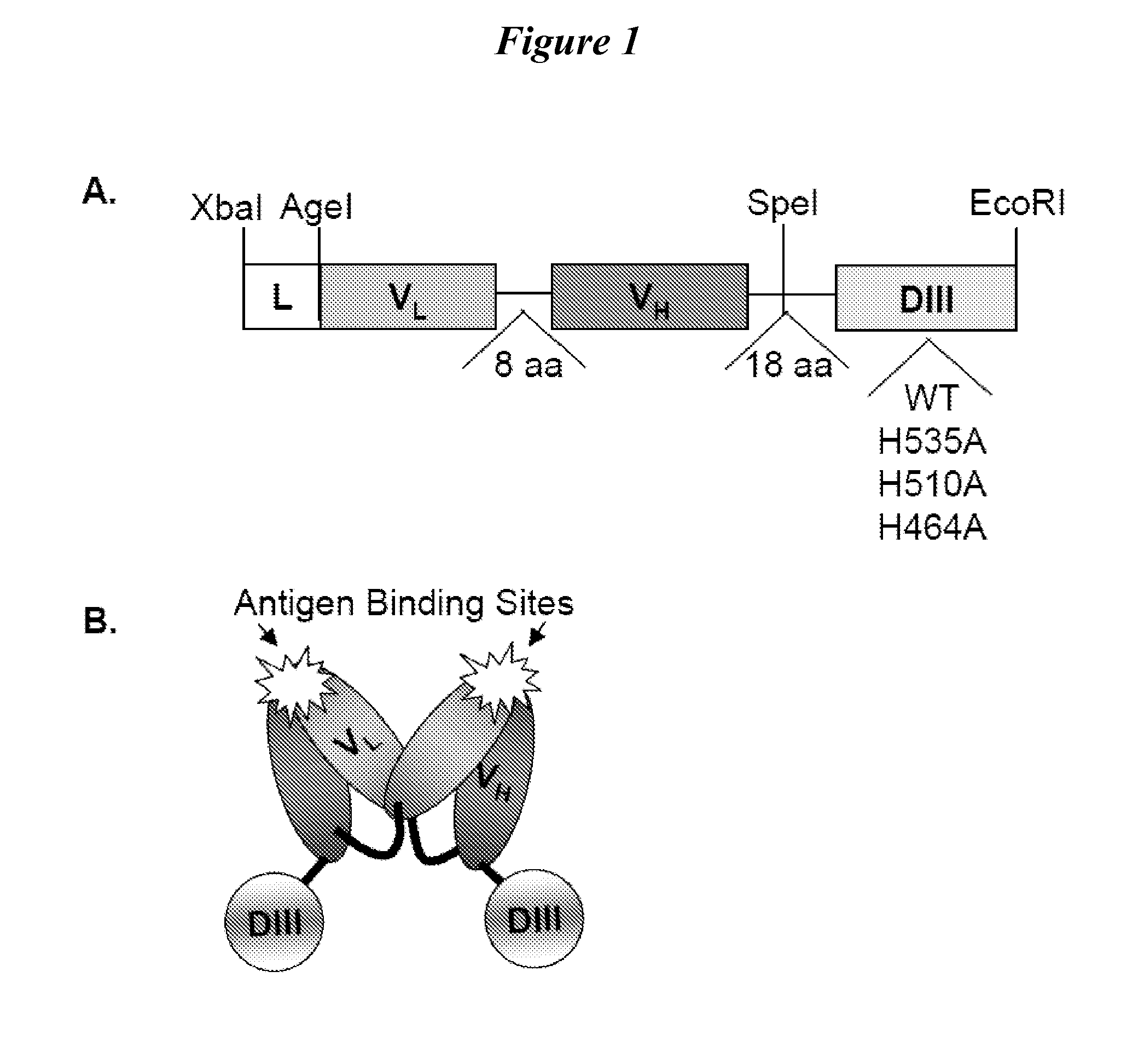
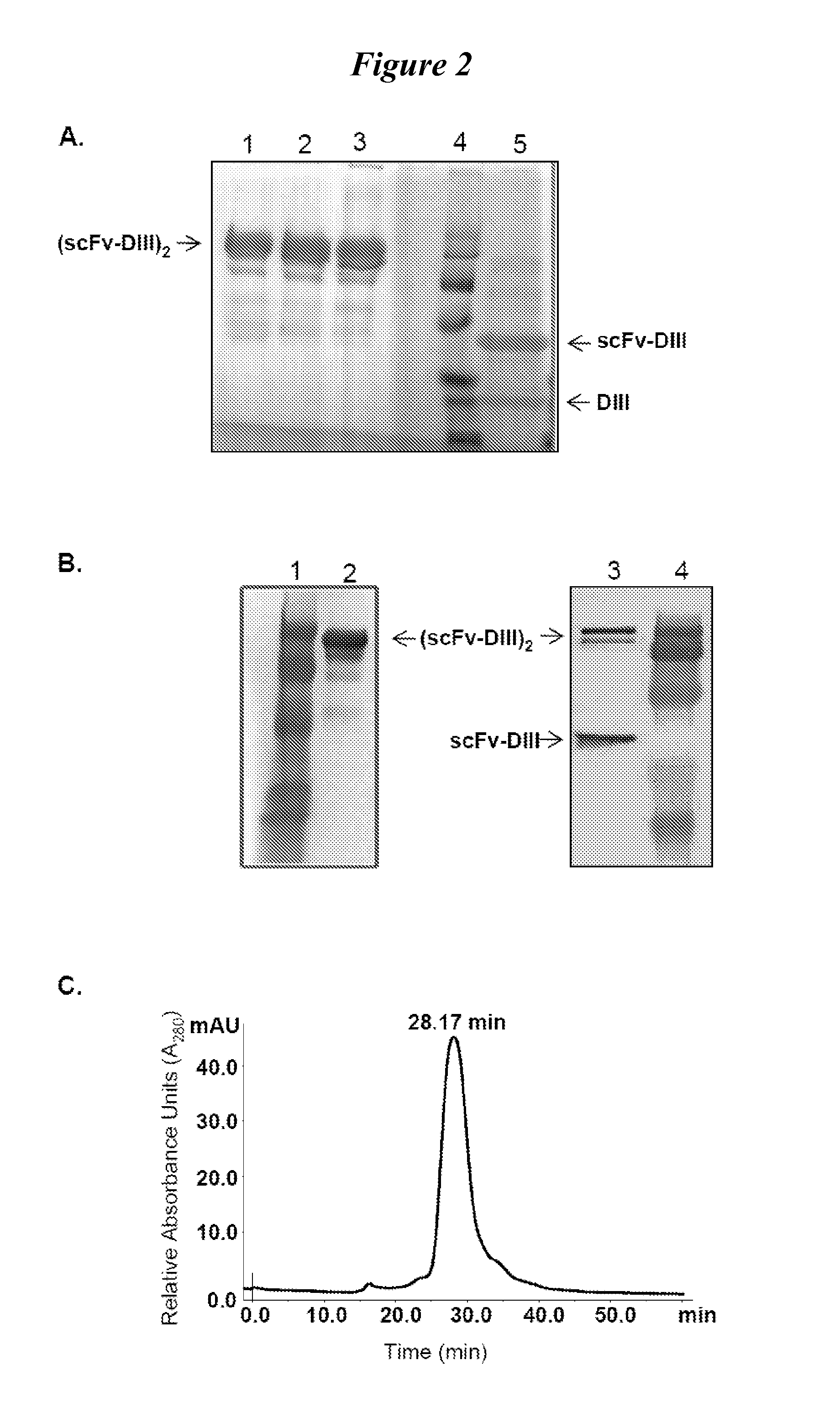
This invention provides constructs comprising a protein scaffold, wherein the scaffold comprises Domain III, Domain IIIa, or Domain IIIb of human serum albumin or a polypeptide having substantial sequence identity to the Domain III, the Domain IIIa, or the Domain IIIb; and a targeting moiety in covalent linkage to the protein scaffold; and a therapeutic moiety and / or an imaging moiety in covalent linkage to the protein scaffold. The scaffold can be modified to tune the serum pharmacokinetics of the construct. In addition to methods of making the constructs, therapeutic, imaging and diagnostic uses of the constructs are also provided.
Owner:RGT UNIV OF CALIFORNIA
Humanized Viral Vectors and Methods of Use Thereof
ActiveUS20170128594A1Reduce clearanceReduced responseVectorsPeptide/protein ingredientsGene transferHuman proteins
The present invention provides humanized viral vectors and methods of use thereof for delivery of transgenes or therapeutic nucleic acids to human subjects. Humanized viral vectors are modified from known viral vectors such as those based on AAV by coating their surface with a human protein such as human serum albumin and optionally a lipid coating or formulation, so that the foreign or non-human nature of the vector is masked. The coating is performed in a manner that reduces or prevents binding of antibodies to the vector surface, thereby reducing or preventing antibody-mediated clearance of vector, but still allowing the vector to transduce target cells and achieve therapeutic gene transfer. Such humanized vectors therefore evade pre-existing immune surveillance, reduce immune responses, and achieve therapeutic gene transfer in the presence of pre-existing antibodies to the viral vector.
Owner:THE CHILDRENS HOSPITAL OF PHILADELPHIA
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com