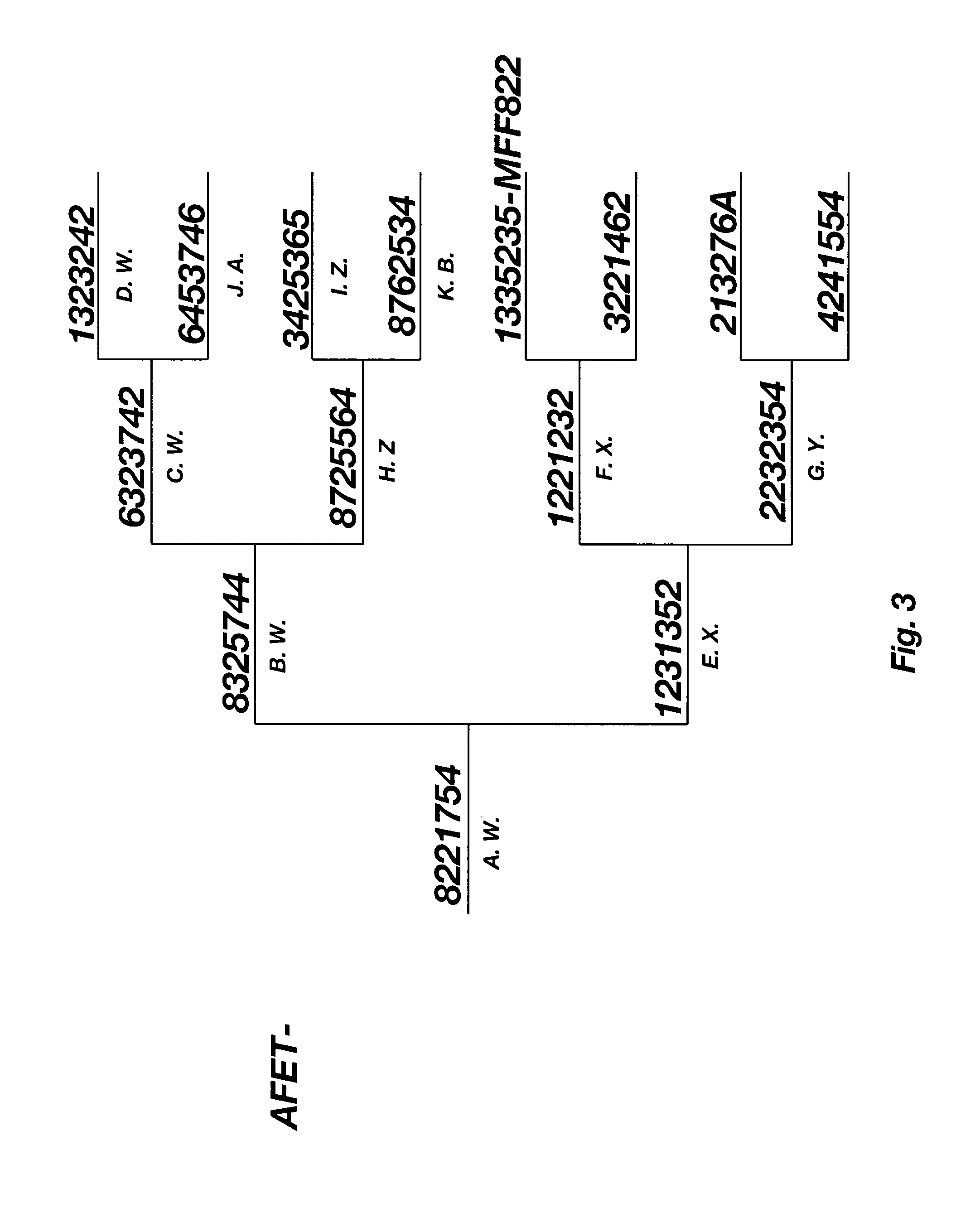
Patents
Literature
323results about "Evolutionary biology" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
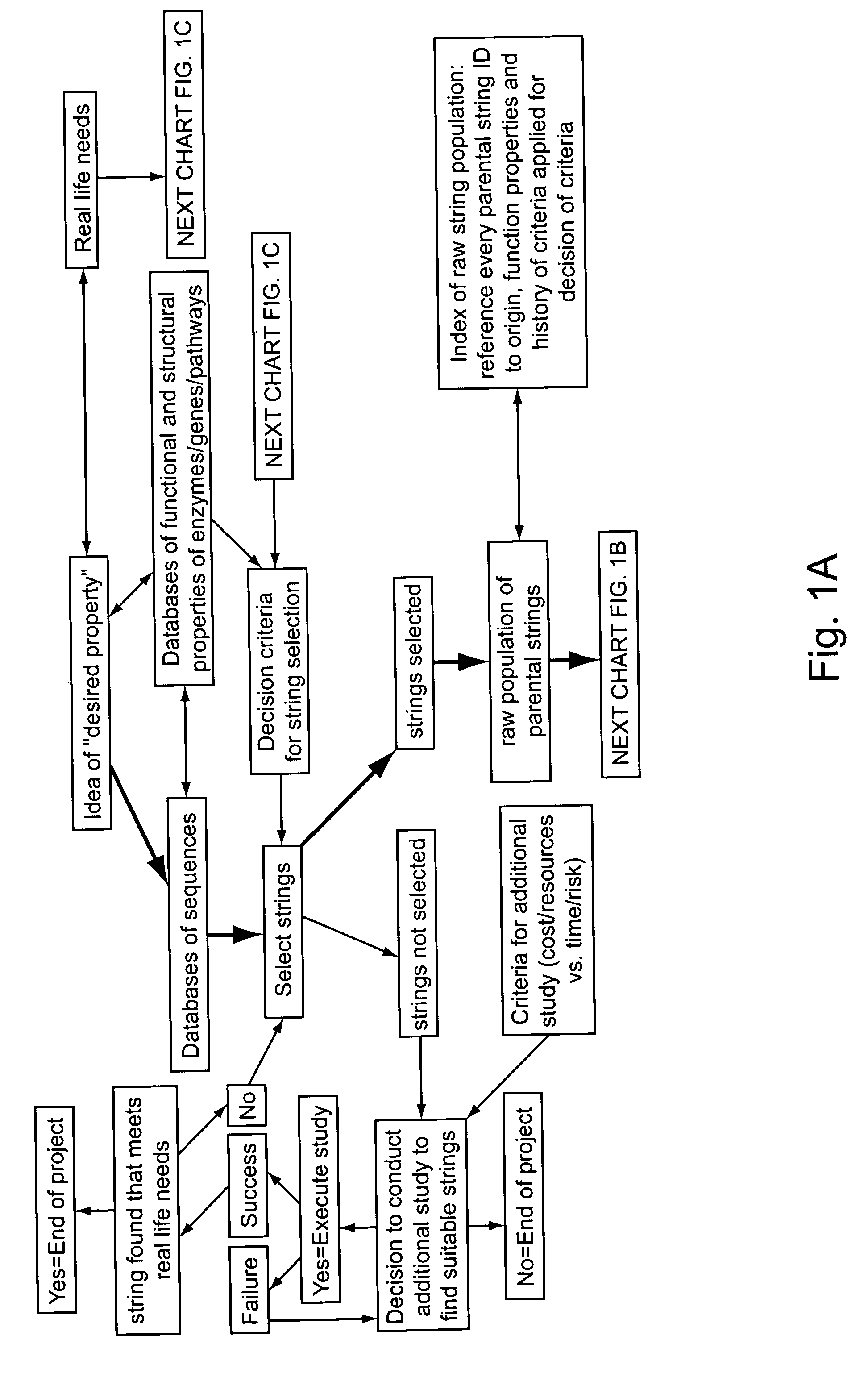
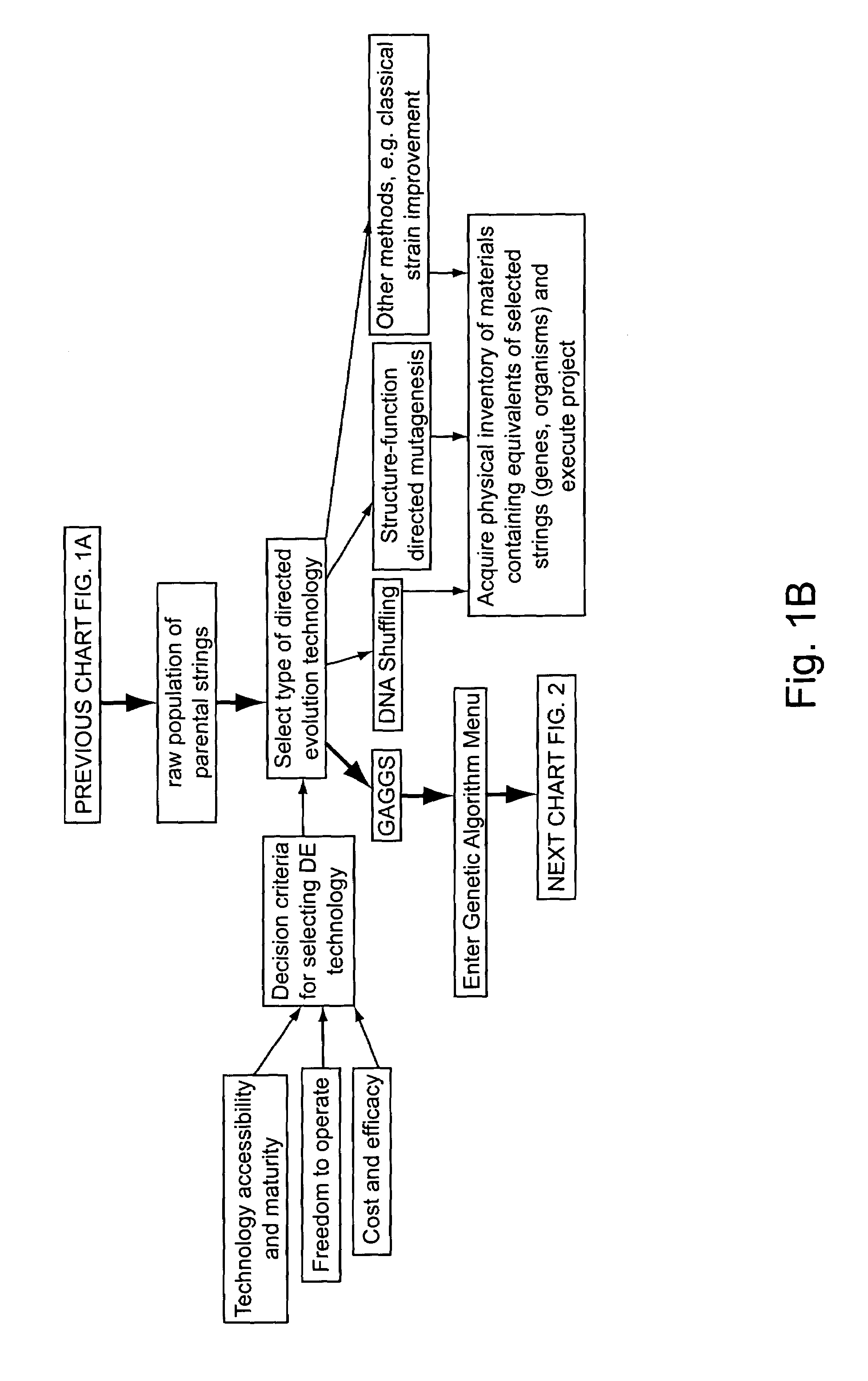
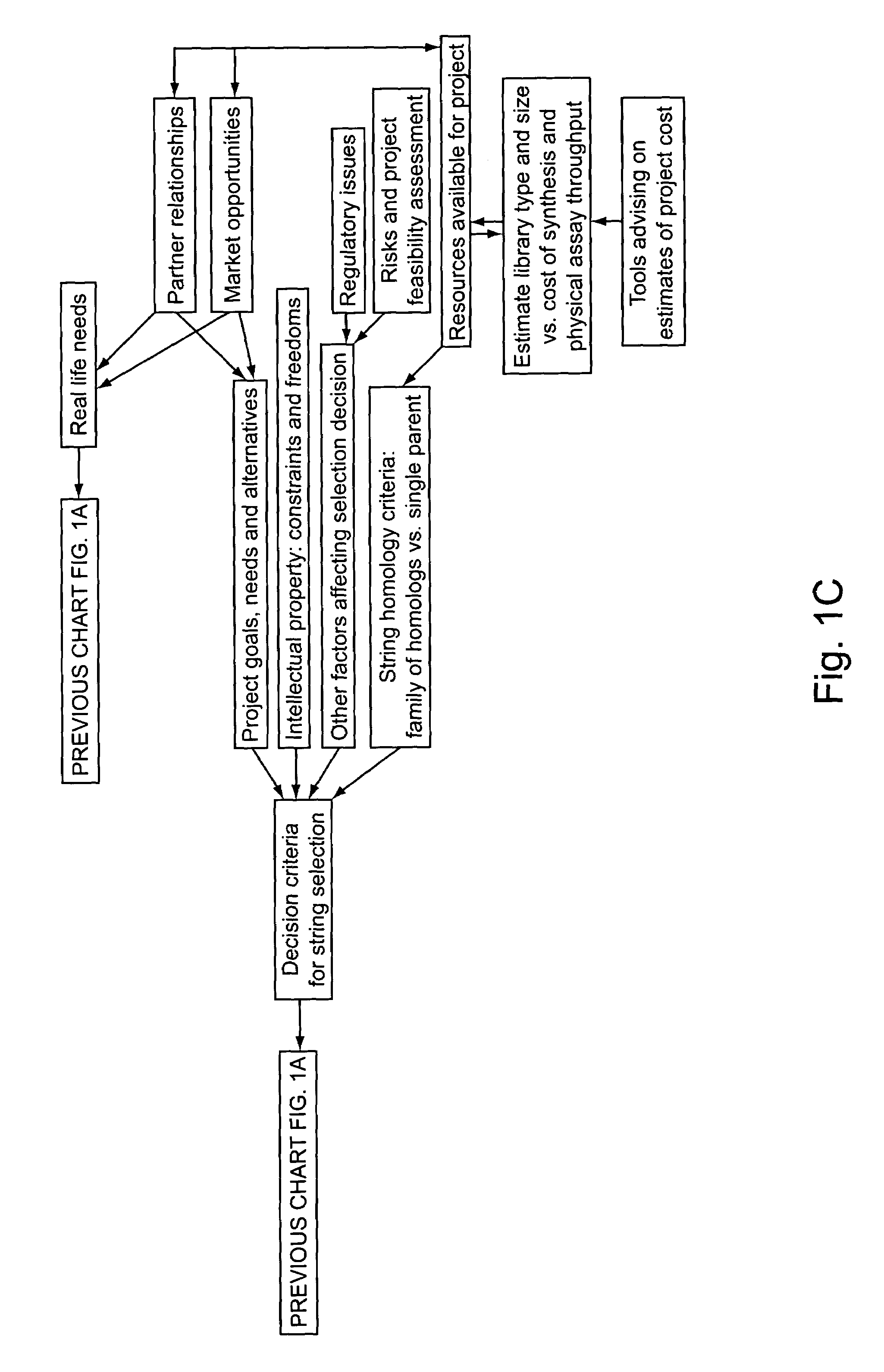
Methods for making character strings, polynucleotides and polypeptides having desired characteristics
InactiveUS7024312B1Simplifies overall synthesis strategyLow levelPeptide/protein ingredientsBiostatisticsPolynucleotideIn silico
“In silico” nucleic acid recombination methods, related integrated systems utilizing genetic operators and libraries made by in silico shuffling methods are provided.
Owner:CODEXIS MAYFLOWER HLDG LLC
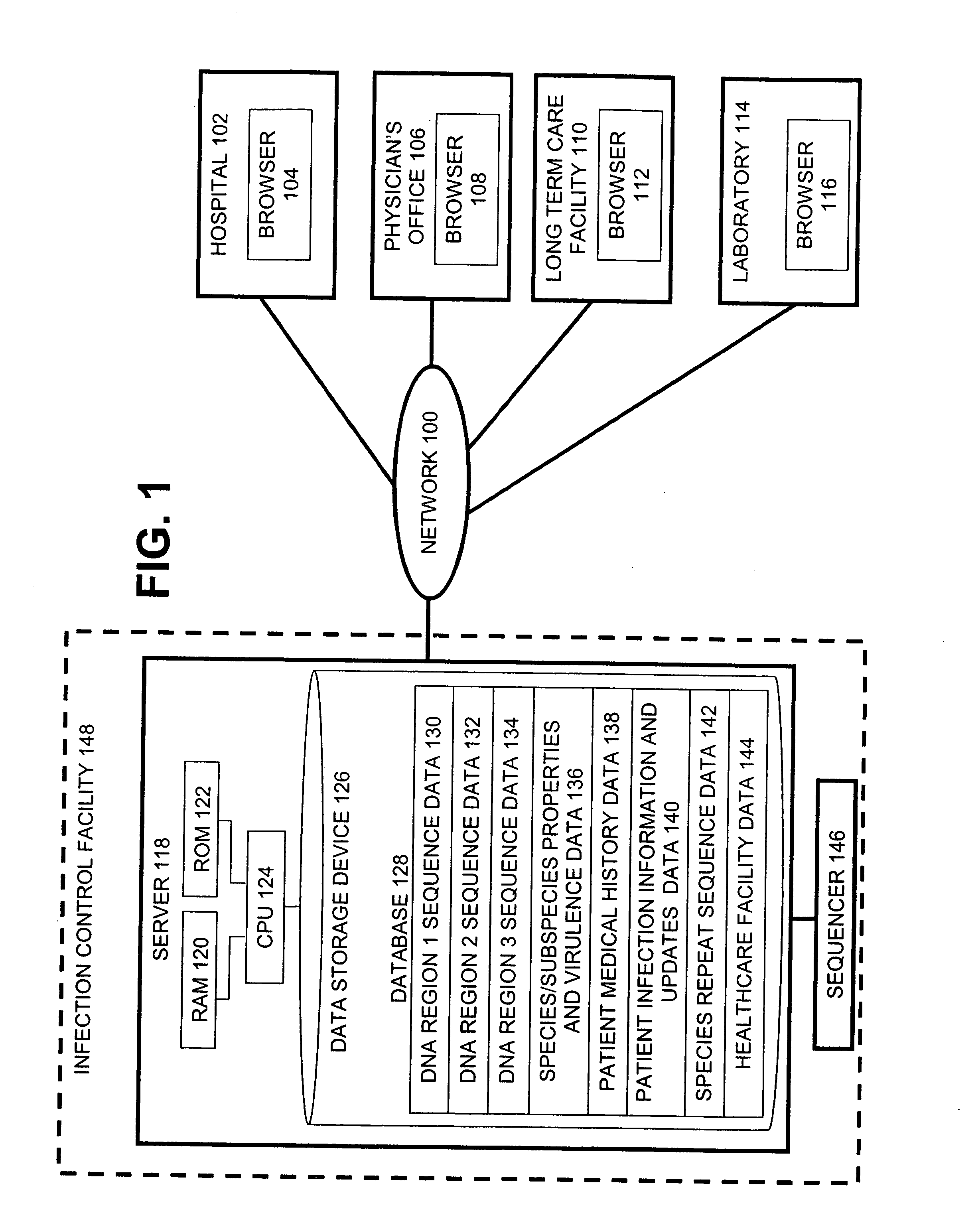
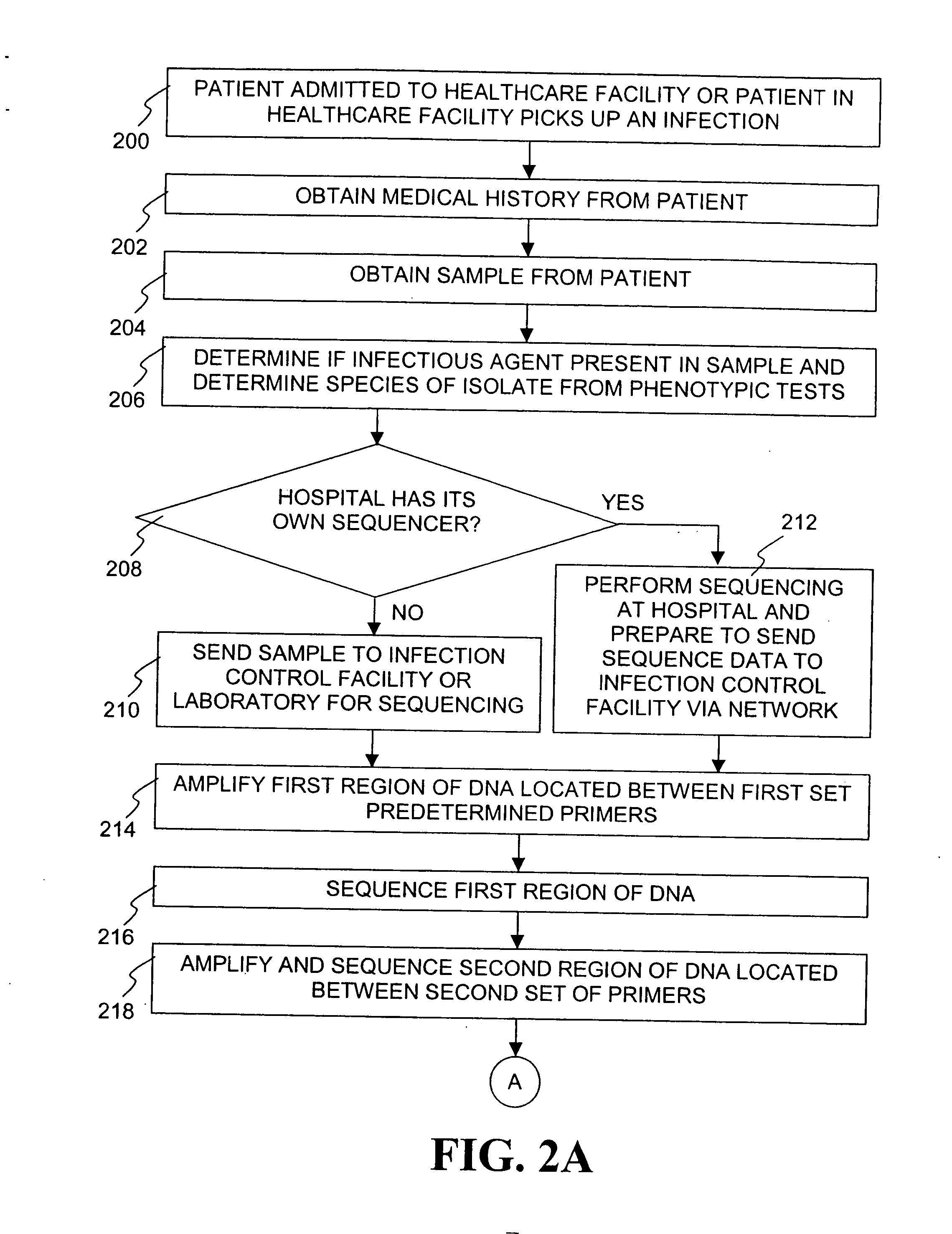
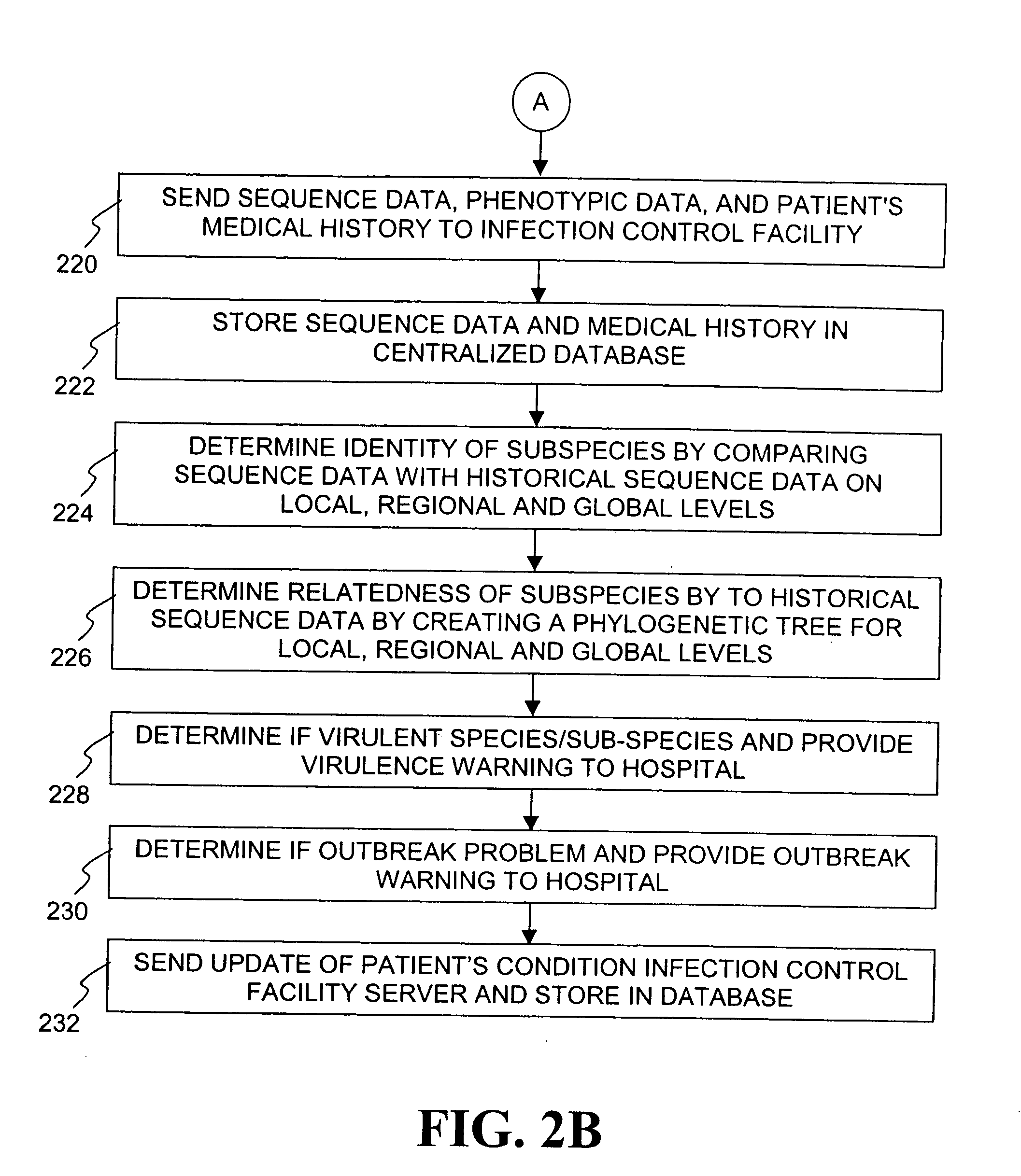
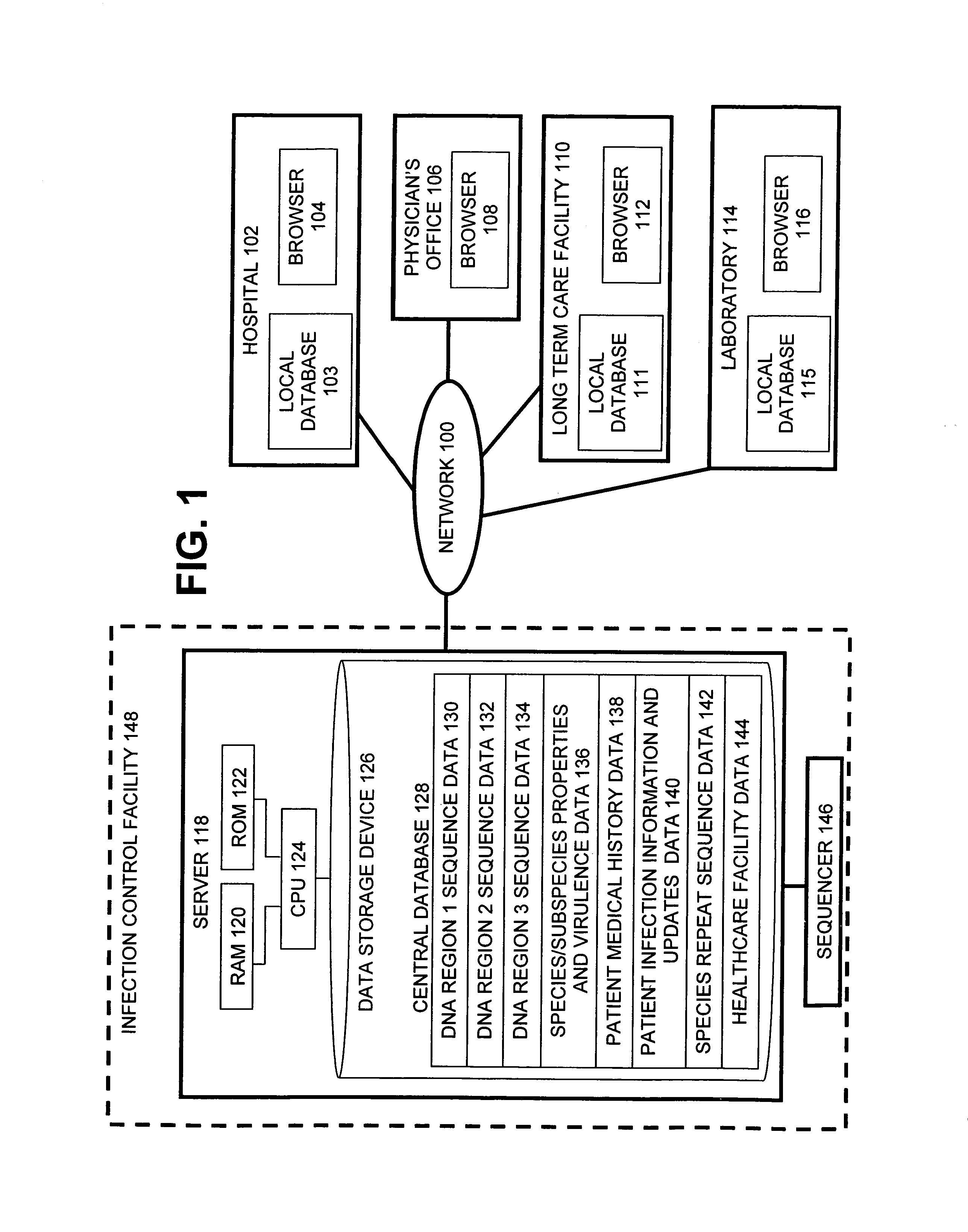
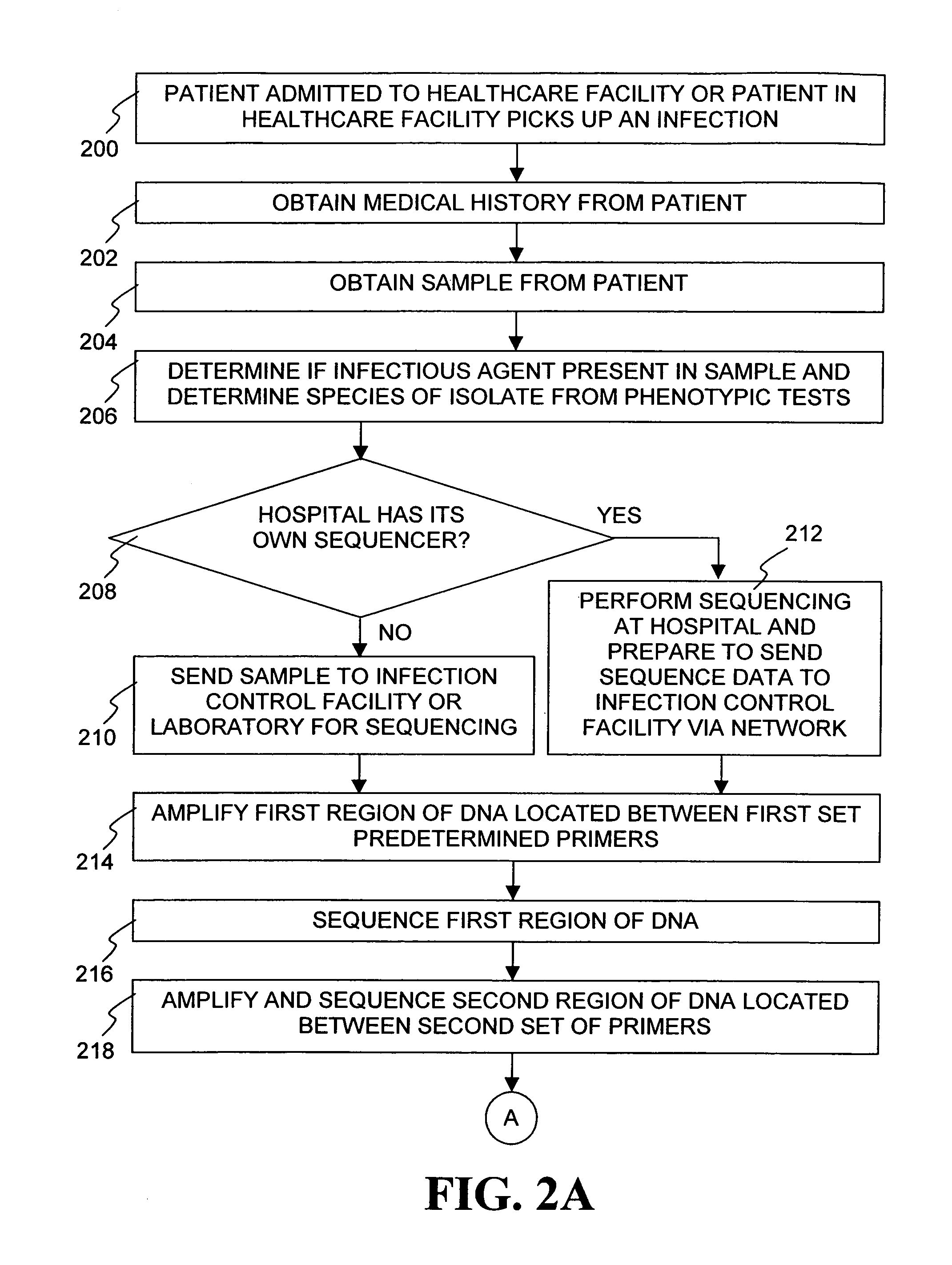
Method for tracking and controlling infections
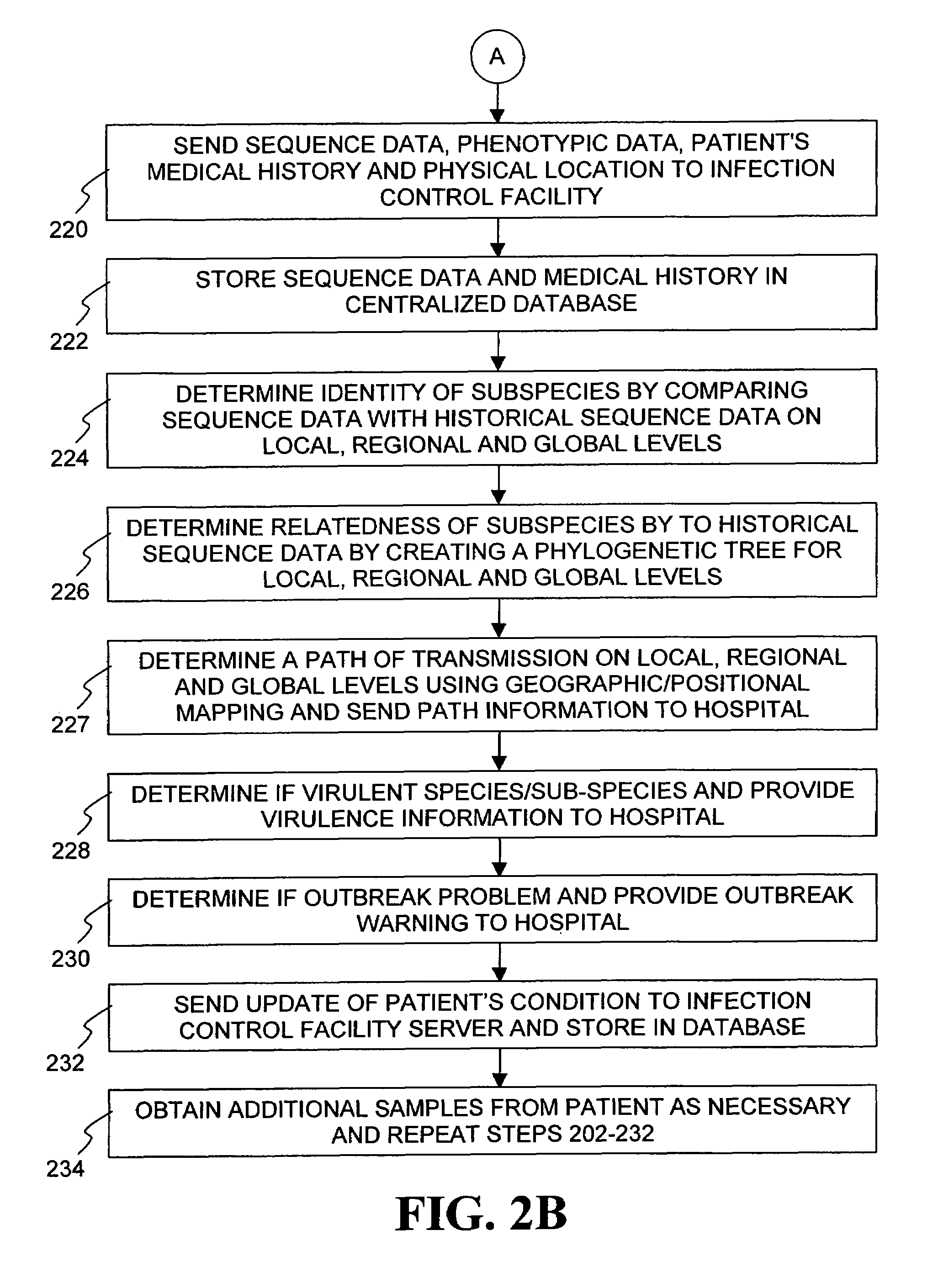
InactiveUS20060020391A1Computer-assisted medical data acquisitionMedical imagesPhylogenetic relatednessMicroorganism
The present invention is a system and method for performing real-time infection control over a computer network. The method comprises obtaining a sample of a microorganism at a health care facility, sequencing a first region of a nucleic acid from the microorganism sample, comparing the first sequenced region with historical sequence data stored in a database, determining a measure of phylogenetic relatedness between the microorganism sample and historical samples stored in the database, and providing infection control information based on the phylogenetic relatedness determination to the health care facility, thereby allowing the health care facility to use the infection control information to control or prevent the spread of an infection.
Owner:EGENOMICS INC
System and method for tracking and controlling infections
InactiveUS7349808B1Digital data processing detailsAnalogue computers for chemical processesMicroorganismPhylogenetic relatedness
The present invention is a system and method for performing real-time infection control over a computer network. The method comprises obtaining a sample of a microorganism at a health care facility, sequencing a first region of a nucleic acid from the microorganism sample, comparing the first sequenced region with historical sequence data stored in a database, determining a measure of phylogenetic relatedness between the microorganism sample and historical samples stored in the database, and providing infection control information based on the phylogenetic relatedness determination to the health care facility, thereby allowing the health care facility to use the infection control information to control or prevent the spread of an infection.
Owner:EGENOMICS INC +1
Method for redesign of microbial production systems
ActiveUS20050079482A1Solve complexityIncreased production objectiveMicrobiological testing/measurementProteomicsMicroorganismBiological body
Owner:PENN STATE RES FOUND
Methods for obtaining and using haplotype data
InactiveUS20050191731A1Accurate predictionReduce costs and risksData visualisationBiostatisticsHaplotypeCrowds
Methods, computer program(s) and database(s) to analyze and make use of gene haplotype information. These include methods, program, and database to find and measure the frequency of haplotypes in the general population; methods, program, and database to find correlation's between an individual's haplotypes or genotypes and a clinical outcome; methods, program, and database to predict an individual's haplotypes from the individual's gen type for a gene; and methods, program, and database to predict an individual's clinical response to a treatment based on the individual's genotype or haplotype.
Owner:JUDSON RICHARD S +3
Gene recombination and hybrid protein development
InactiveUS20020045175A1Preserve stabilityPreserve desired propertyMicrobiological testing/measurementBiological testingBiopolymerHybrid protein
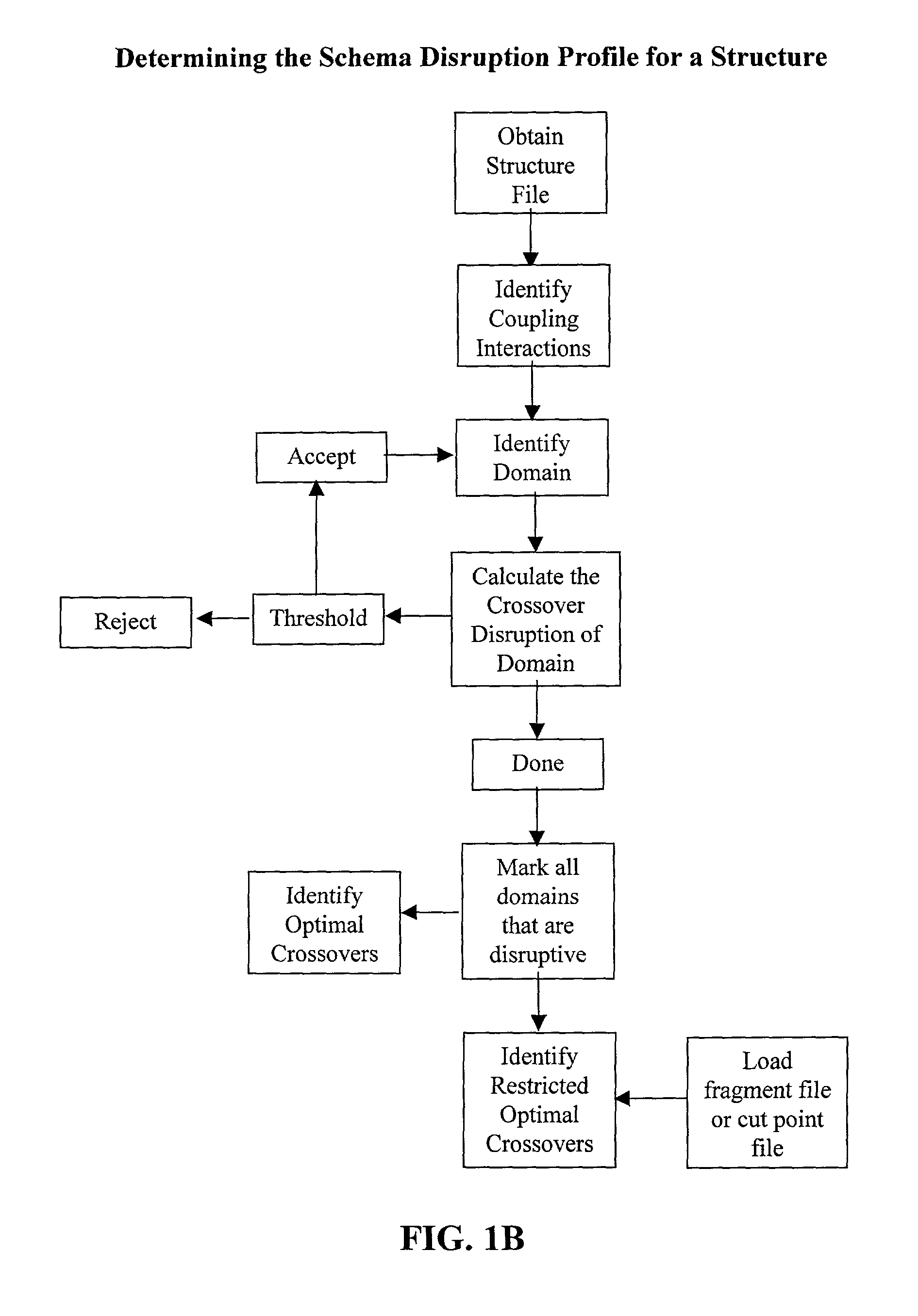
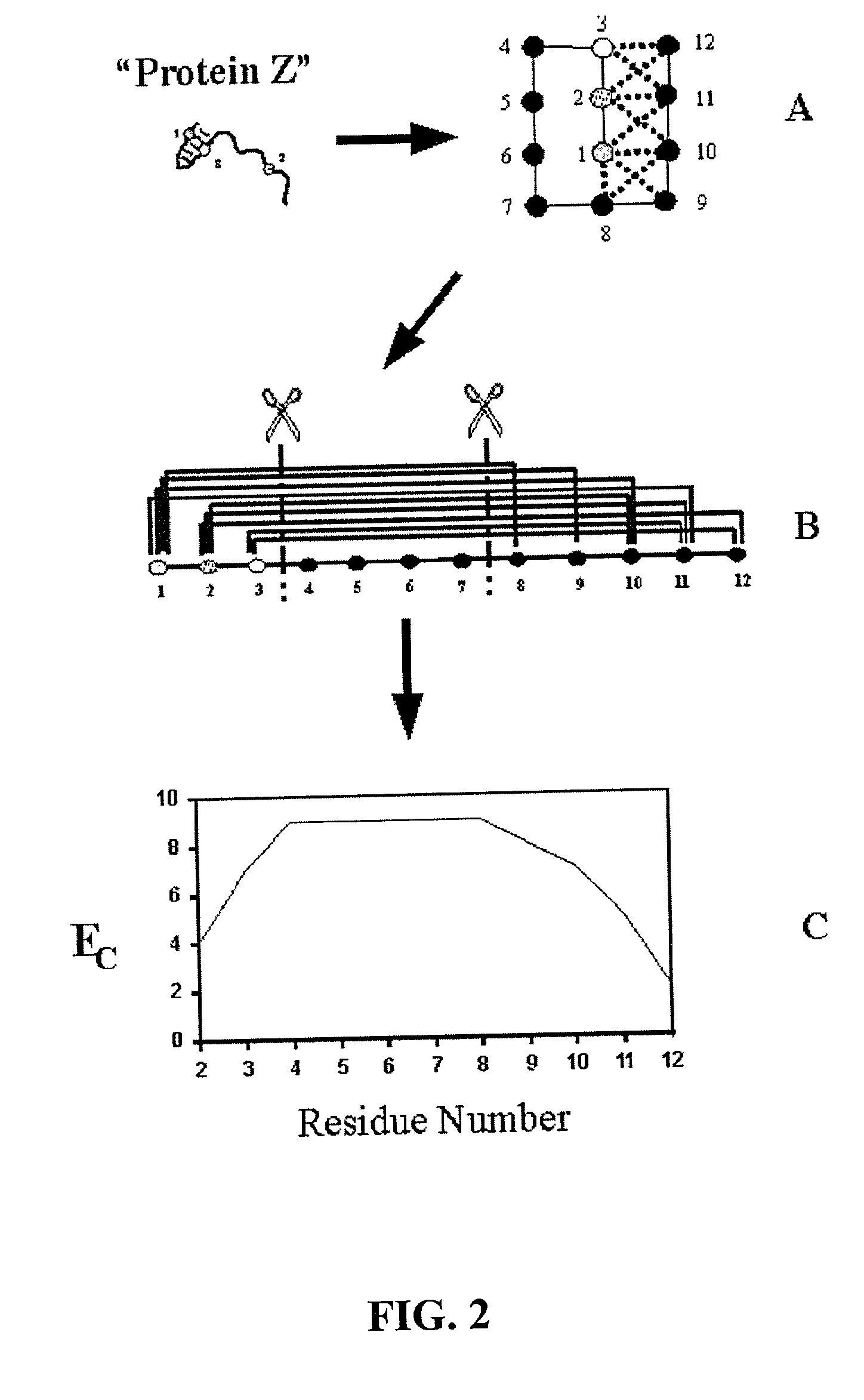
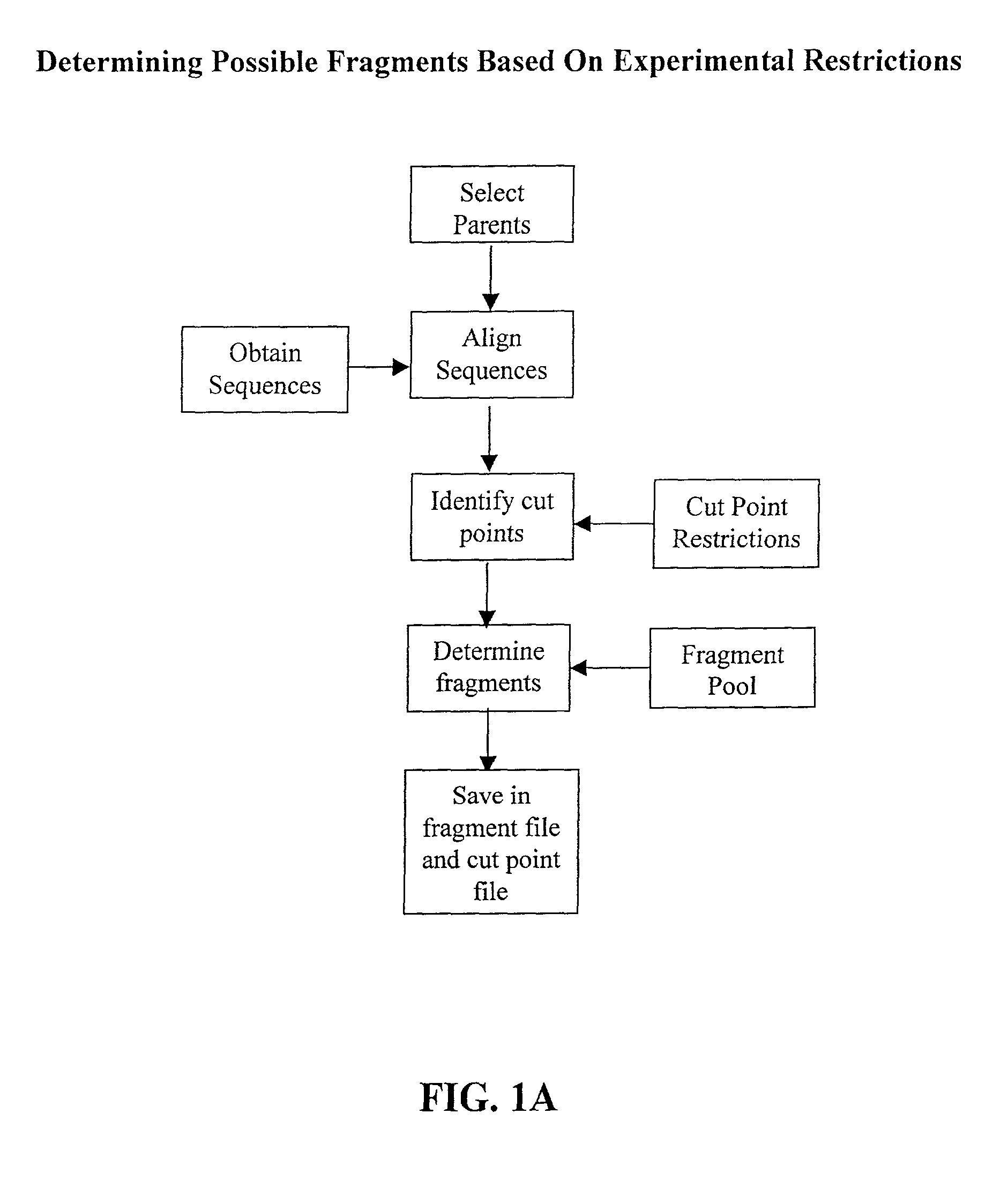
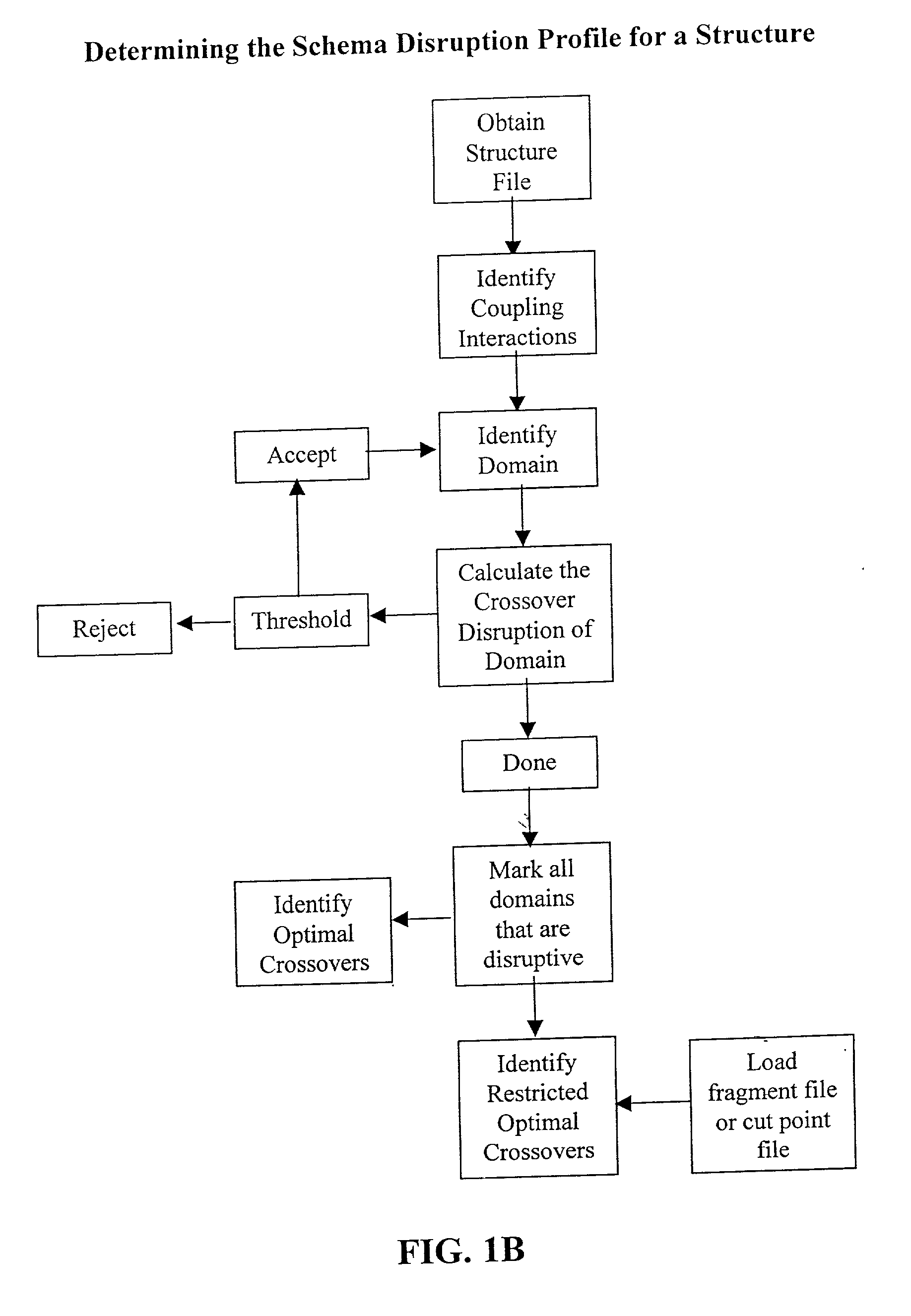
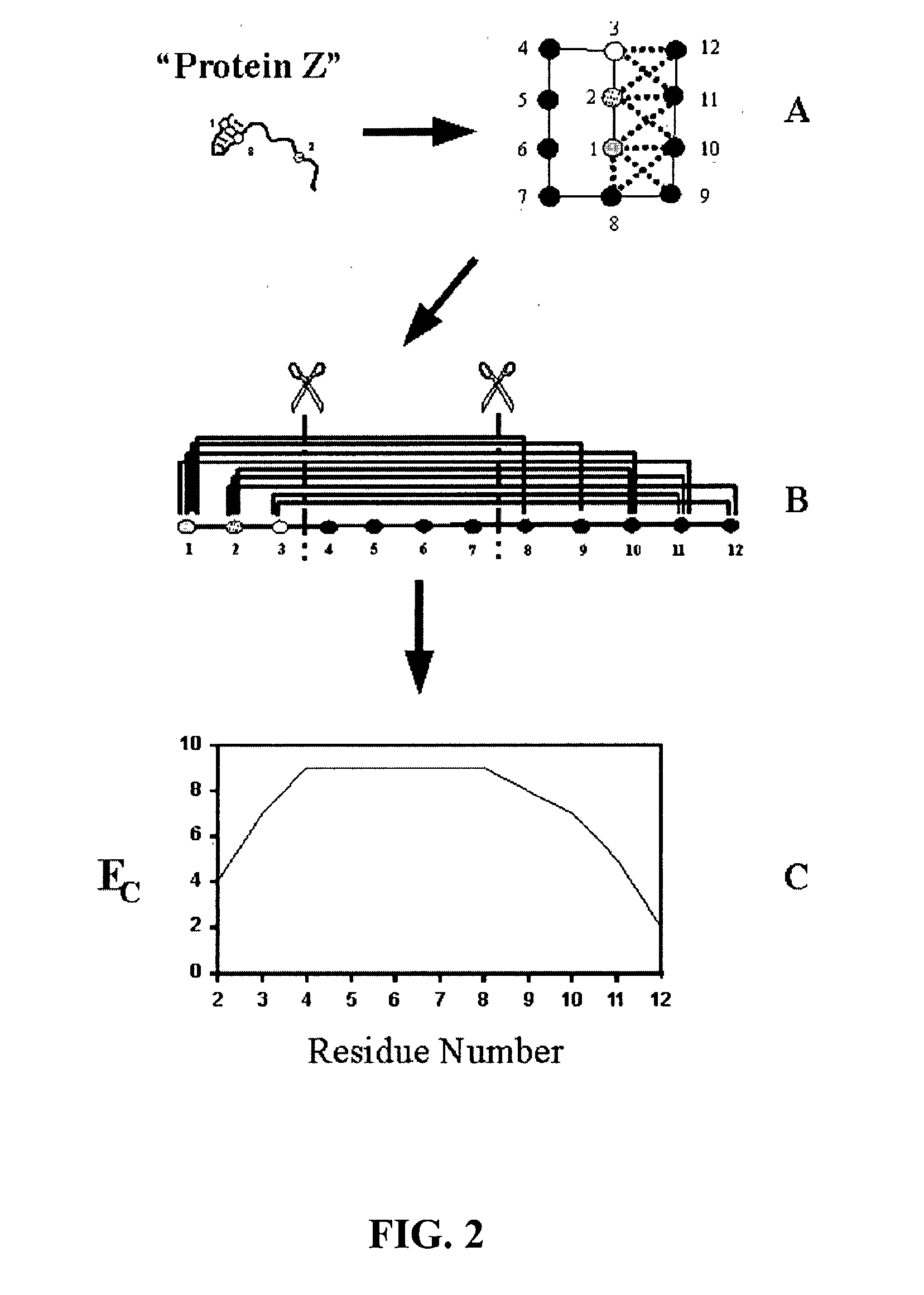
The invention relates to improved methods for directed evolution of polymers, including directed evolution of nucleic acids and proteins. Specifically, the methods of the invention include analytical methods for identifying "crossover locations" in a polymer. Crossovers at these locations are less likely to disrupt desirable properties of the protein, such as stability or functionality. The invention further provides improved methods for directed evolution wherein the polymer is selectively recombined at the identified "crossover locations". Crossover disruption profiles can be used to identify preferred crossover locations. Structural domains of a biopolymer can also be identified and analyzed, and domains can be organized into schema. Schema disruption profiles can be calculated, for example based on conformational energy or interatomic distances, and these can be used to identify preferred or candidate crossover locations. Computer systems for implementing analytical methods of the invention are also provided.
Owner:CALIFORNIA INST OF TECH
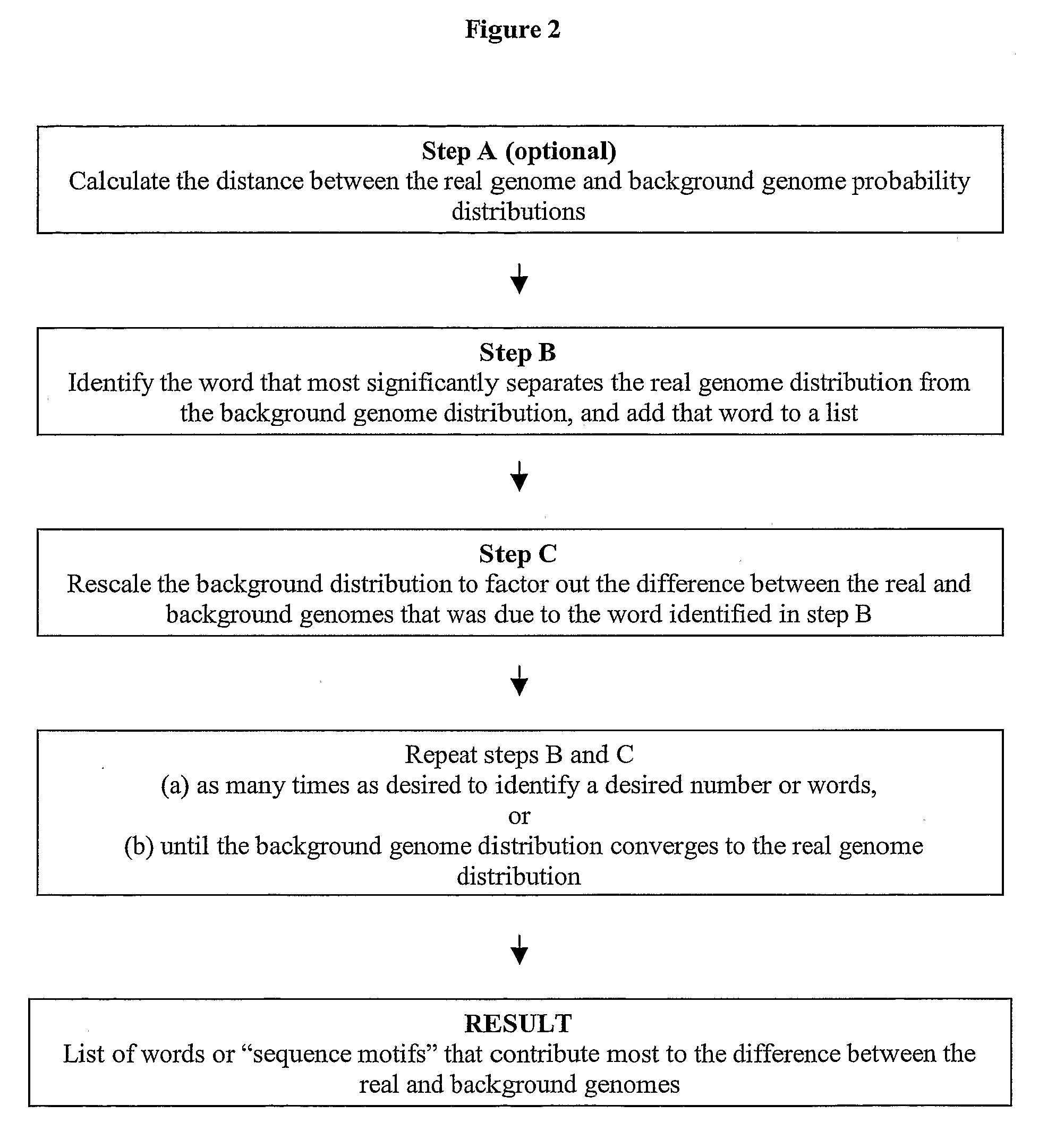
Methods for identifying sequence motifs, and applications thereof
InactiveUS20090208955A1Reduce in quantityIncrease the number ofMicrobiological testing/measurementProteomicsBinding siteInstability
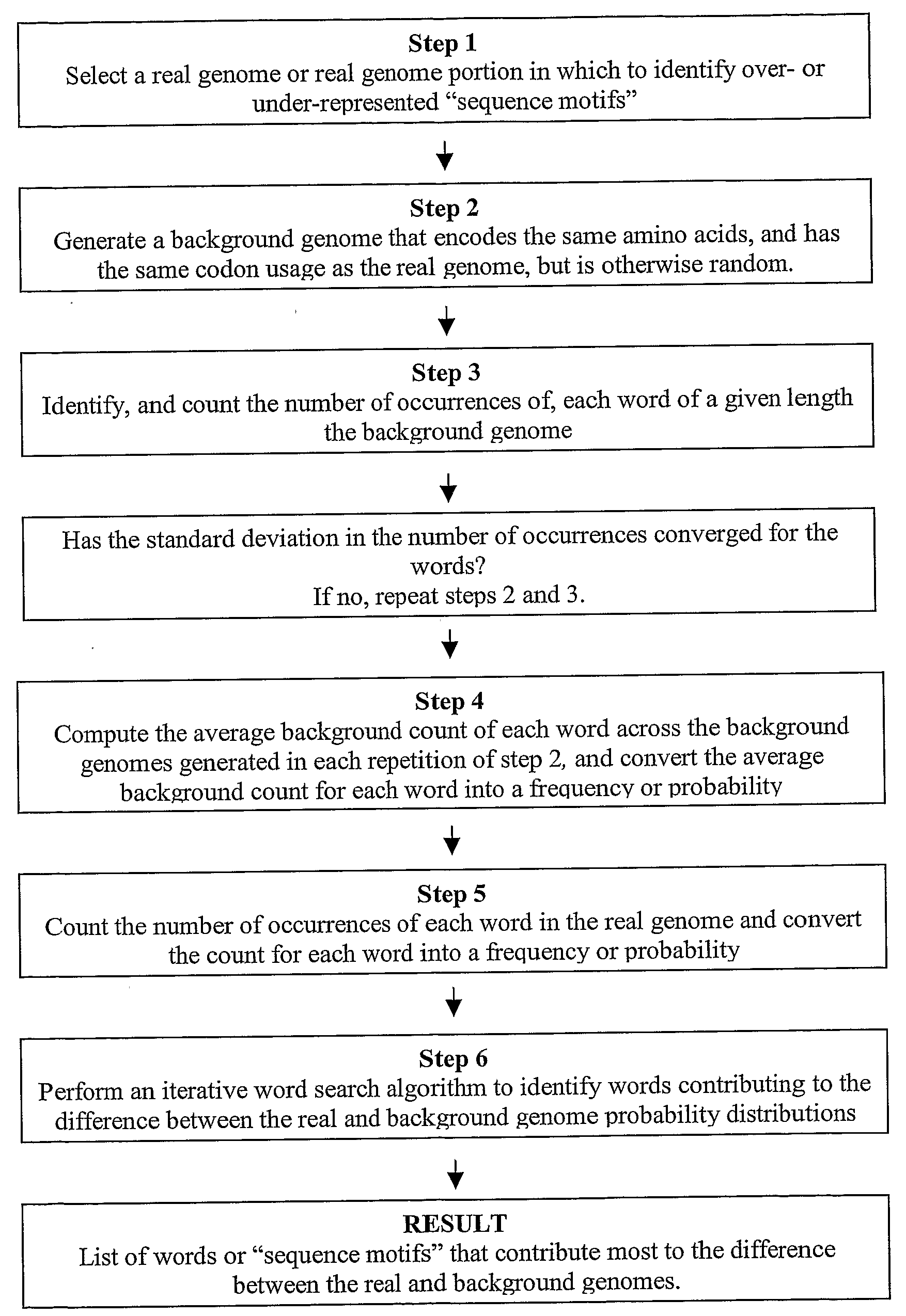
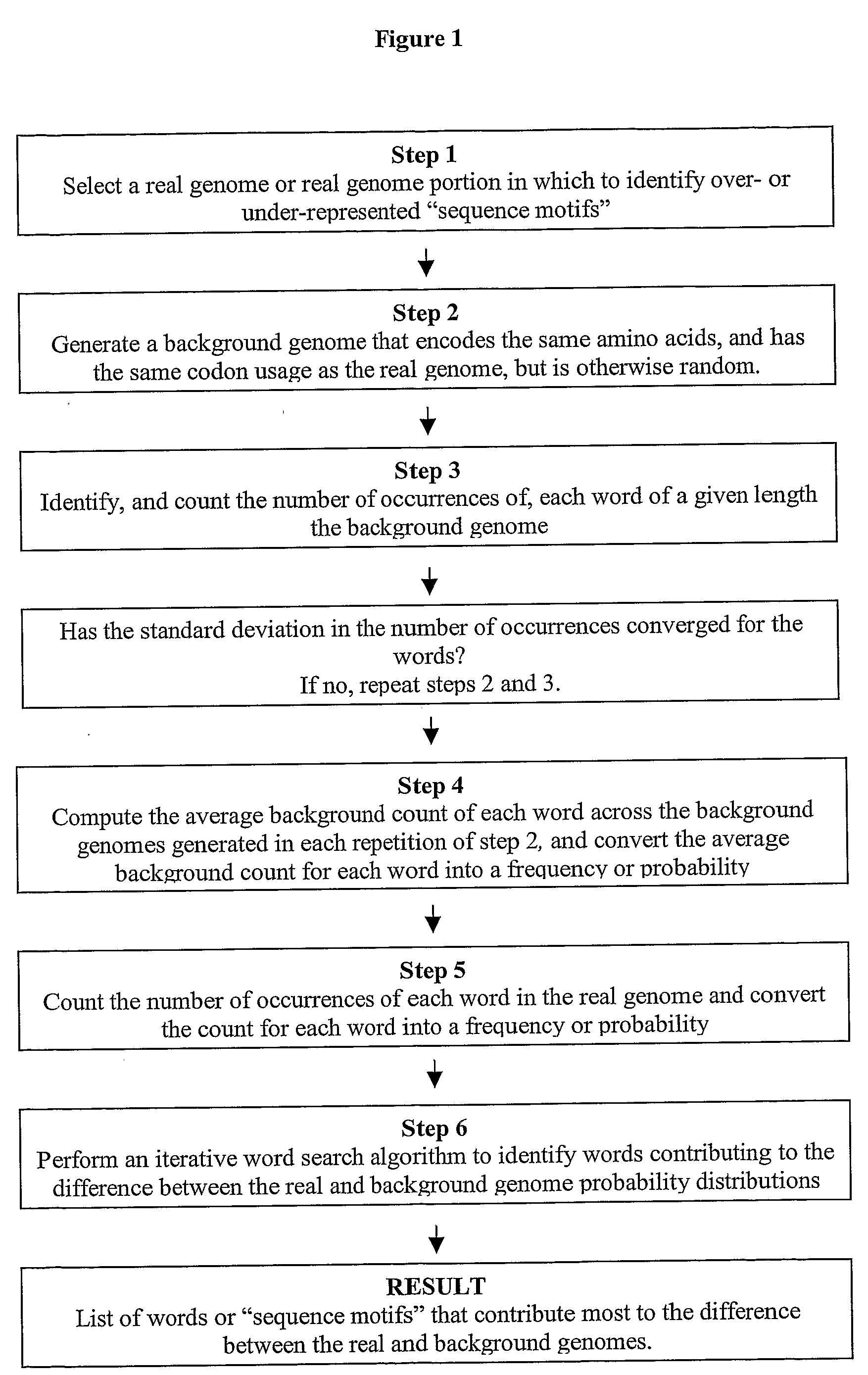
The present invention relates to methods and algorithms that can be used to identify sequence motifs that are either under- or over-represented in a given nucleotide sequence as compared to the frequency of those sequences that would be expected to occur by chance, or that are either under- or over-represented as compared to the frequency of those sequences that occur in other nucleotide sequences, and to methods of scoring sequences based on the occurrence of these sequence motifs. Such sequence motifs may be biologically significant, for example they may constitute transcription factor binding sites, mRNA stability / instability signals, epigenetic signals, and the like. The methods of the invention can also be used, inter alia, to classify sequences or organisms in terms of their phylogenetic relationships, or to identify the likely host of a pathogenic organism. The methods of the present invention can also be used to optimize expression of proteins.
Owner:INST FOR ADVANCED STUDY
Method and system using systematically varied data libraries
InactiveUS7873477B1Reduce the amount requiredProteomicsBiological testingComputer scienceData library
Owner:CODEXIS MAYFLOWER HLDG LLC
Method for molecular genealogical research
A genealogical research and record keeping system and method for identifying commonalities in haplotypes and other genetic characteristics of two or more individual members of a biological sample. Chromosomal fragments identical by descent identify family ties between siblings, parents and children and ancestors and progeny across many generations. It is particularly useful in corroborating and improving the accuracy of genealogical data, and identifying previously unknown genetic relationships.
Owner:ANCESTRY COM DNA
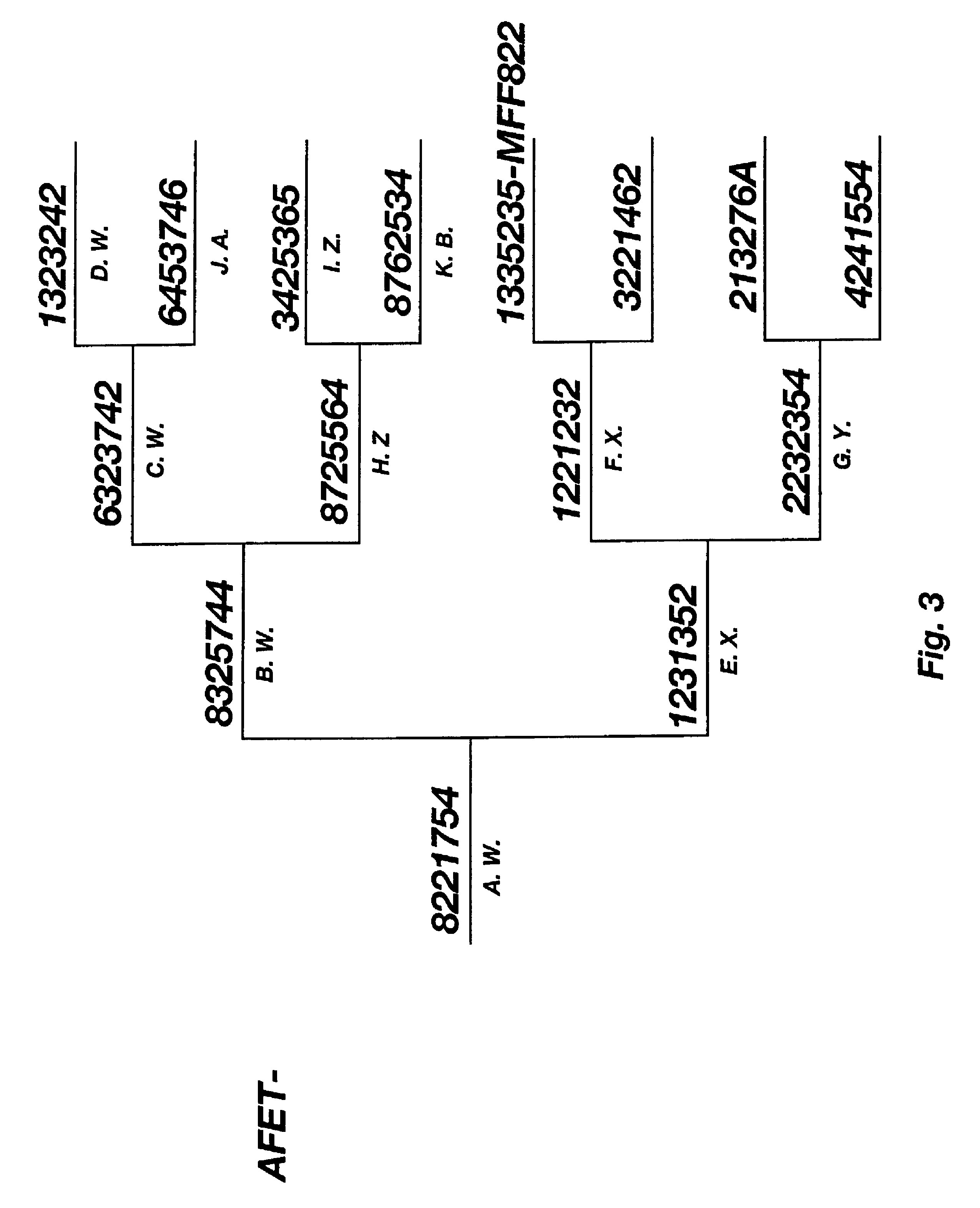
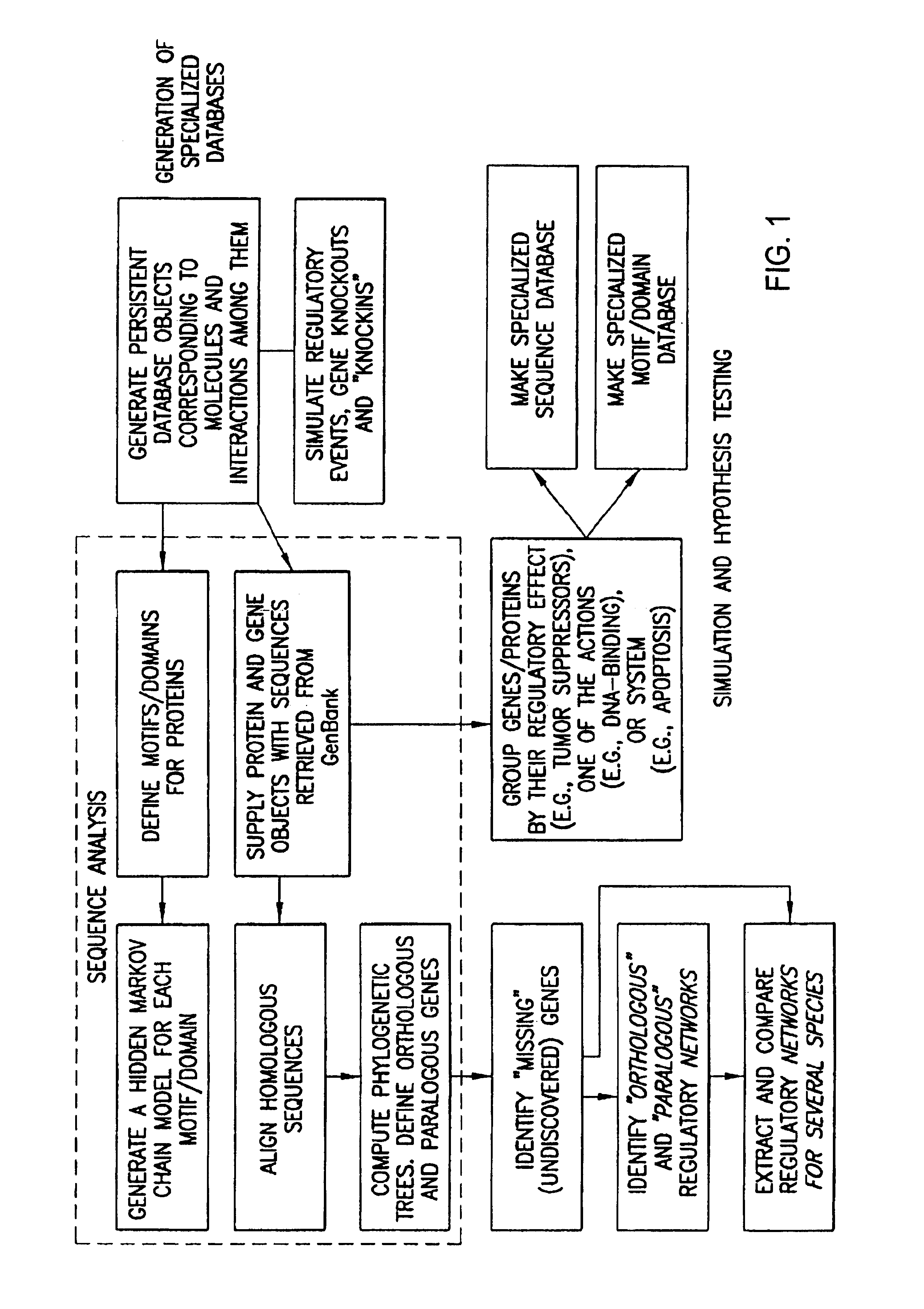
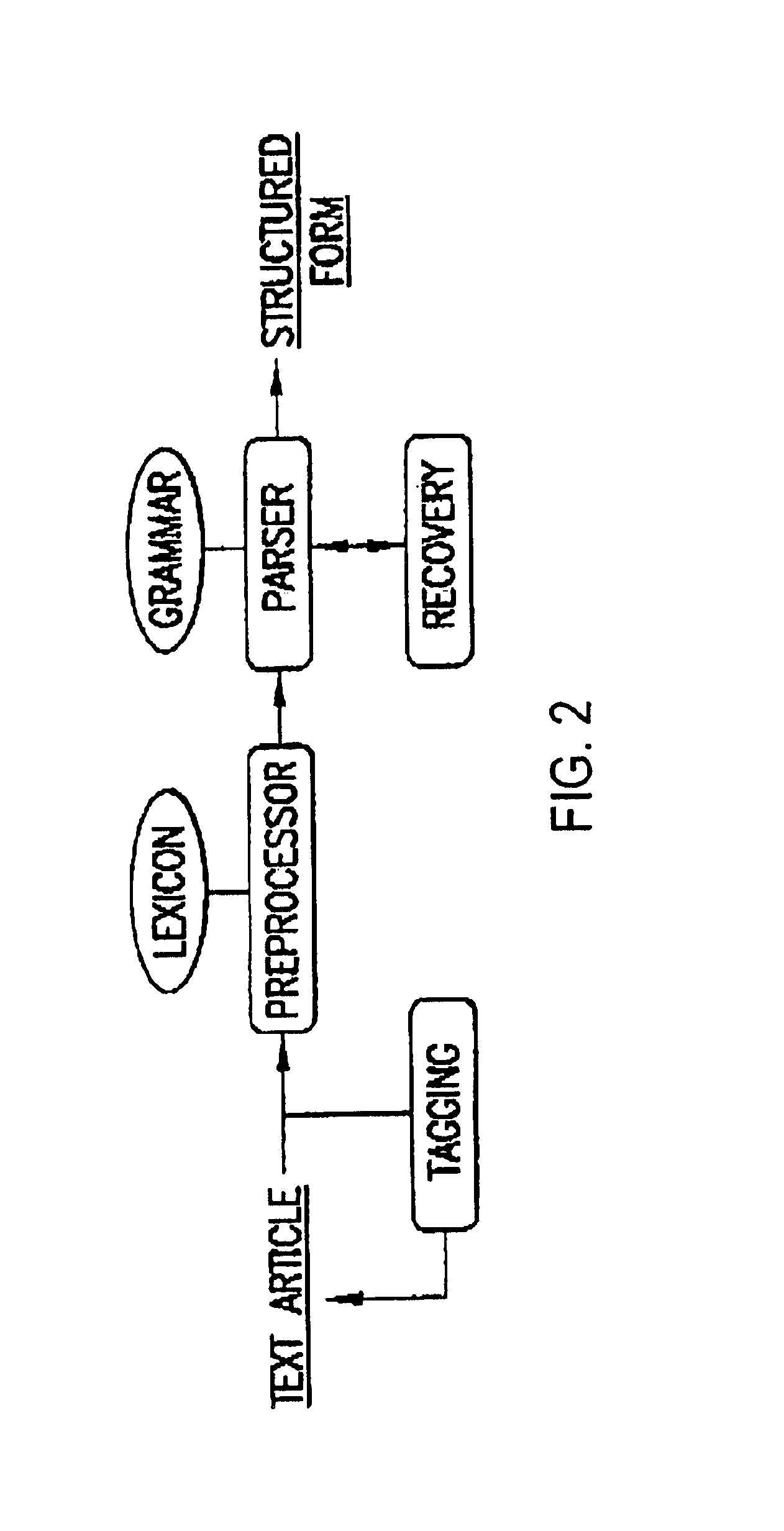
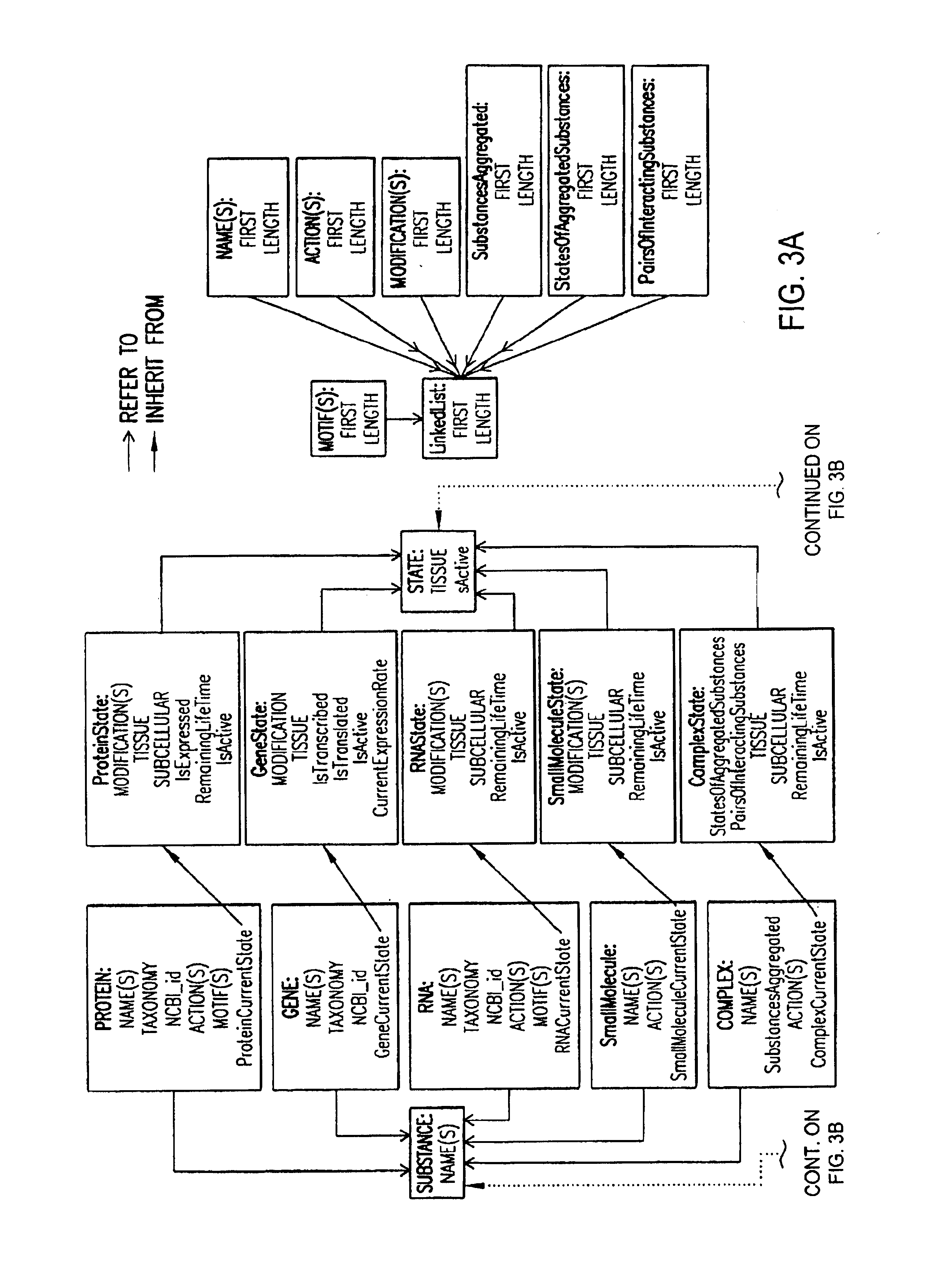
Methods for extracting information on interactions between biological entities from natural language text data
The present invention relates to methods for identifying novel genes comprising: (i) generating one and / or more specialized databases containing information on gene / protein structure, function and / or regulatory interactions; and (ii) searching the specialized databases for homology or for a particular motif and thereby identifying a putative novel gene of interest. The invention may further comprise performing simulation and hypothesis testing to identify or confirm that the putative gene is a novel gene of interest. The present invention also relates to natural language processing and extraction of relational information associated with genes and proteins that are found in genomics journal articles. To enable access to information in textual form, the natural language processing system of the present invention provides a method for extracting and structuring information found in the literature in a form appropriate for subsequent applications.
Owner:THE TRUSTEES OF COLUMBIA UNIV IN THE CITY OF NEW YORK
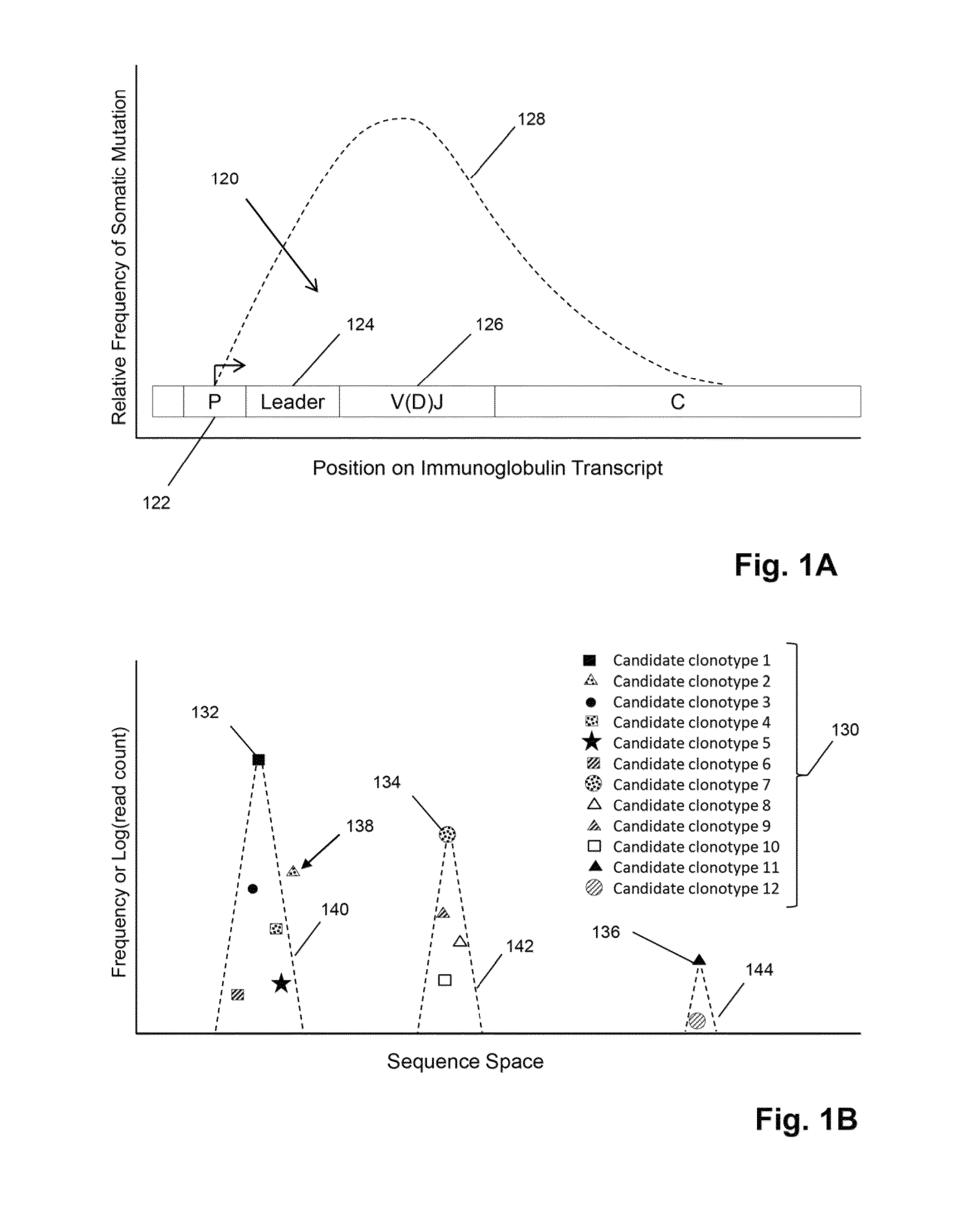
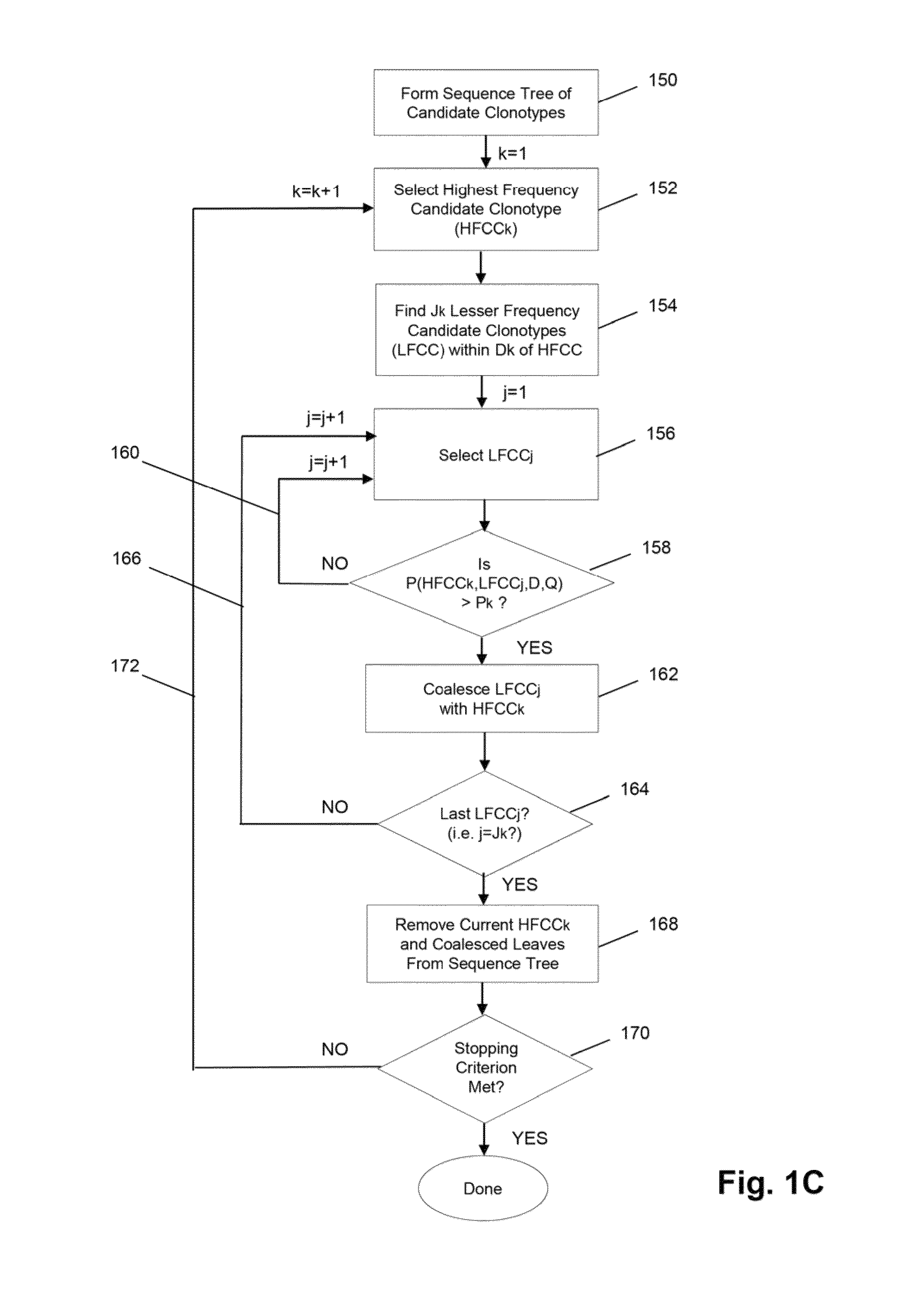
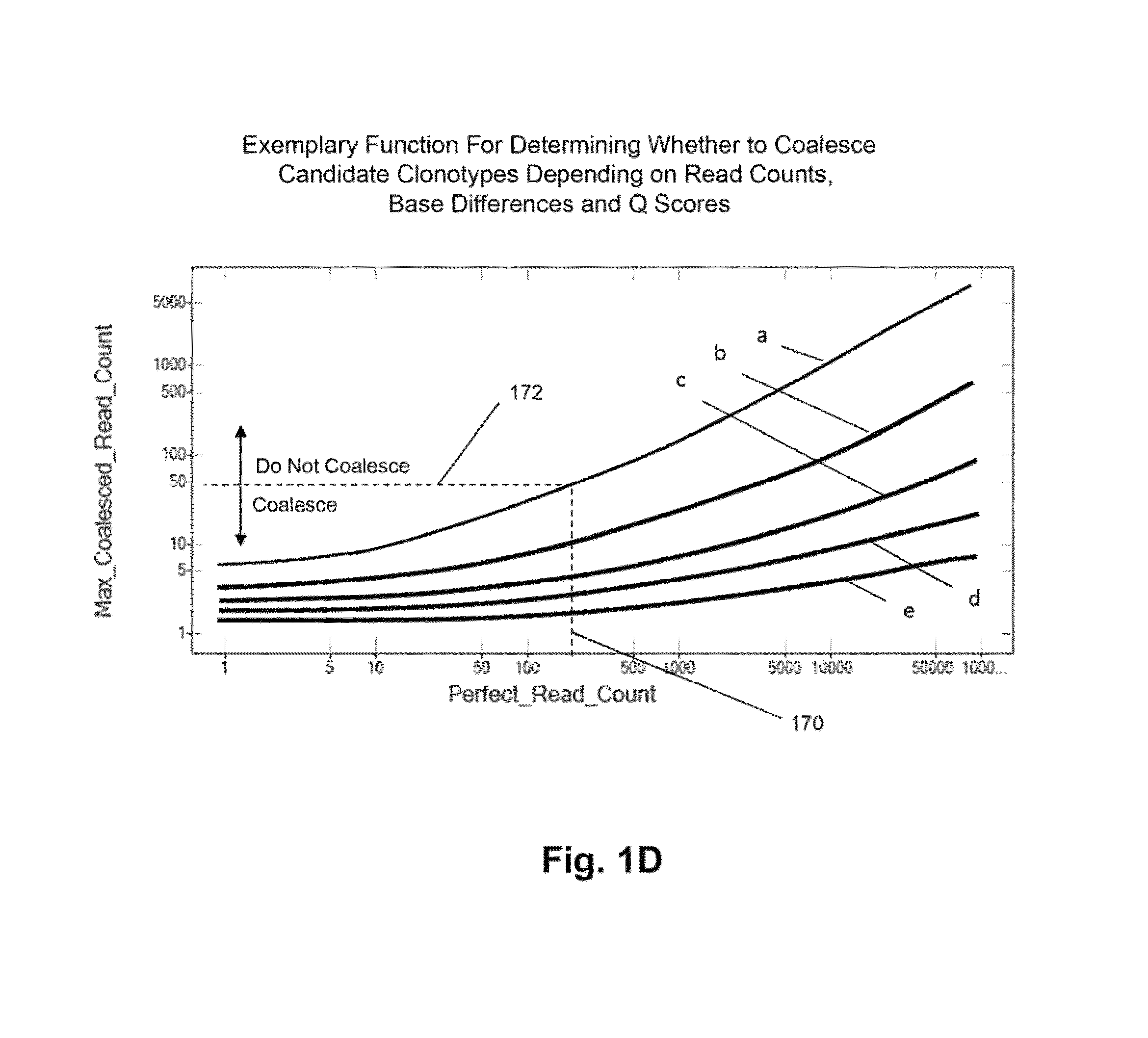
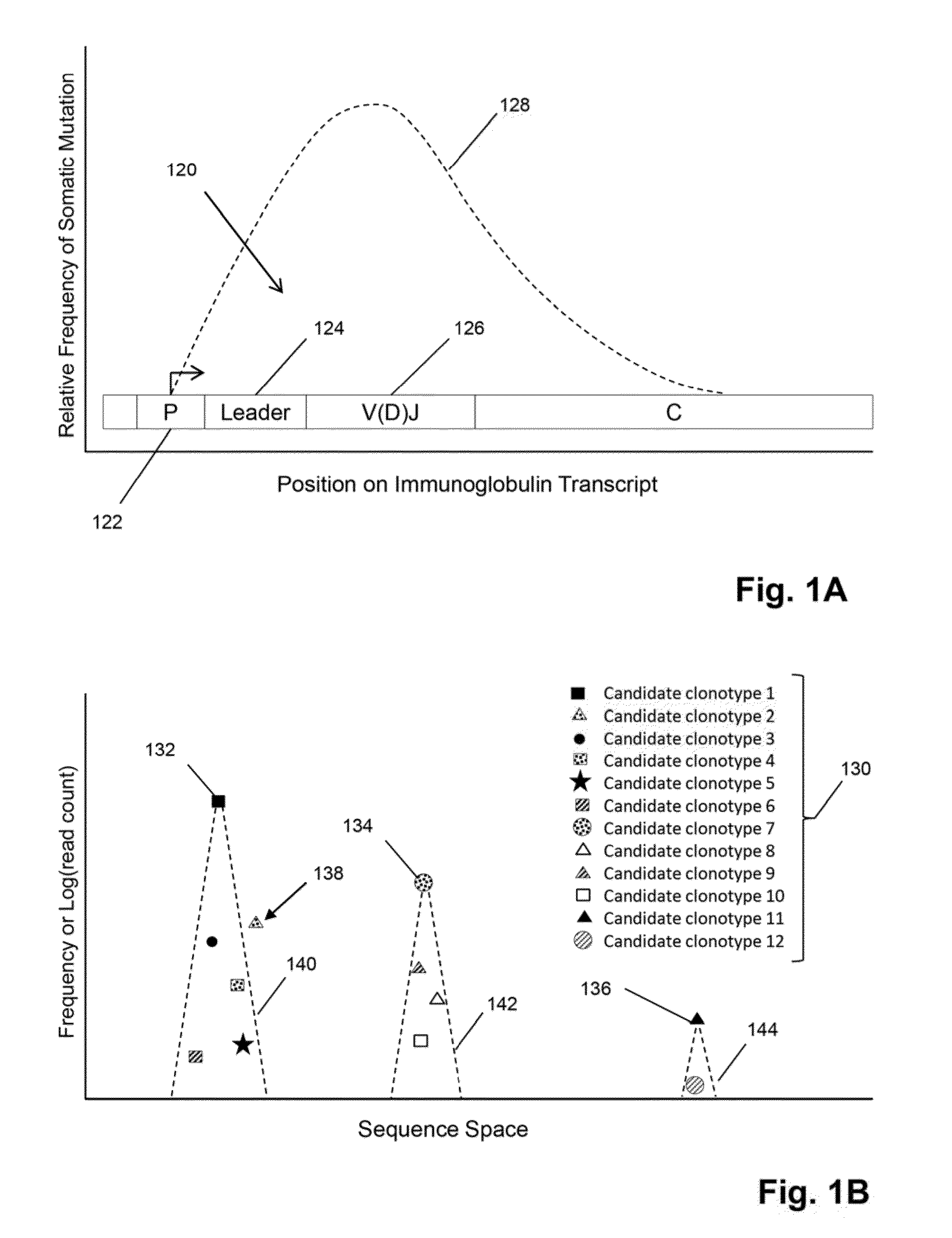
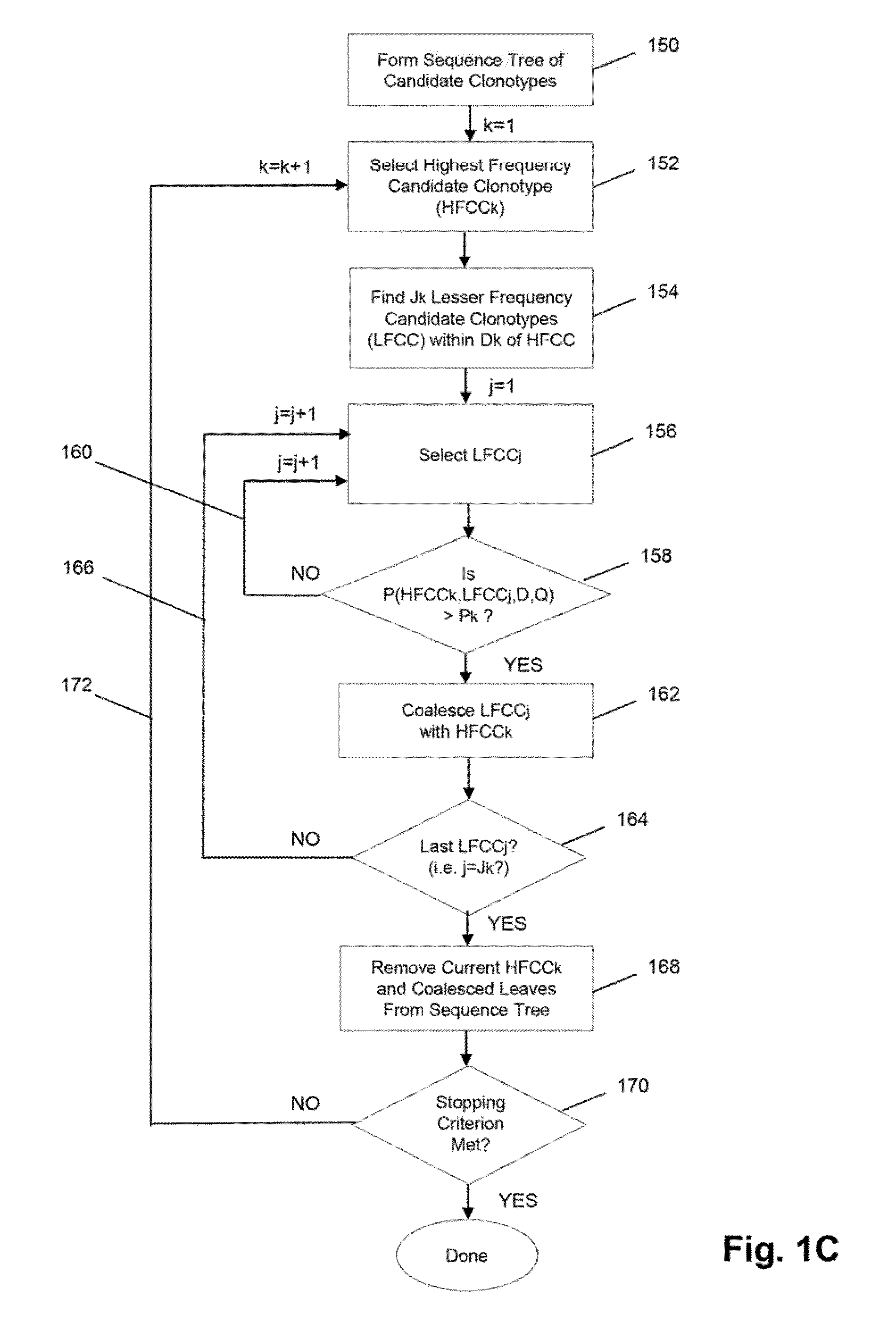
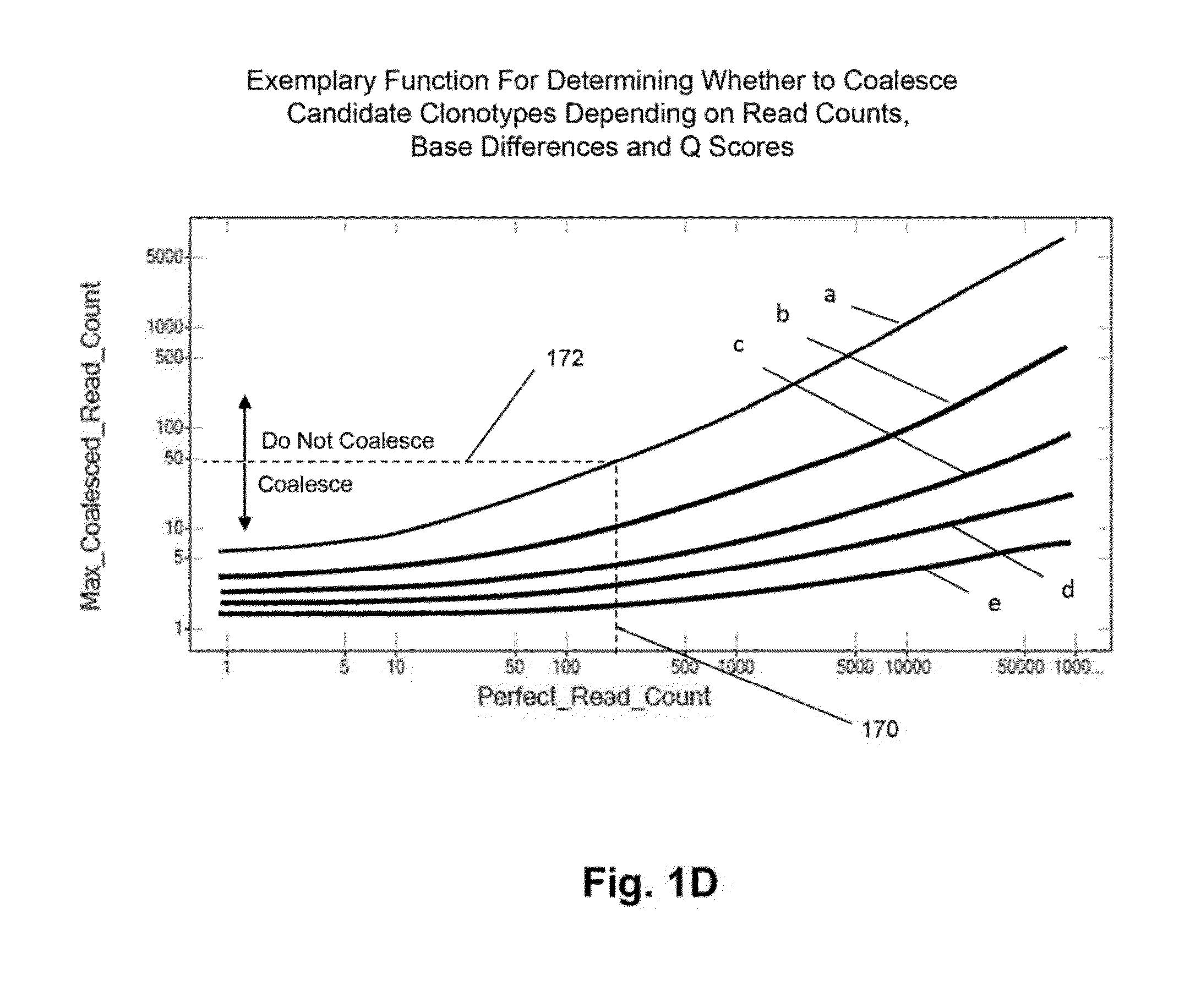
Method of determining clonotypes and clonotype profiles
ActiveUS9043160B1Efficiently carrying out sequence comparisonQuick sortingDigital data processing detailsMicrobiological testing/measurementObservational errorNucleic acid sequencing
The invention is directed to methods for determining clonotypes and clonotype profiles in assays for analyzing immune repertoires by high throughput nucleic acid sequencing of somatically recombined immune molecules. In one aspect, the invention comprises generating a clonotype profile from an individual by generating sequence reads from a sample of recombined immune molecules; forming from the sequence reads a sequence tree representing candidate clonotypes each having a frequency; coalescing with a highest frequency candidate clonotype any lesser frequency candidate clonotypes whenever such lesser frequency is below a predetermined value and whenever a sequence difference therebetween is below a predetermined value to form a clonotype. After such coalescence, the candidate clonotypes is removed from the sequence tree and the process is repeated. This approach permits rapid and efficient differentiation of candidate clonotypes with genuine sequence differences from those with experimental or measurement errors, such as sequencing errors.
Owner:ADAPTIVE BIOTECH
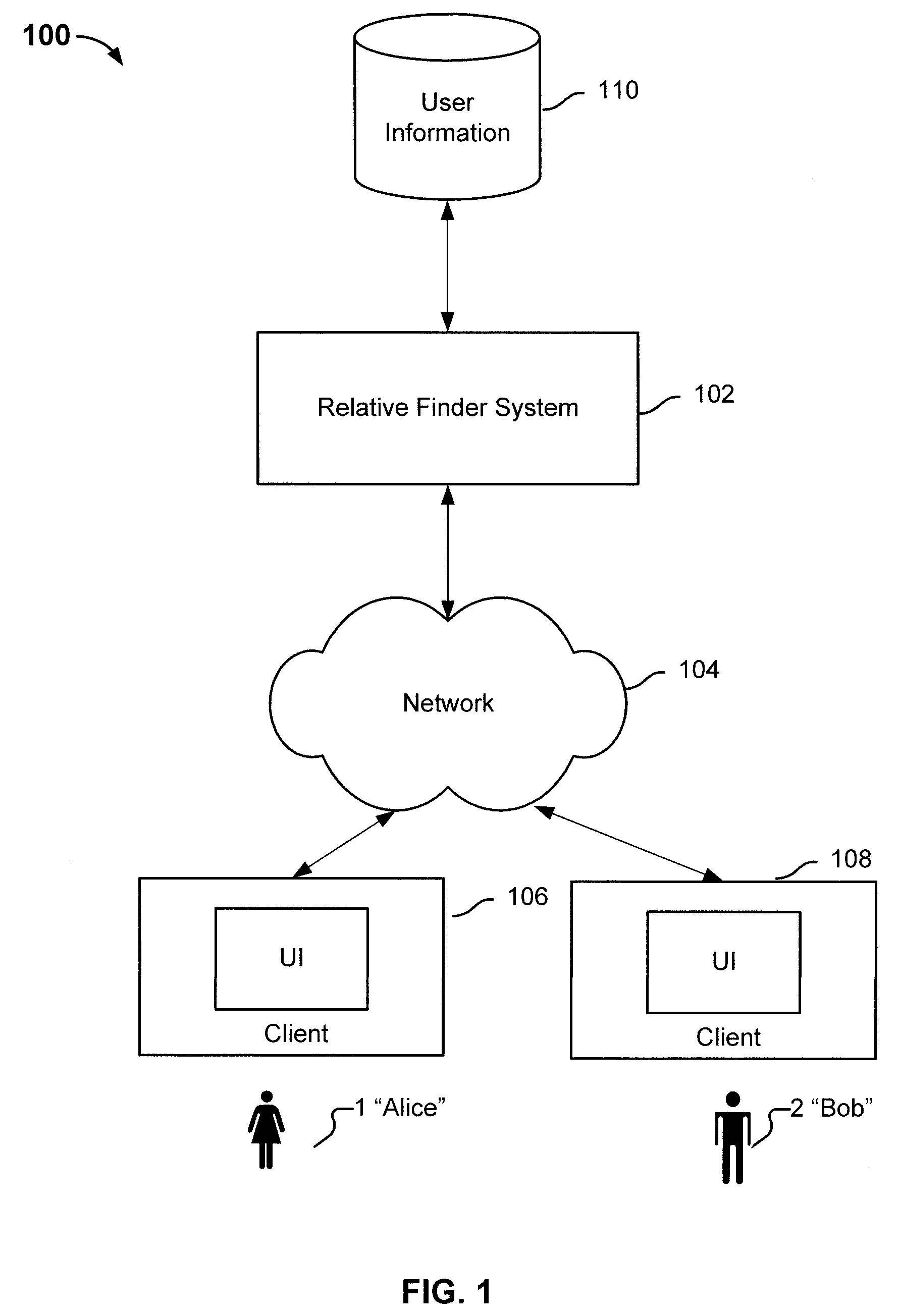
Finding relatives in a database
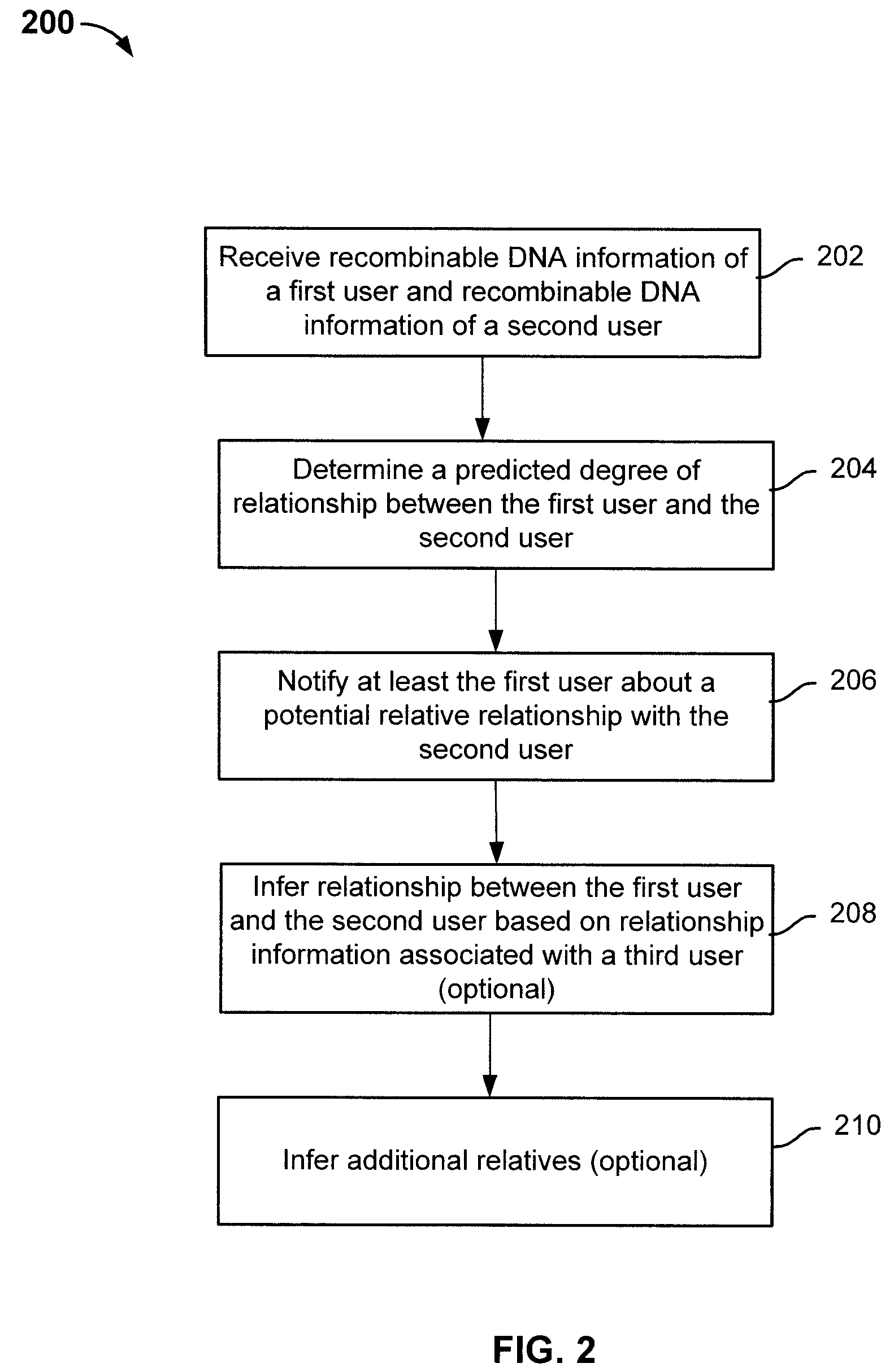
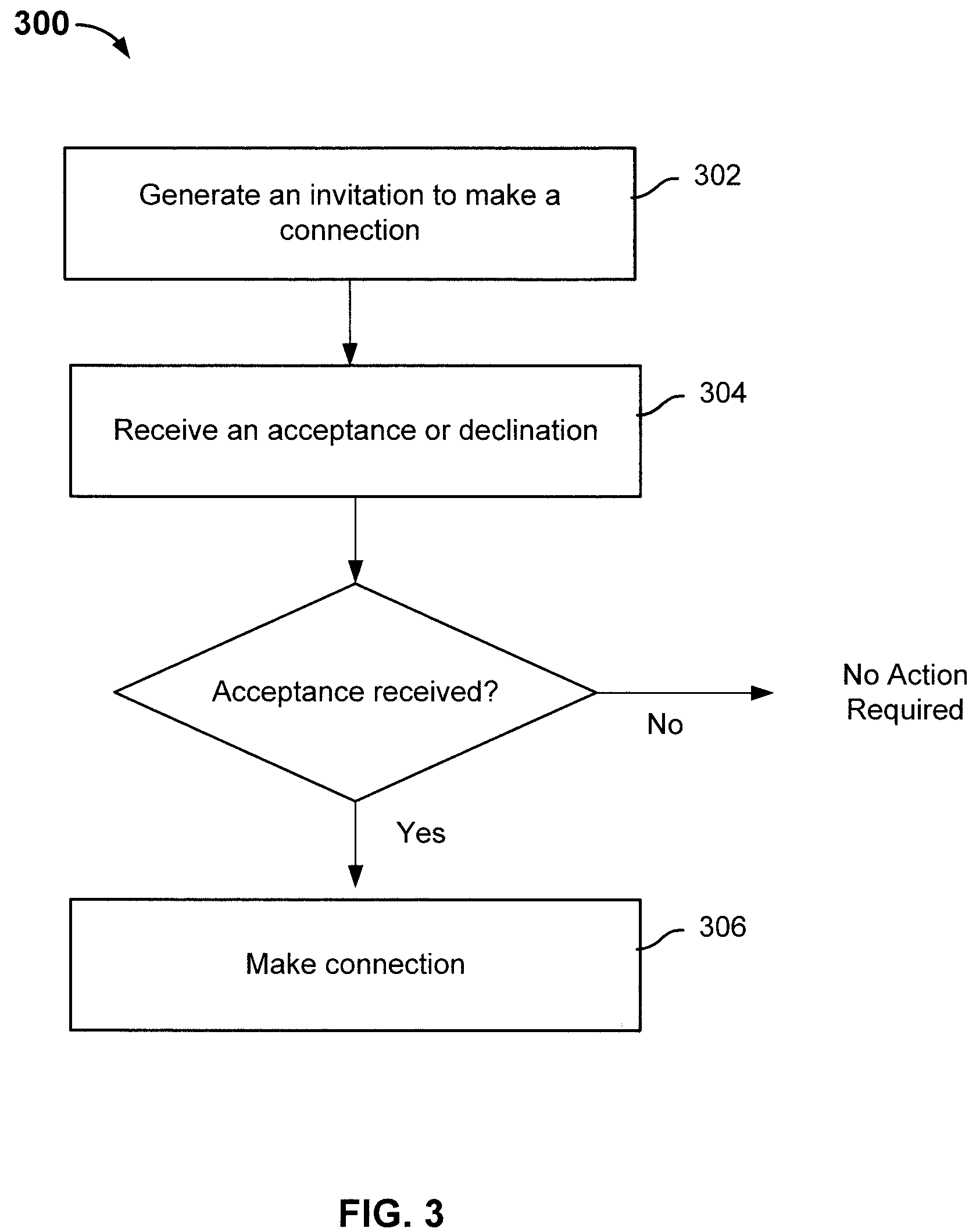
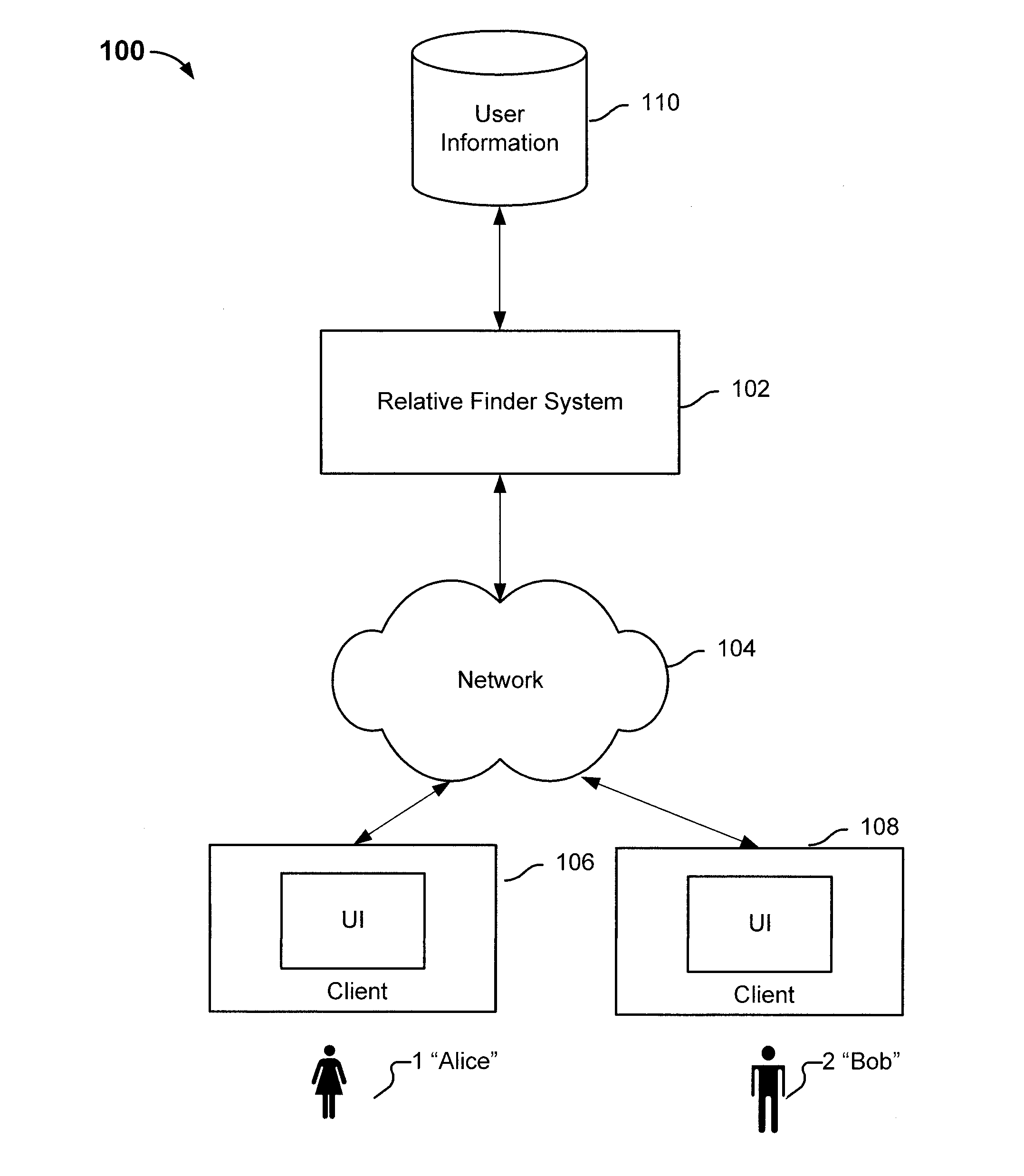
ActiveUS8463554B2Digital data processing detailsAnalogue computers for chemical processesHuman–computer interactionRecombinant DNA
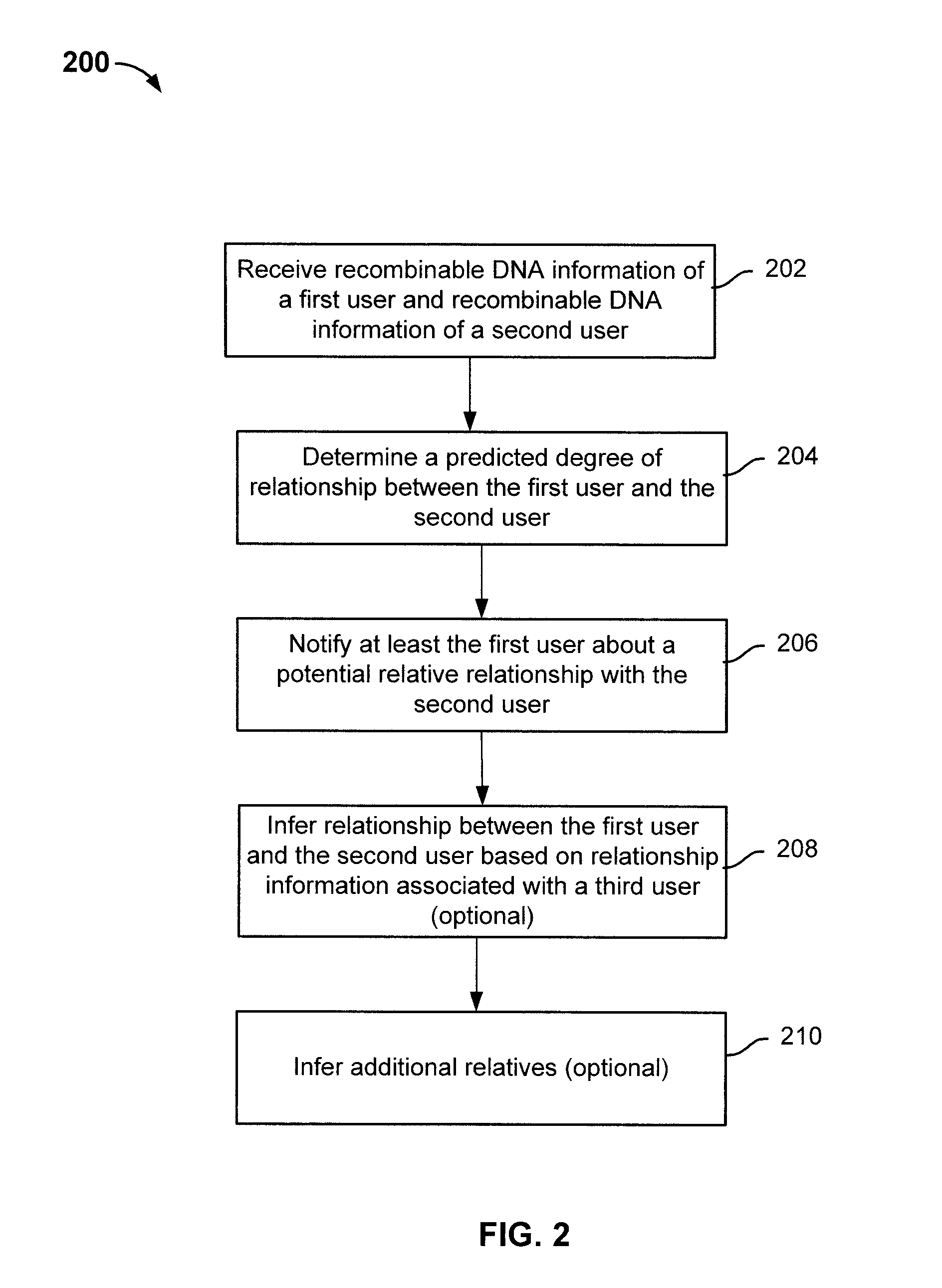
Determining relative relationship includes receiving recombinable deoxyribonucleic acid (DNA) information of a first user and recombinable DNA information of a second user, determining, based at least in part on the recombinable DNA information of the first user and recombinable DNA information of the second user, a predicted degree of relationship between the first user and the second user, and in the event that the expected degree of relationship between the first user and the second user at least meets the threshold, notifying at least the first user about a relative relationship with the second user.
Owner:23ANDME
Method of determining clonotypes and clonotype profiles
ActiveUS20150167080A1Quick sortingEfficiently carrying out sequence comparisonsMicrobiological testing/measurementLibrary member identificationAssayNucleic acid sequencing
Owner:ADAPTIVE BIOTECH
Display device and method for driving the same
ActiveUS20120229431A1Accurate detectionAdvertisingCathode-ray tube indicators3d imageAttention Concentration
In order to display 3D images by a parallax barrier method, a display screen and the eyes of a viewer need to have a specific positional relation. An object is to provide a display device with an extended area where the viewer can perceive 3D images with the naked eye. Attention is focused on the position of the viewer with respect to pixels provided in a display device and a mode of a parallax barrier provided between the viewer and the pixels. This leads to a structure in which the position of the viewer with respect to pixels is specified by using an ultrasonic wave to change a mode of a parallax barrier in accordance with the position of the viewer, thereby achieving the above object.
Owner:SEMICON ENERGY LAB CO LTD
Gene recombination and hybrid protein development
InactiveUS20030032059A1Preserve stability and desired propertyComputationally tractableBiological testingSequence analysisBiopolymerProtein insertion
The invention relates to improved methods for directed evolution of polymers, including directed evolution of nucleic acids and proteins. Specifically, the methods of the invention include analytical methods for identifying "crossover locations" in a polymer. Crossovers at these locations are less likely to disrupt desirable properties of the protein, such as stability or functionality. The invention further provides improved methods for directed evolution wherein the polymer is selectively recombined at the identified "crossover locations". Crossover disruption profiles can be used to identify preferred crossover locations. Structural domains of a biopolymer can also be identified and analyzed, and domains can be organized into schema. Schema disruption profiles can be calculated, for example based on conformational energy or interatomic distances, and these can be used to identify preferred or candidate crossover locations. Computer systems for implementing analytical methods of the invention are also provided.
Owner:WANG ZHEN GANG +3
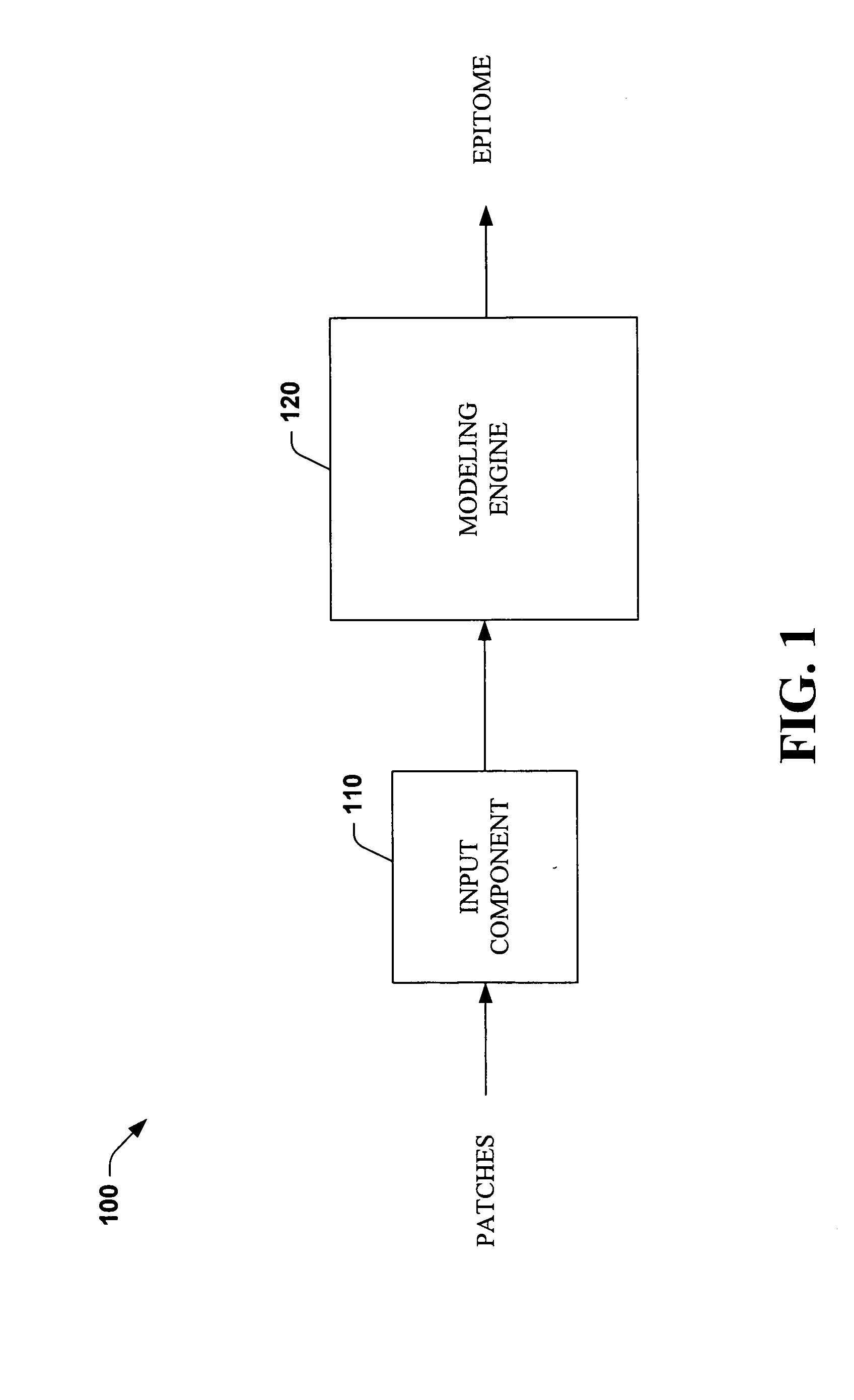
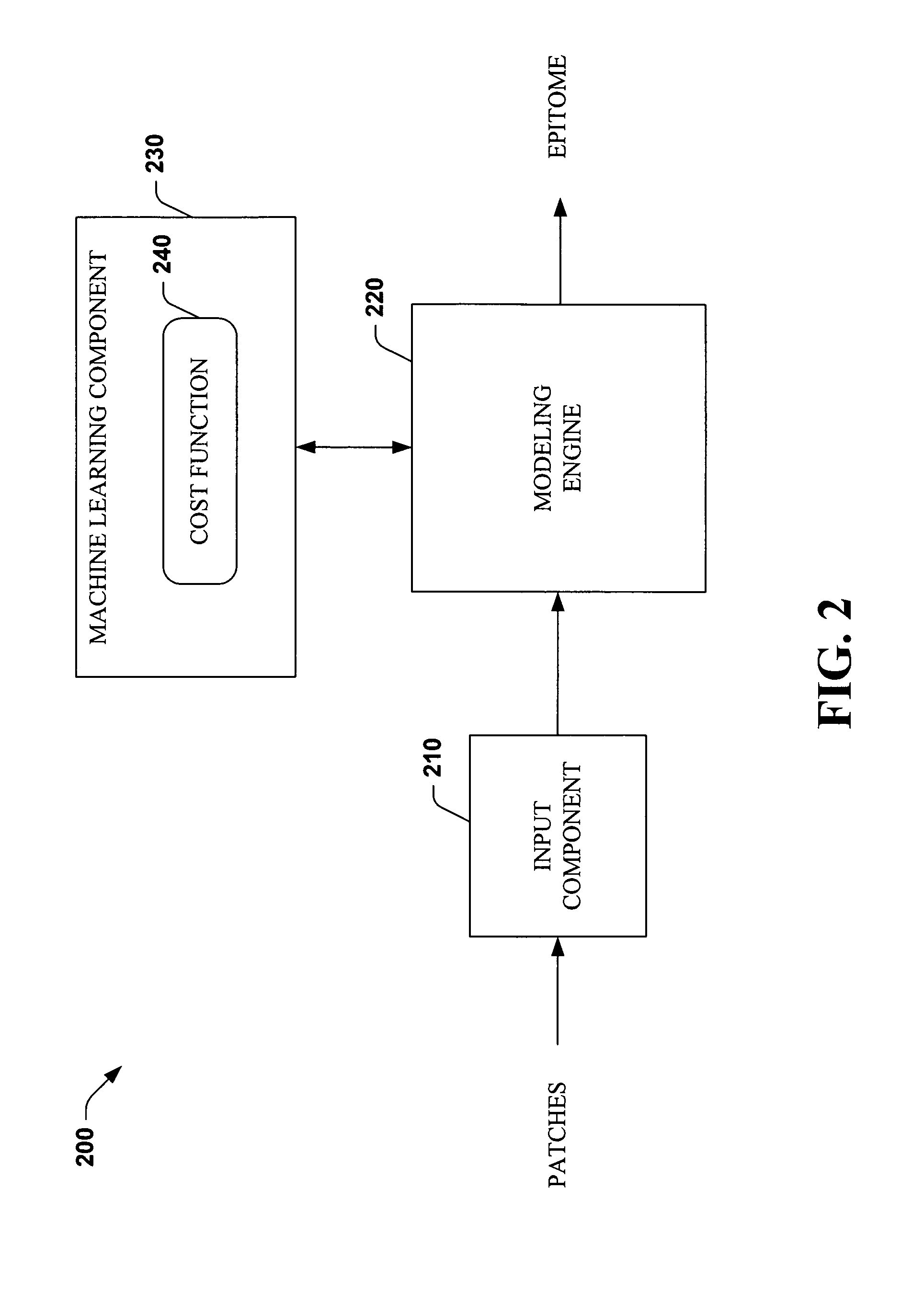
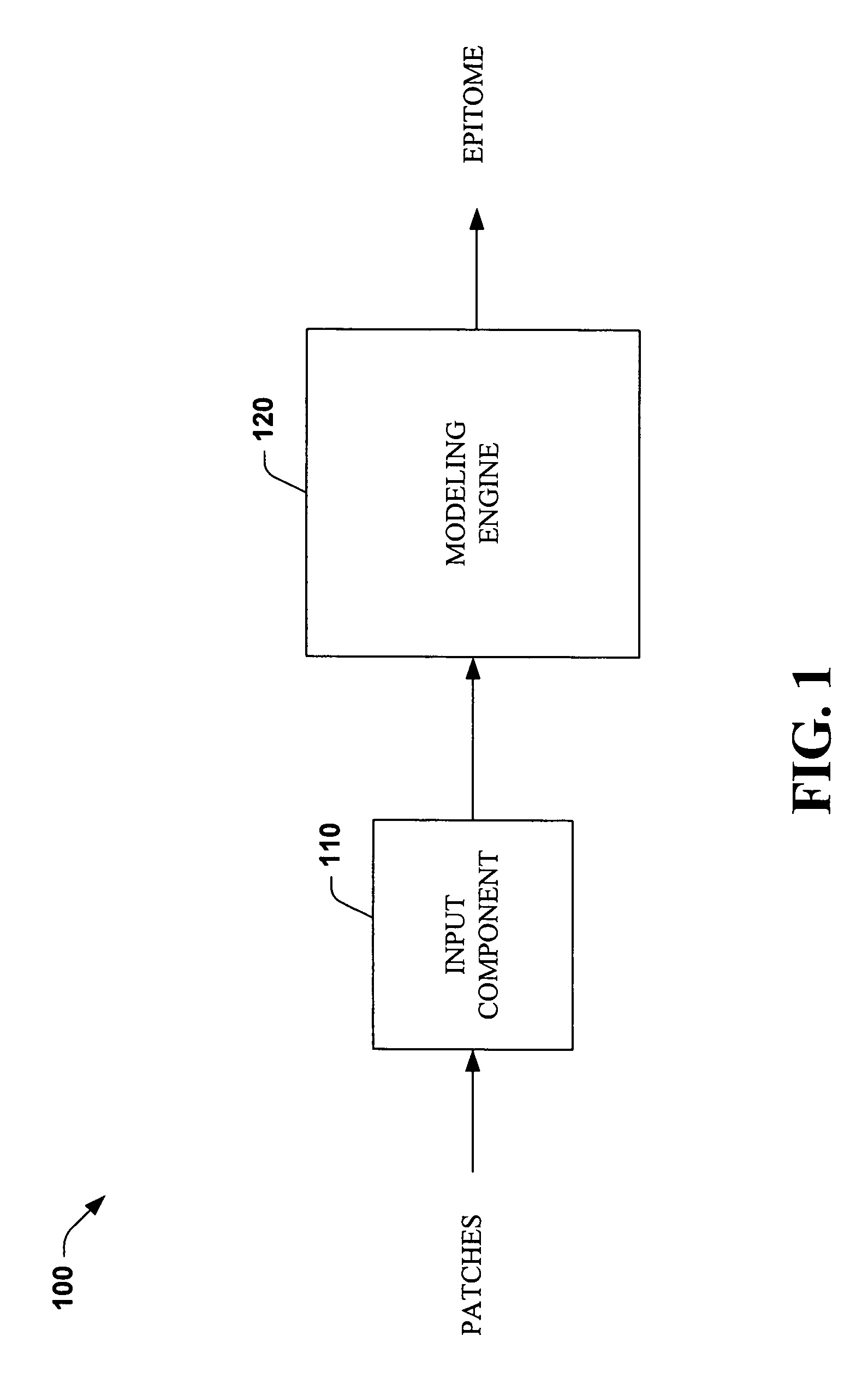
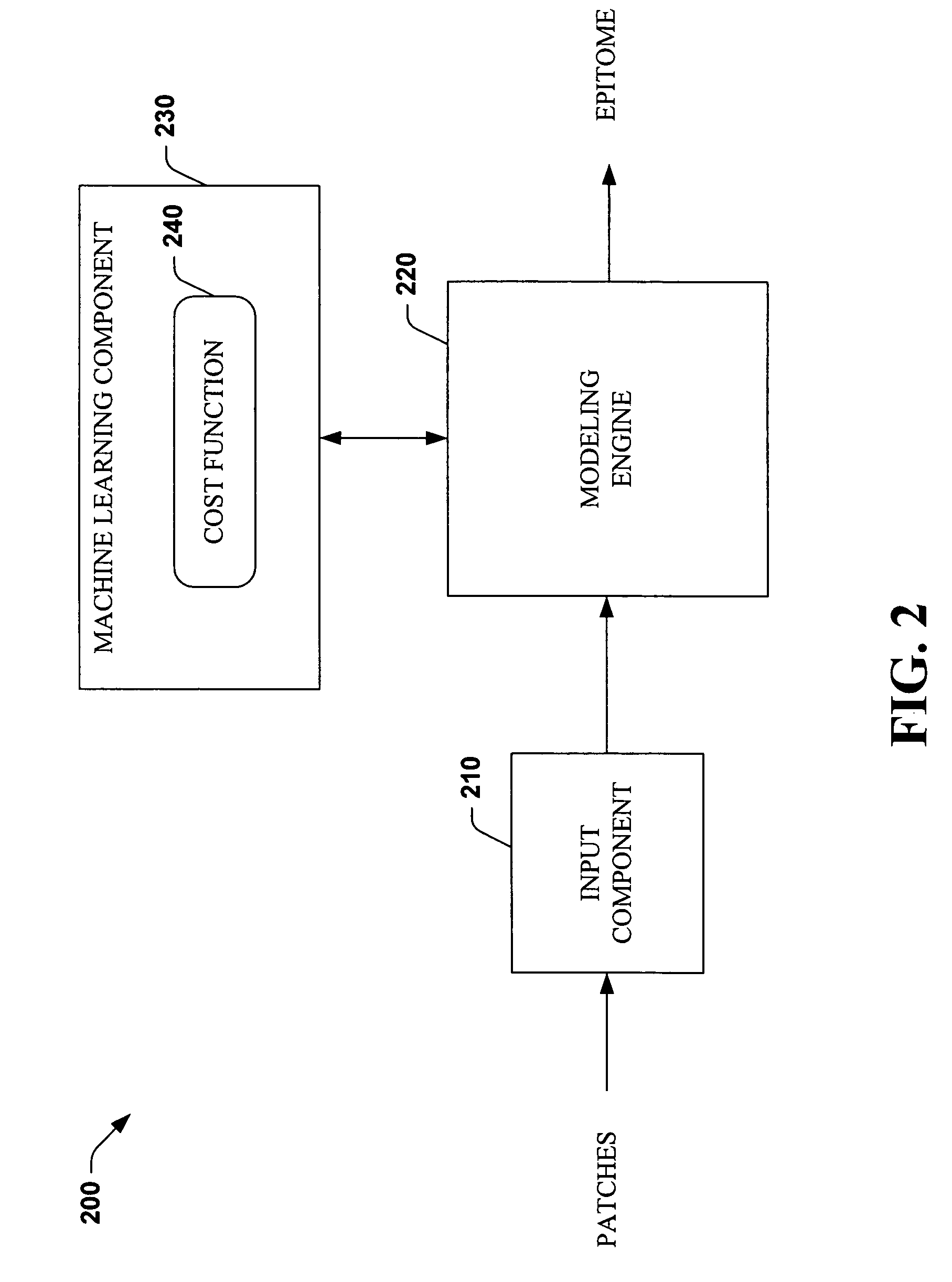
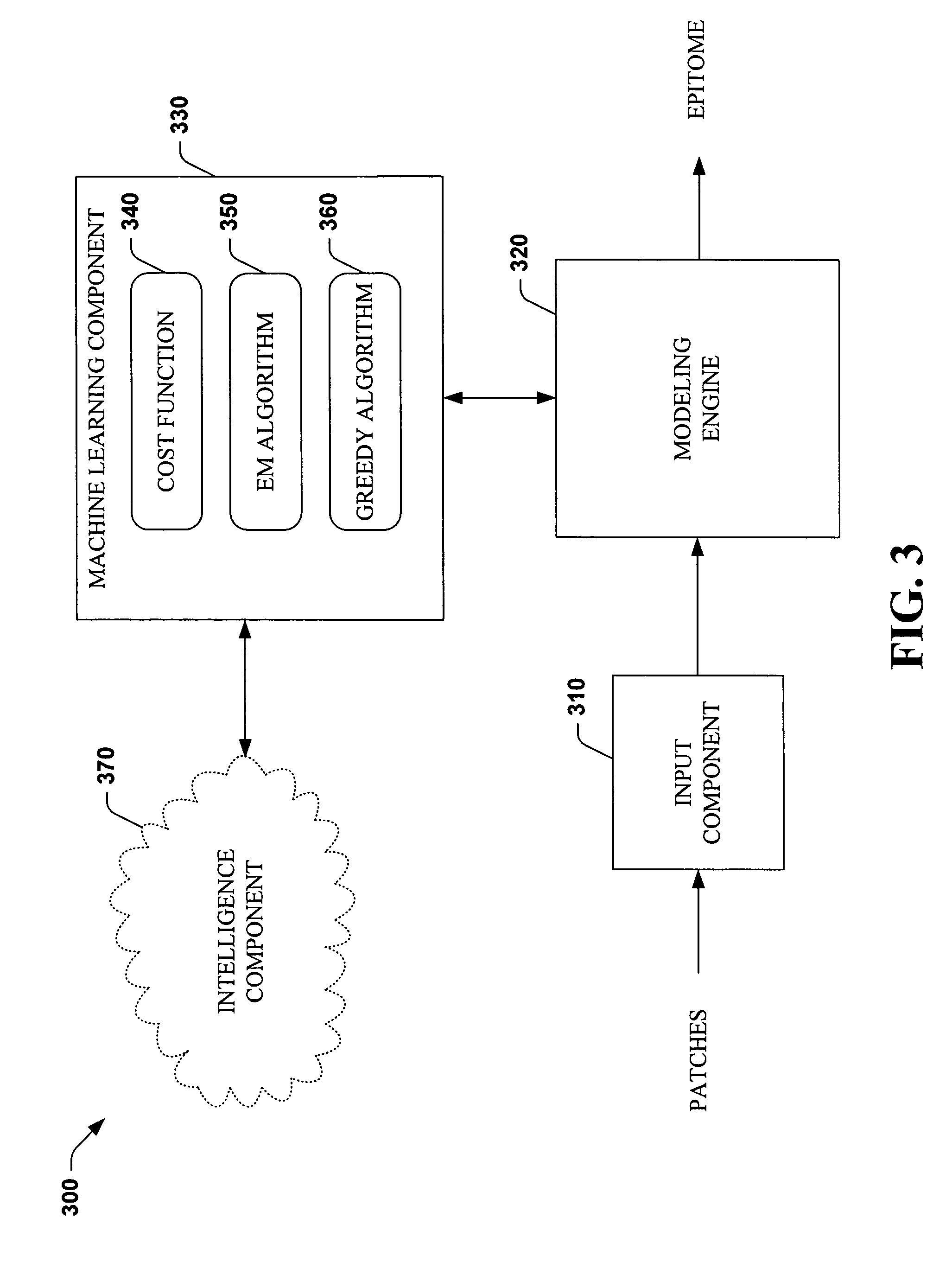
Systems and methods that utilize machine learning algorithms to facilitate assembly of aids vaccine cocktails
Owner:MICROSOFT TECH LICENSING LLC
Systems and methods that utilize machine learning algorithms to facilitate assembly of aids vaccine cocktails
The subject invention provides systems and methods that facilitate AIDS vaccine cocktail assembly via machine learning algorithms such as a cost function, a greedy algorithm, an expectation-maximization (EM) algorithm, etc. Such assembly can be utilized to generate vaccine cocktails for species of pathogens that evolve quickly under immune pressure of the host. For example, the systems and methods of the subject invention can be utilized to facilitate design of T cell vaccines for pathogens such HIV. In addition, the systems and methods of the subject invention can be utilized in connection with other applications, such as, for example, sequence alignment, motif discovery, classification, and recombination hot spot detection. The novel techniques described herein can provide for improvements over traditional approaches to designing vaccines by constructing vaccine cocktails with higher epitope coverage, for example, in comparison with cocktails of consensi, tree nodes and random strains from data.
Owner:MICROSOFT TECH LICENSING LLC
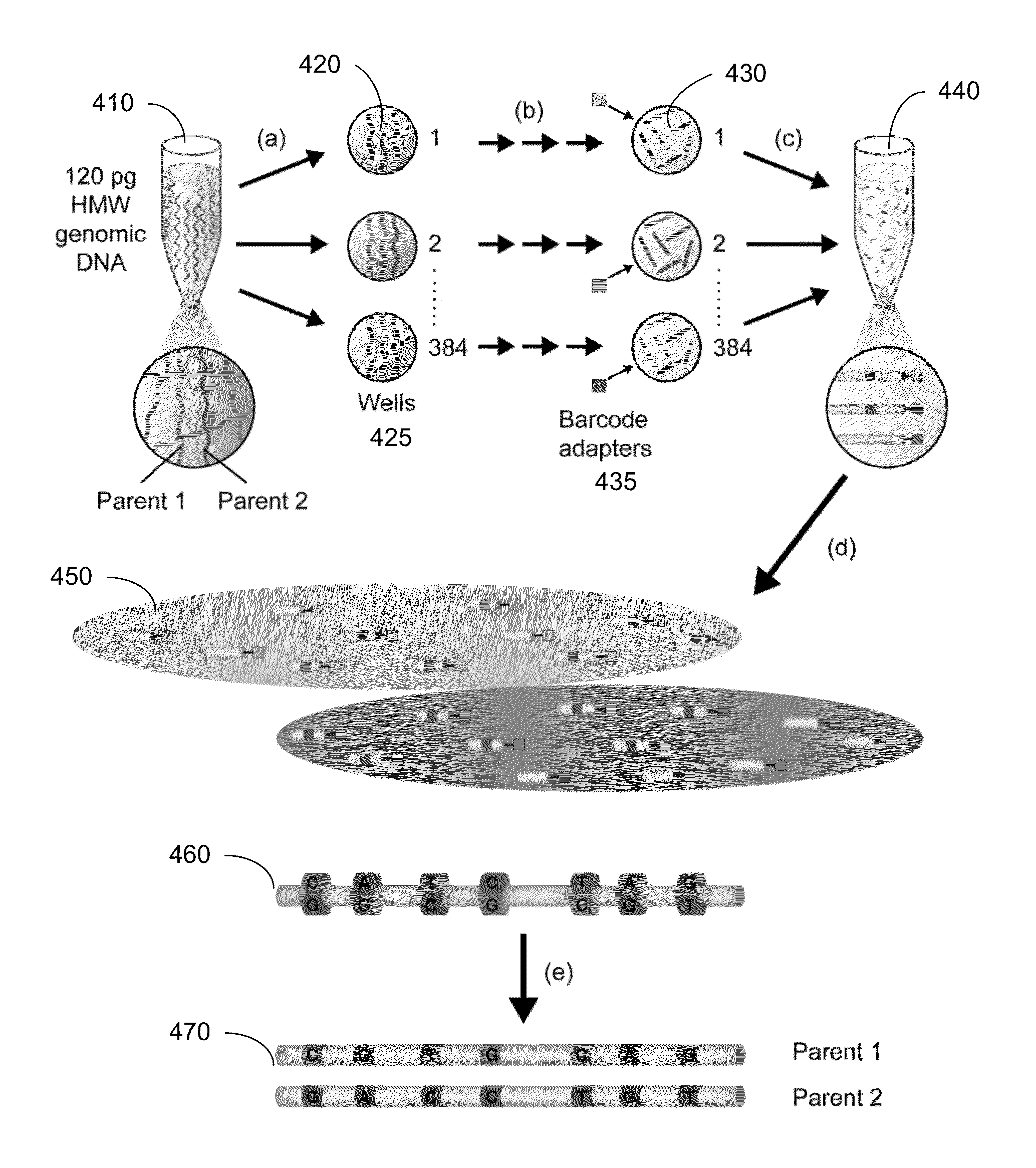
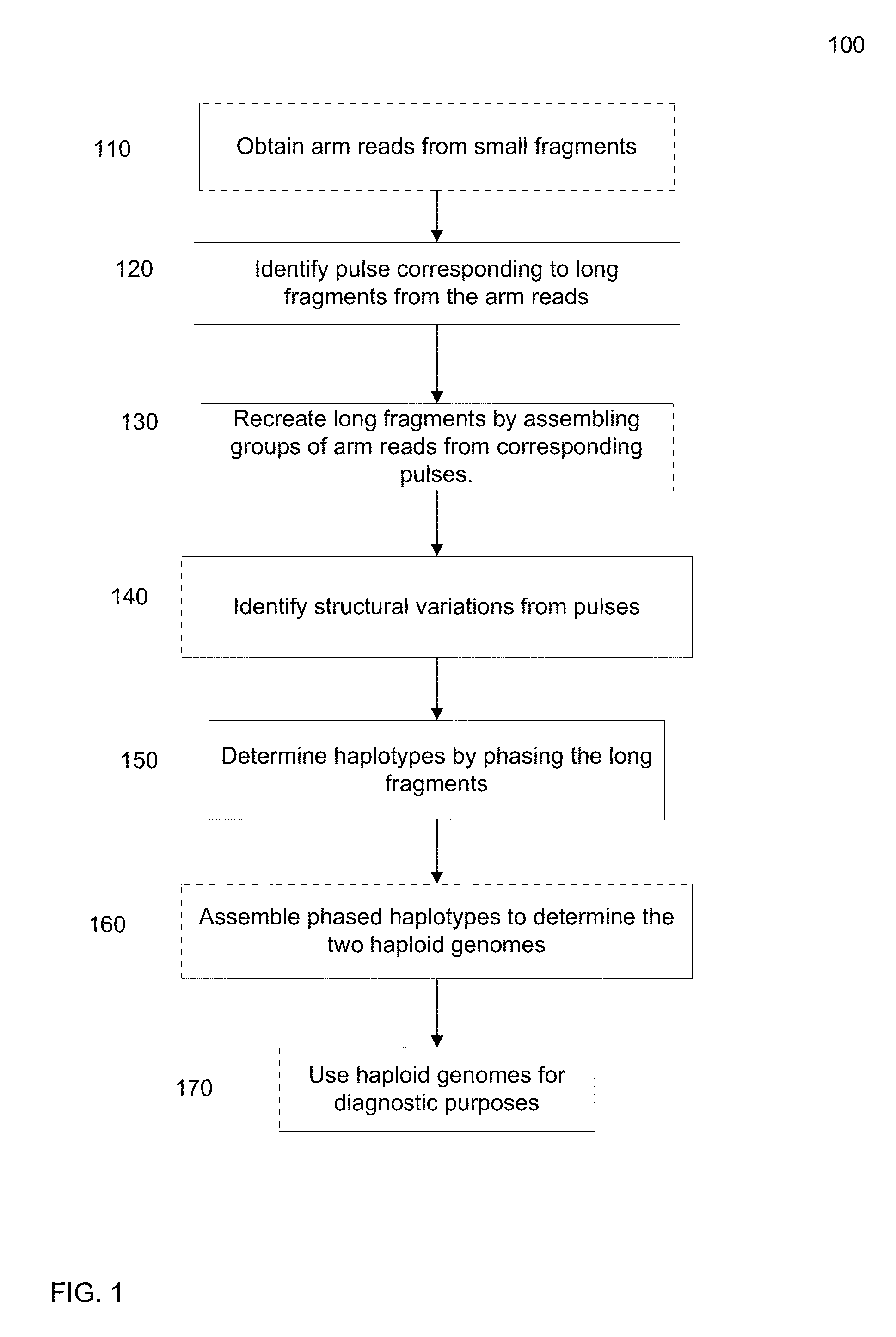
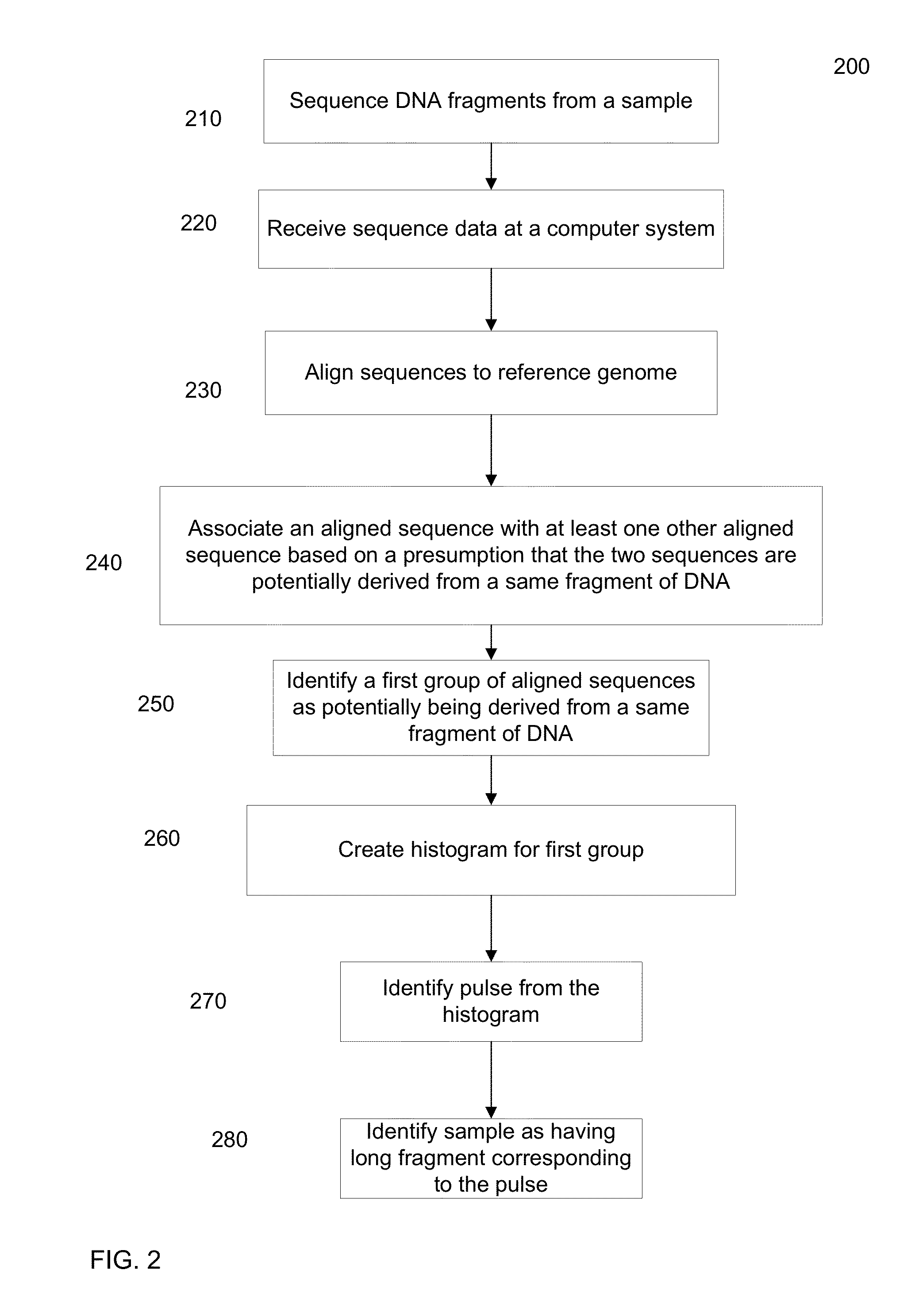
Identification of DNA fragments and structural variations
Various short reads can be grouped and identified as coming from a same long DNA fragment (e.g., by using wells with a relatively low-concentration of DNA). A histogram of the genomic coverage of a group of short reads can provide the edges of the corresponding long fragment (pulse). The knowledge of these pulses can provide an ability to determine the haploid genome and to identify structural variations.
Owner:COMPLETE GENOMICS INC
Method for redesign of microbial production systems
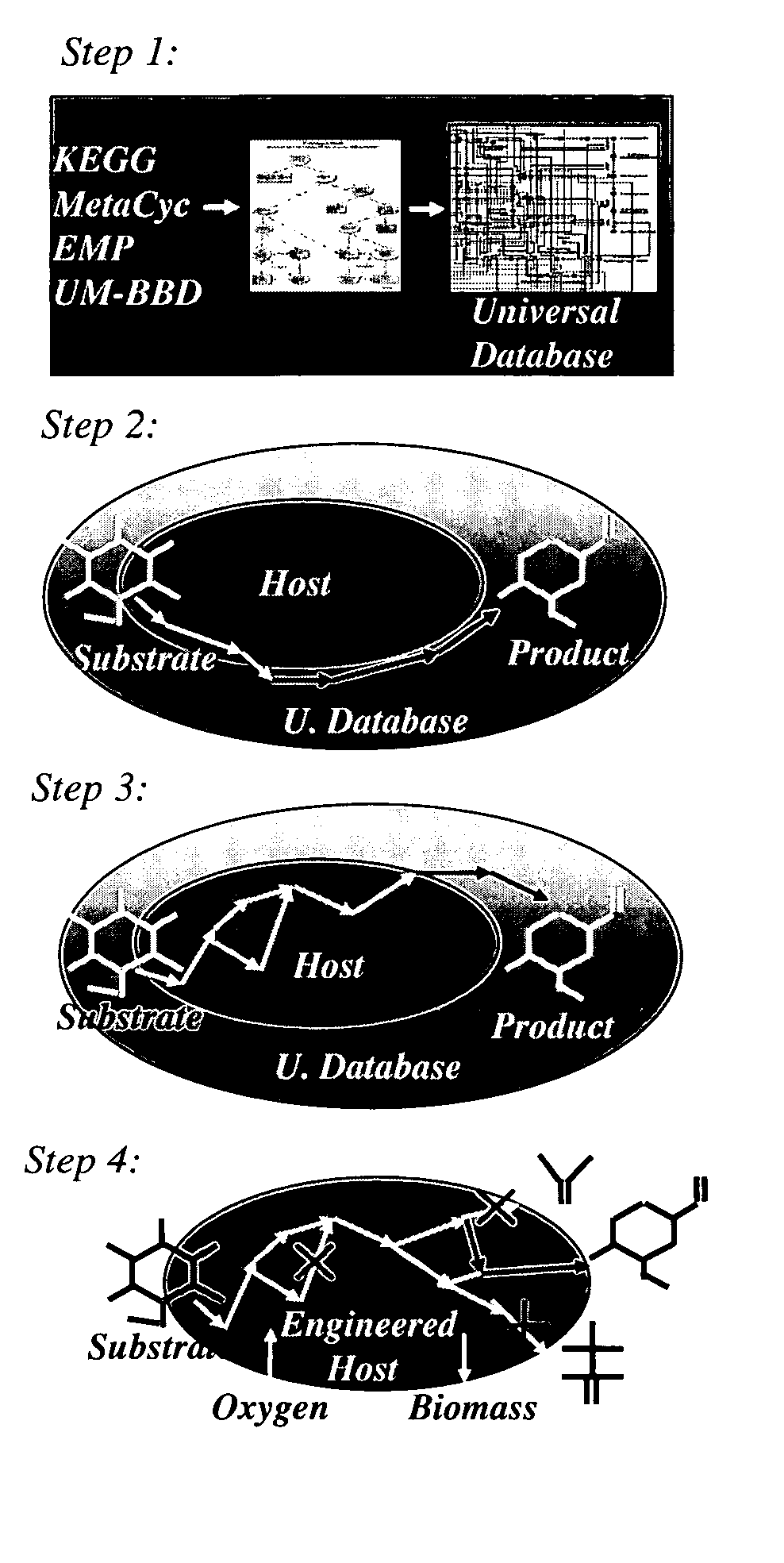
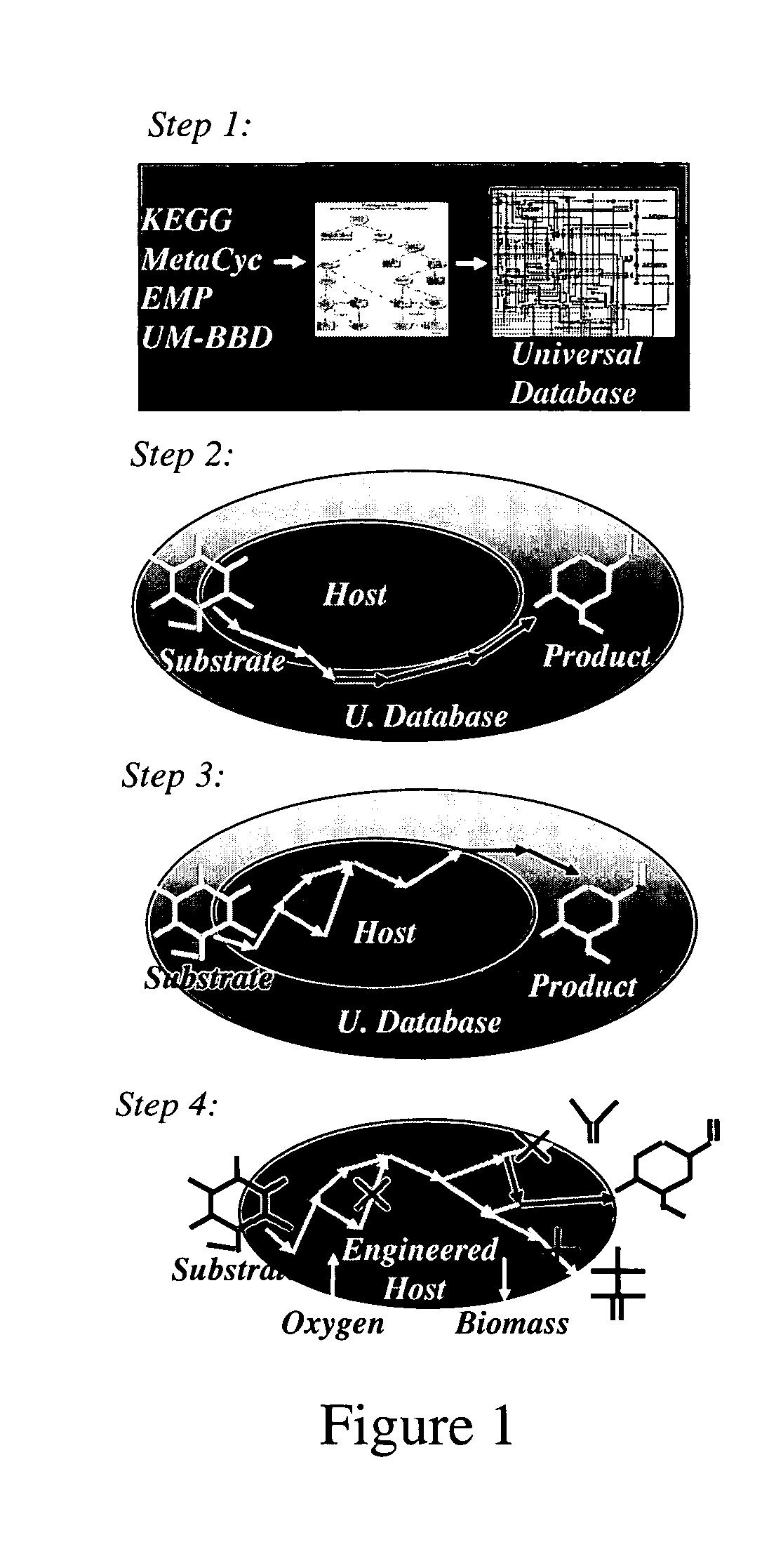
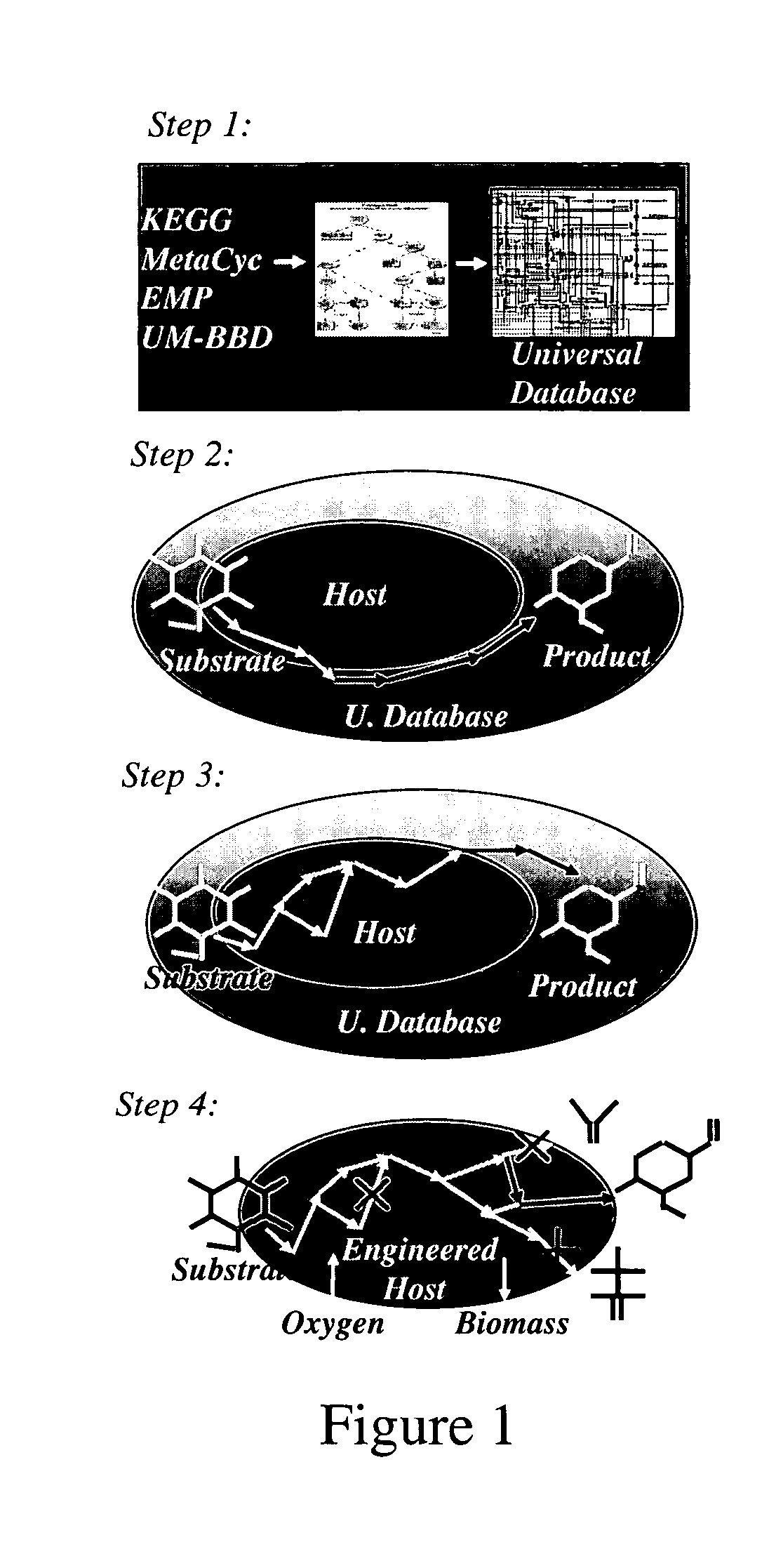
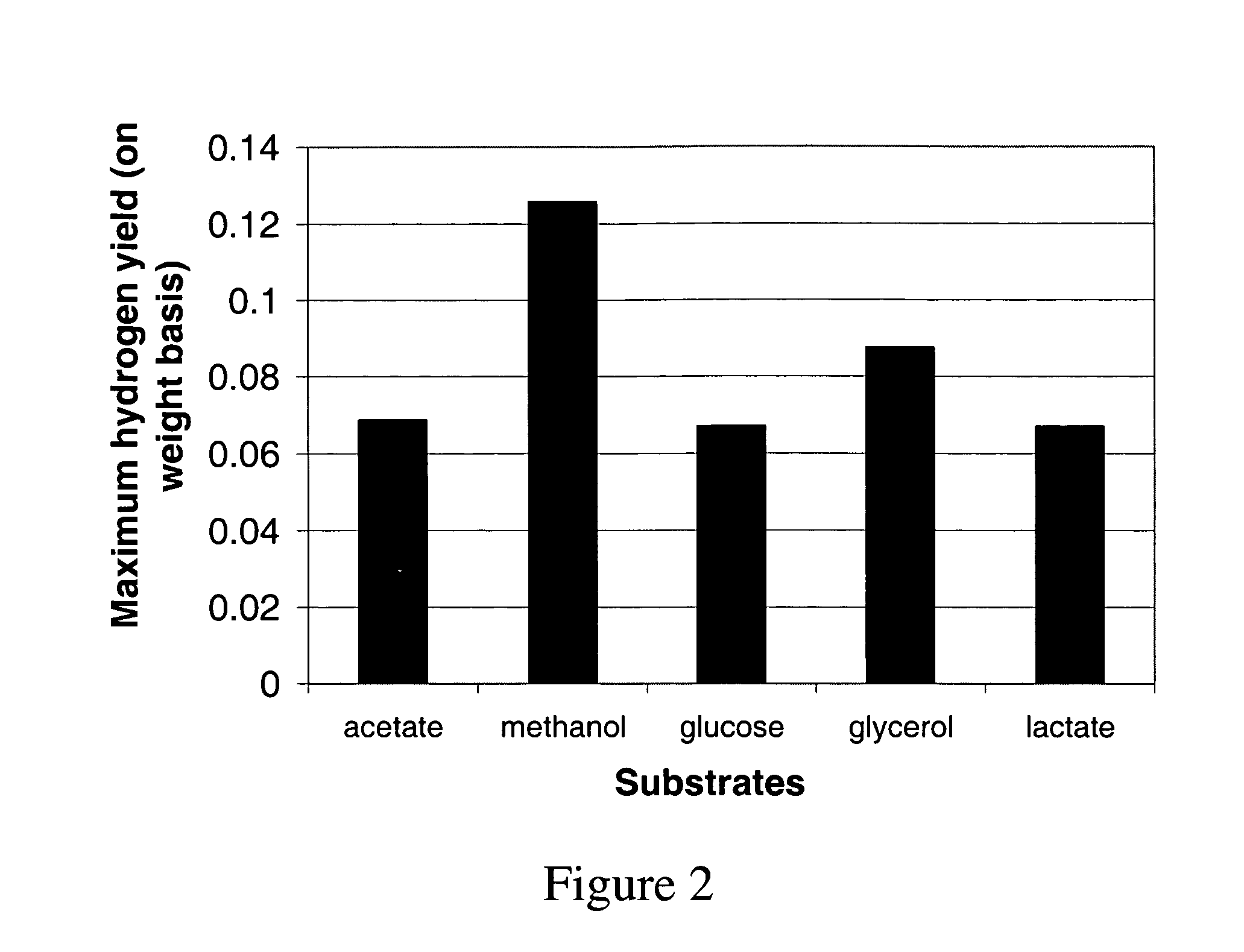
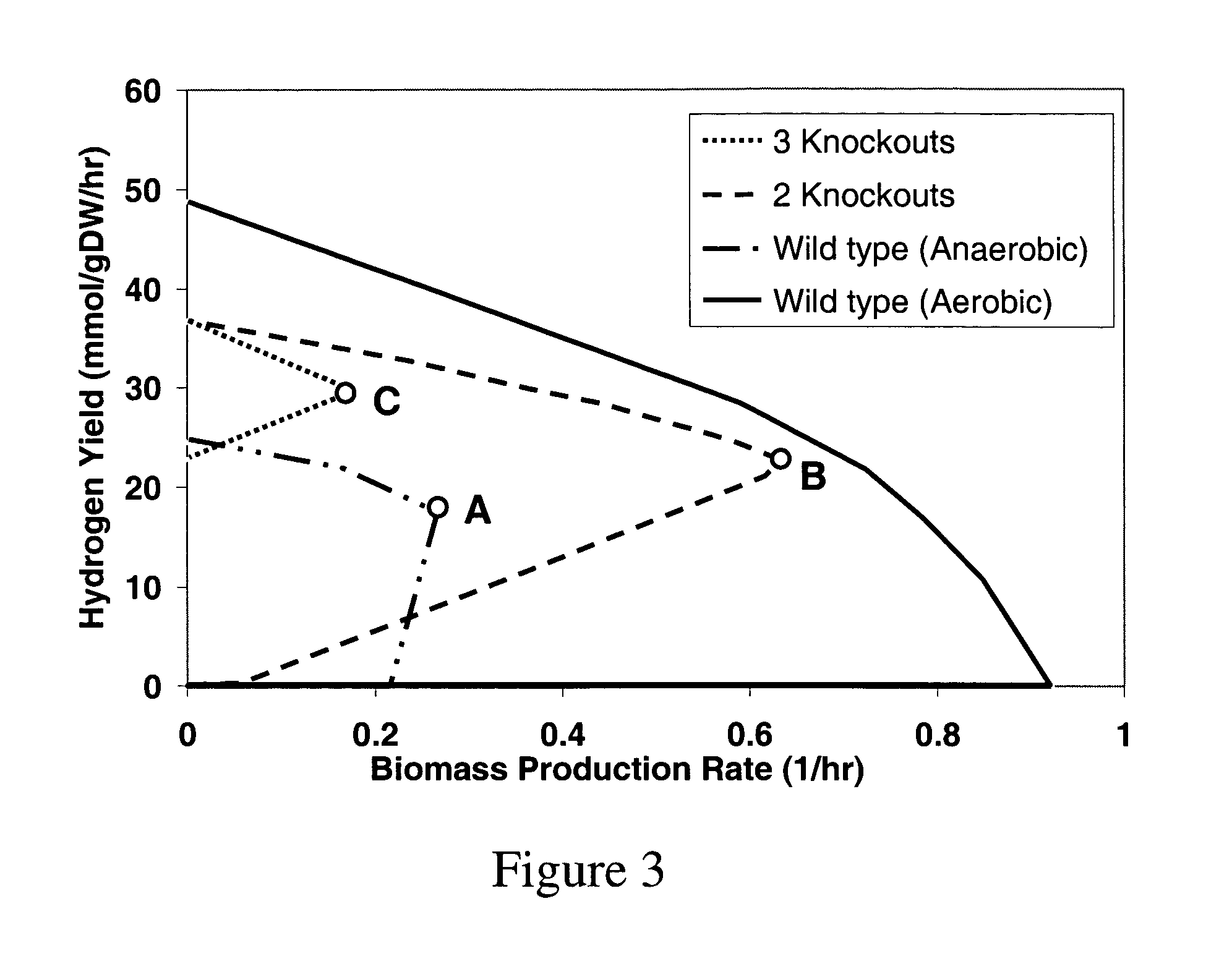
A computer-assisted method for identifying functionalities to add to an organism-specific metabolic network to enable a desired biotransformation in a host includes accessing reactions from a universal database to provide stoichiometric balance, identifying at least one stoichiometrically balanced pathway at least partially based on the reactions and a substrate to minimize a number of non-native functionalities in the production host, and incorporating the at least one stoichiometrically balanced pathway into the host to provide the desired biotransformation. A representation of the metabolic network as modified can be stored.
Owner:PENN STATE RES FOUND
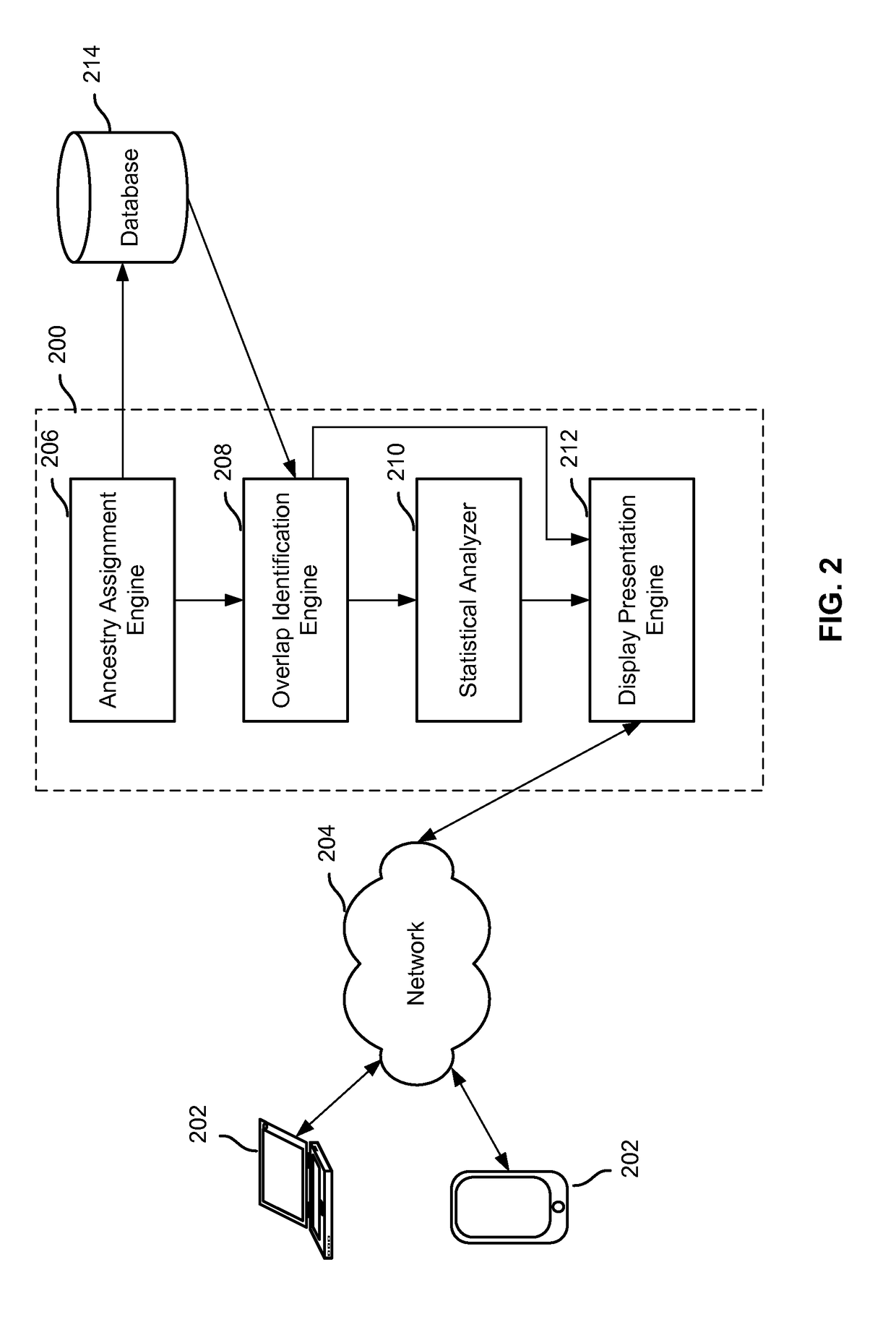
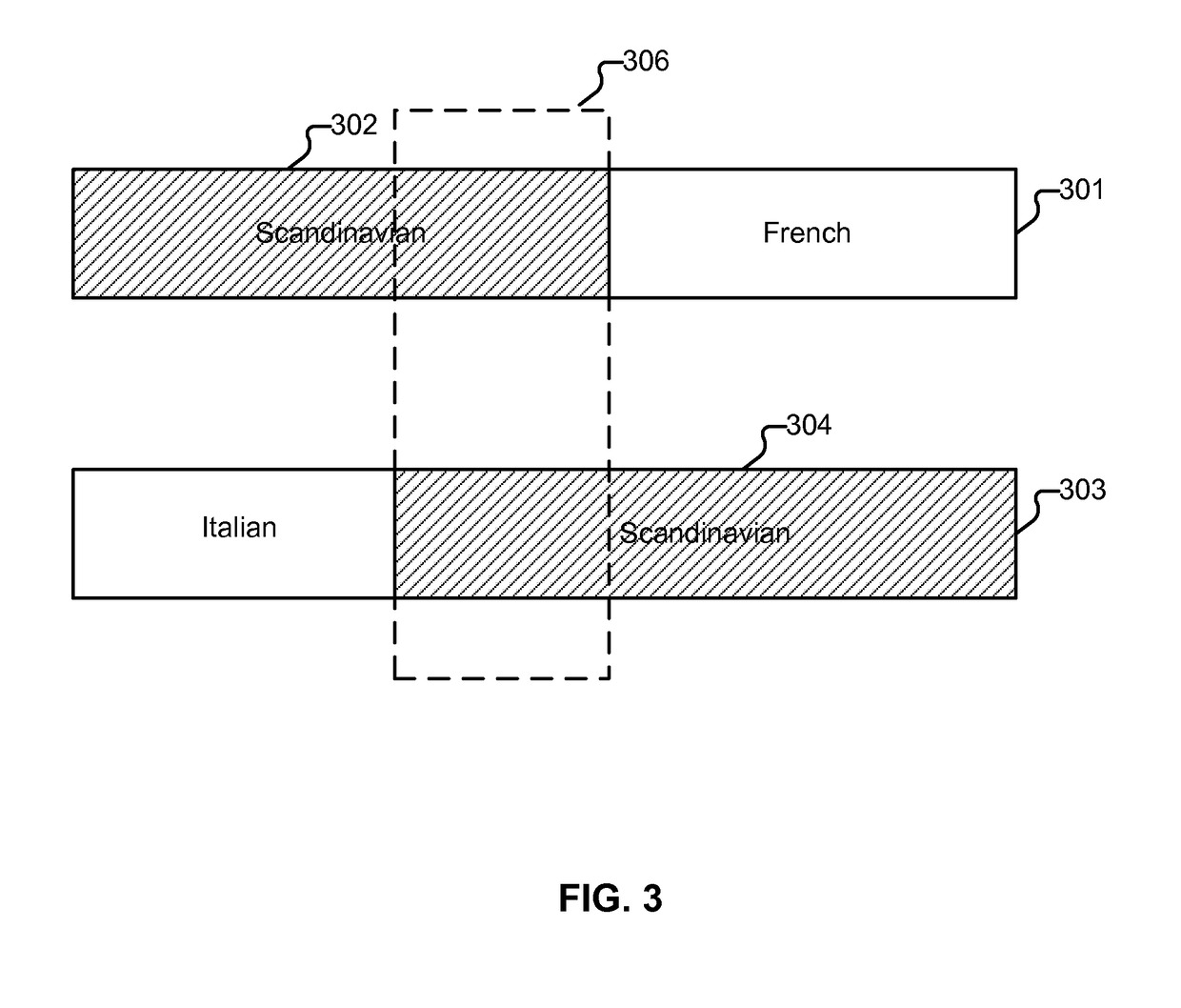
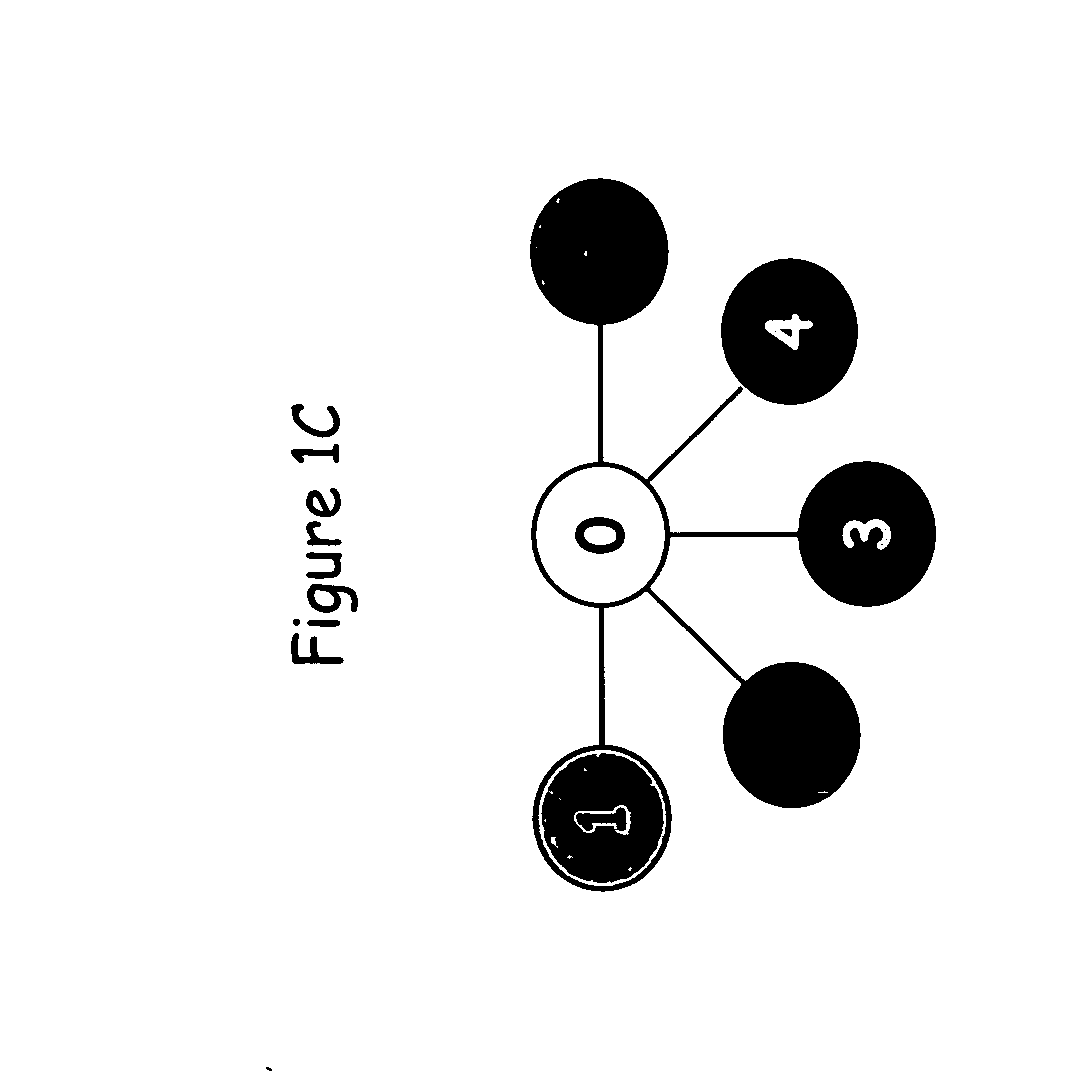
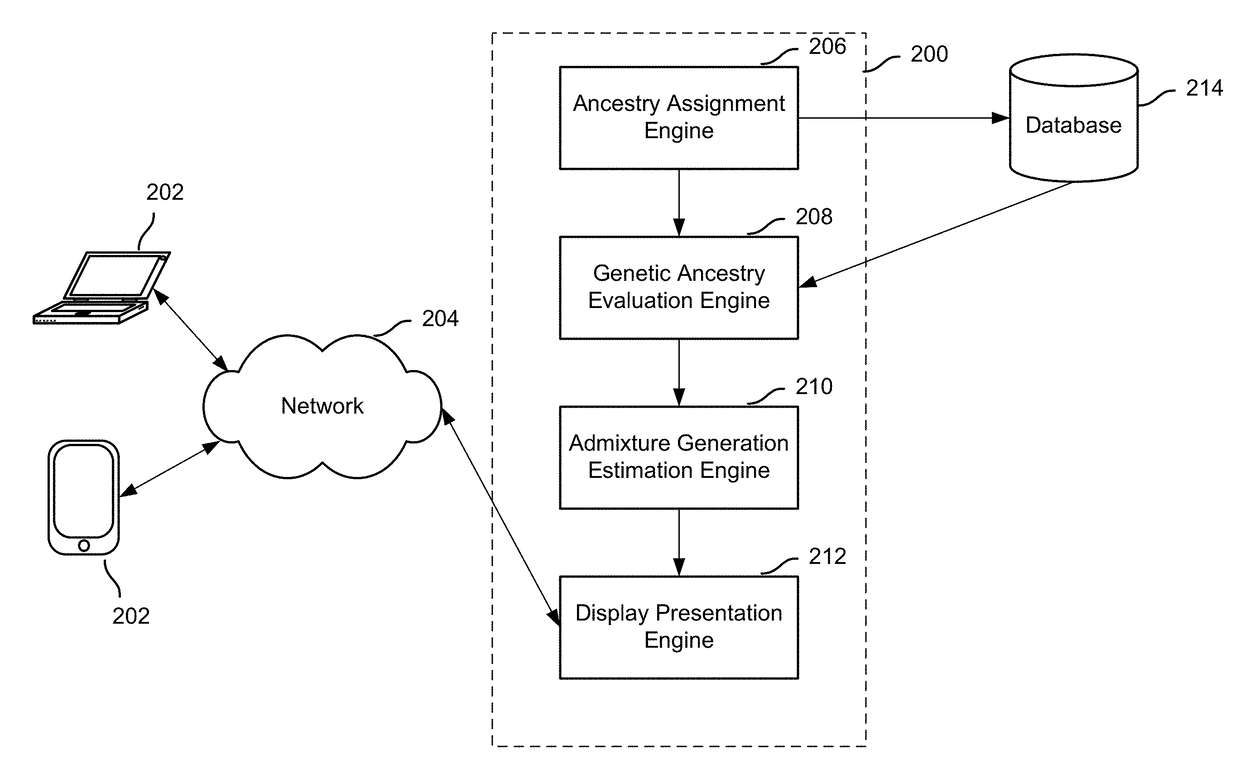
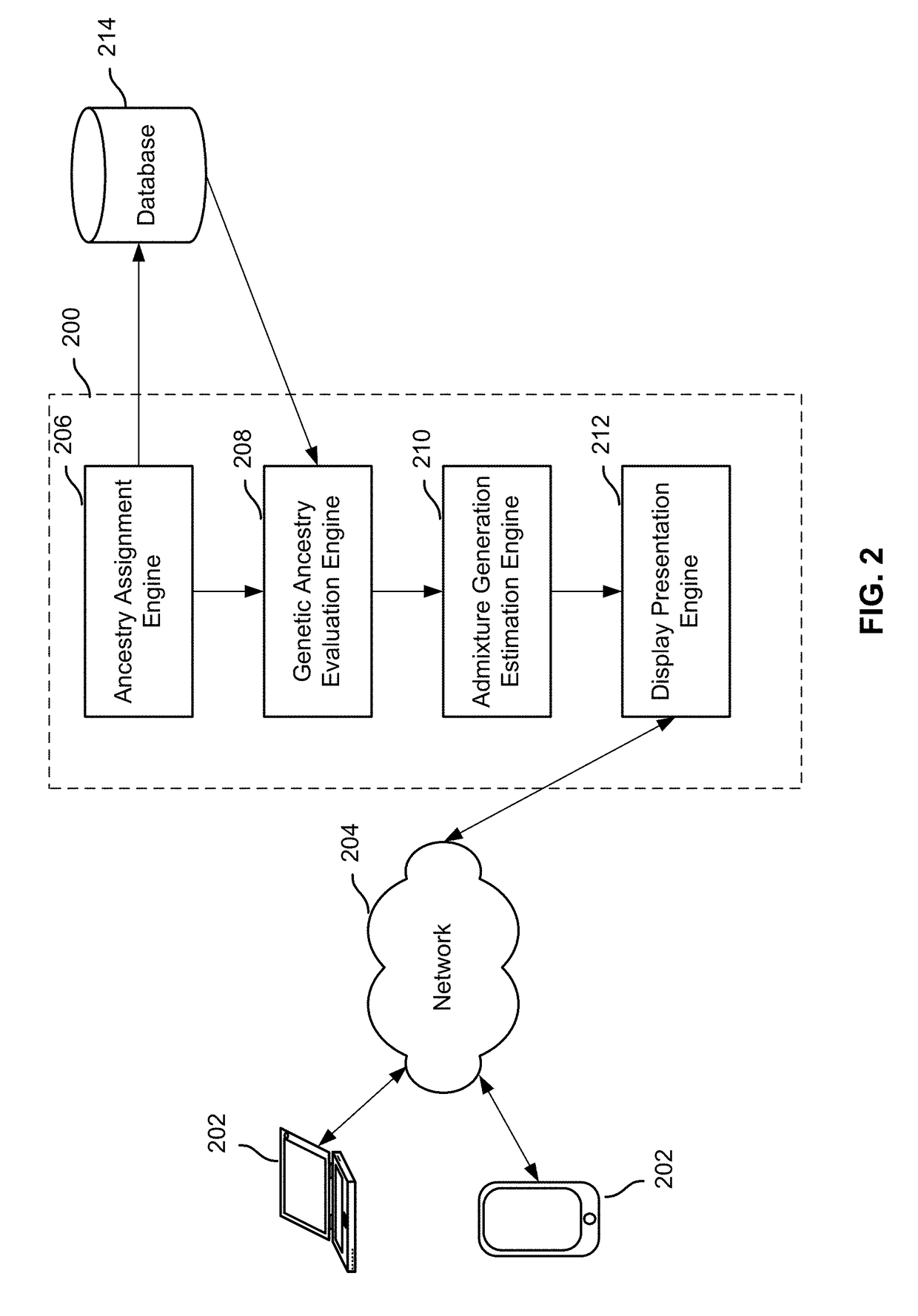
Display of estimated parental contribution to ancestry
Estimating parental contribution of ancestry includes: obtaining a set of ancestry assignment data associated with an individual's genotype data, at least some of the ancestry assignment data indicating that one or more segments of the individual's genotype data is deemed to be associated with a specific ancestry; determining whether in the individual's genotype data there is at least one confirmed region of overlapping ancestry assignment associated with the specific ancestry; in the event that it is determined that there is at least one confirmed region of overlapping ancestry assignment associated with the specific ancestry: specifying that parental contribution of the specific ancestry is made by both parents of the individual; in the event that it is determined that there is no confirmed region of overlapping ancestry assignment associated with the specific ancestry: statistically determining whether the parental contribution to the specific ancestry is made by only one parent of the individual or by both parents of the individual, the determination being based at least in part on one or more lengths of the one or more segments deemed to be associated with the specific ancestry; and outputting information pertaining to the parental contribution to the specific ancestry.
Owner:23ANDME
Genetic diagnosis using multiple sequence variant analysis
The present invention is in the field of nucleic acid-based genetic analysis. More particularly, it discloses novel insights into the overall structure of genetic variation in all living species. The structure can be revealed with the use of any data set of genetic variants from a particular locus. The invention is useful to define the subset of variations that are most suited as genetic markers to search for correlations with certain phenotypic traits. Additionally, the insights are useful for the development of algorithms and computer programs that convert genotype data into the constituent haplotypes that are laborious and costly to derive in an experimental way. The invention is useful in areas such as (i) genome-wide association studies, (ii) clinical in vitro diagnosis, (iii) plant and animal breeding, (iv) the identification of micro-organisms.
Owner:METHEXIS GENOMICS
Estimation of admixture generation
Admixture generation determination includes: obtaining ancestry assignment information associated with an individual's genotype data, the ancestry assignment information at least indicating that a portion of the individual's genotype data is deemed to be associated with a specific ancestry; determining the individual's genetic ancestry summary data corresponding to the specific ancestry; estimating an admixture generation associated with the specific ancestry, the admixture generation indicating a most recent generation or a most recent generation range from which the individual has at least one non-admixed ancestor of the specific ancestry, the estimation including a maximum likelihood determination based at least in part on the individual's genetic ancestry summary data and a recombination model; and outputting the estimated admixture generation.
Owner:23ANDME
Finding relatives in a database
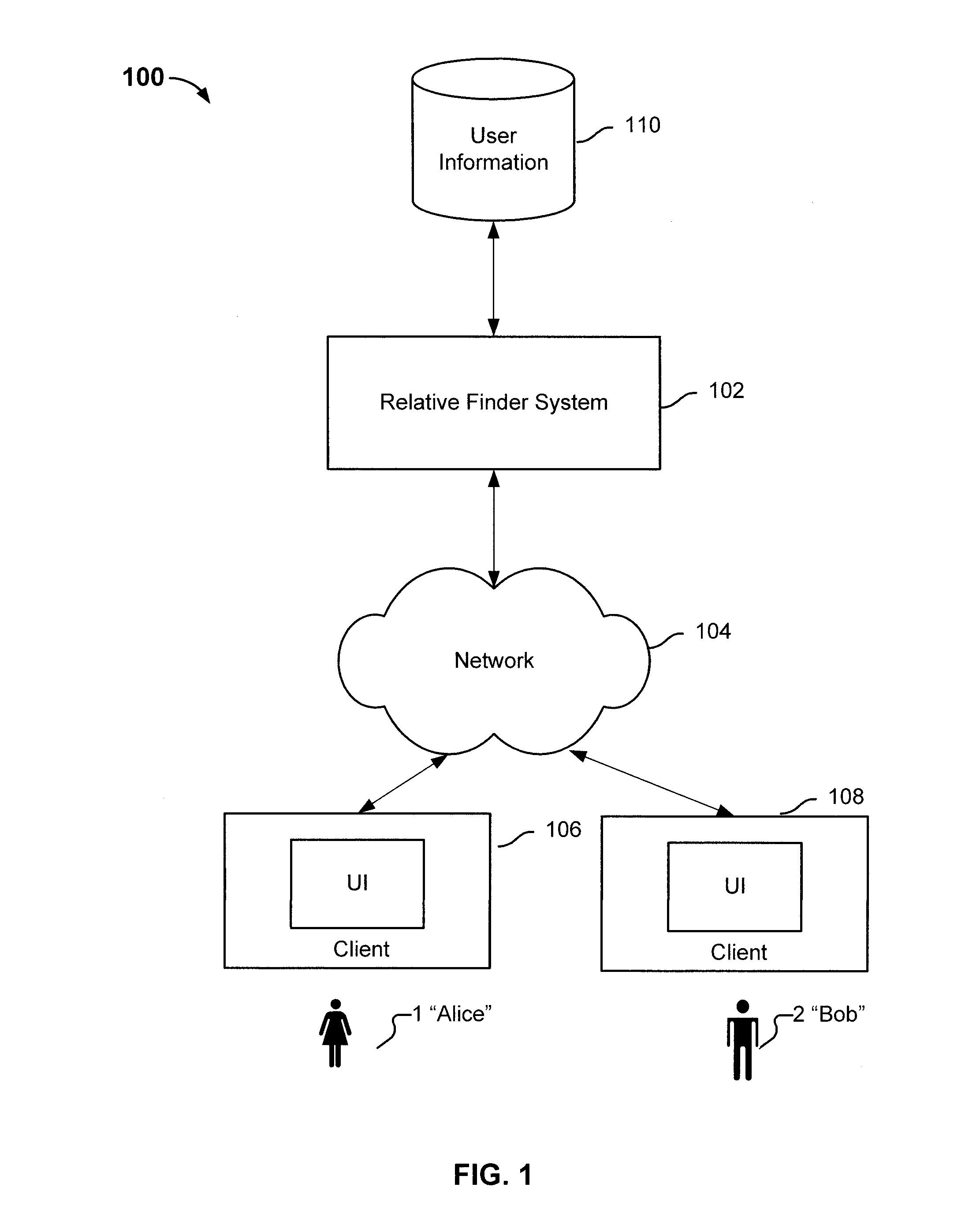
ActiveUS20100223281A1Digital data processing detailsSequence analysisHuman–computer interactionRecombinant DNA
Determining relative relationship includes receiving recombinable deoxyribonucleic acid (DNA) information of a first user and recombinable DNA information of a second user, determining, based at least in part on the recombinable DNA information of the first user and recombinable DNA information of the second user, a predicted degree of relationship between the first user and the second user, and in the event that the expected degree of relationship between the first user and the second user at least meets the threshold, notifying at least the first user about a relative relationship with the second user.
Owner:23ANDME
Methods of Clustering Gene and Protein Sequences
InactiveUS20090327170A1Less computationally intensiveReduce computing loadOrganic active ingredientsPeptide/protein ingredientsAntigenNetwork topology
The invention relates to methods for clustering gene and protein sequences. In particular, it involves generation of networks of sequences where the interconnections are based upon a measure of similarity. The invention also provides methods of optimizing and improving the networks by re-wiring of the network based upon overlap of the nearest neighbors of given pairs of nodes. The invention further provides methods of identifying clusters of sequences within the networks and the optimized networks based upon the topology of the network. The clusters identified represent groups of sequences that are related by function and / or evolution. The invention has particular applicability in annotation of sequences in databases and identification of functional homologs which can be very useful for novel therapeutic and diagnostic targets based upon such targets belonging to a cluster or family that contains a known sequence such as a diagnostic sequence, antigen or other therapeutic target.
Owner:DONATI CLAUDIO +2
Method for determining the presence or absence of different aneuploidies in a sample
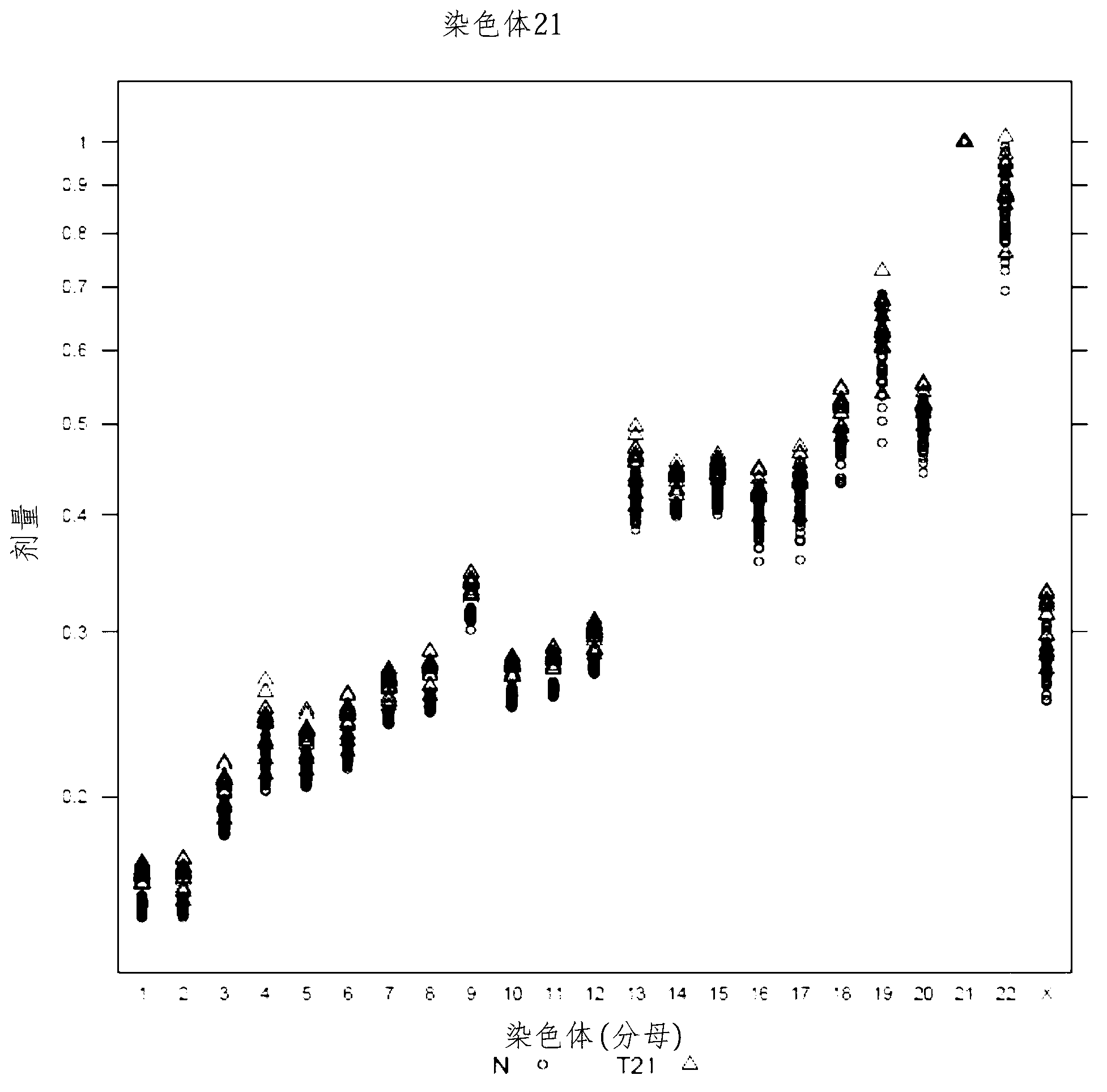
ActiveCN103003447ADiagnose and monitor metastatic progressionMicrobiological testing/measurementData visualisationPresent methodTest sample
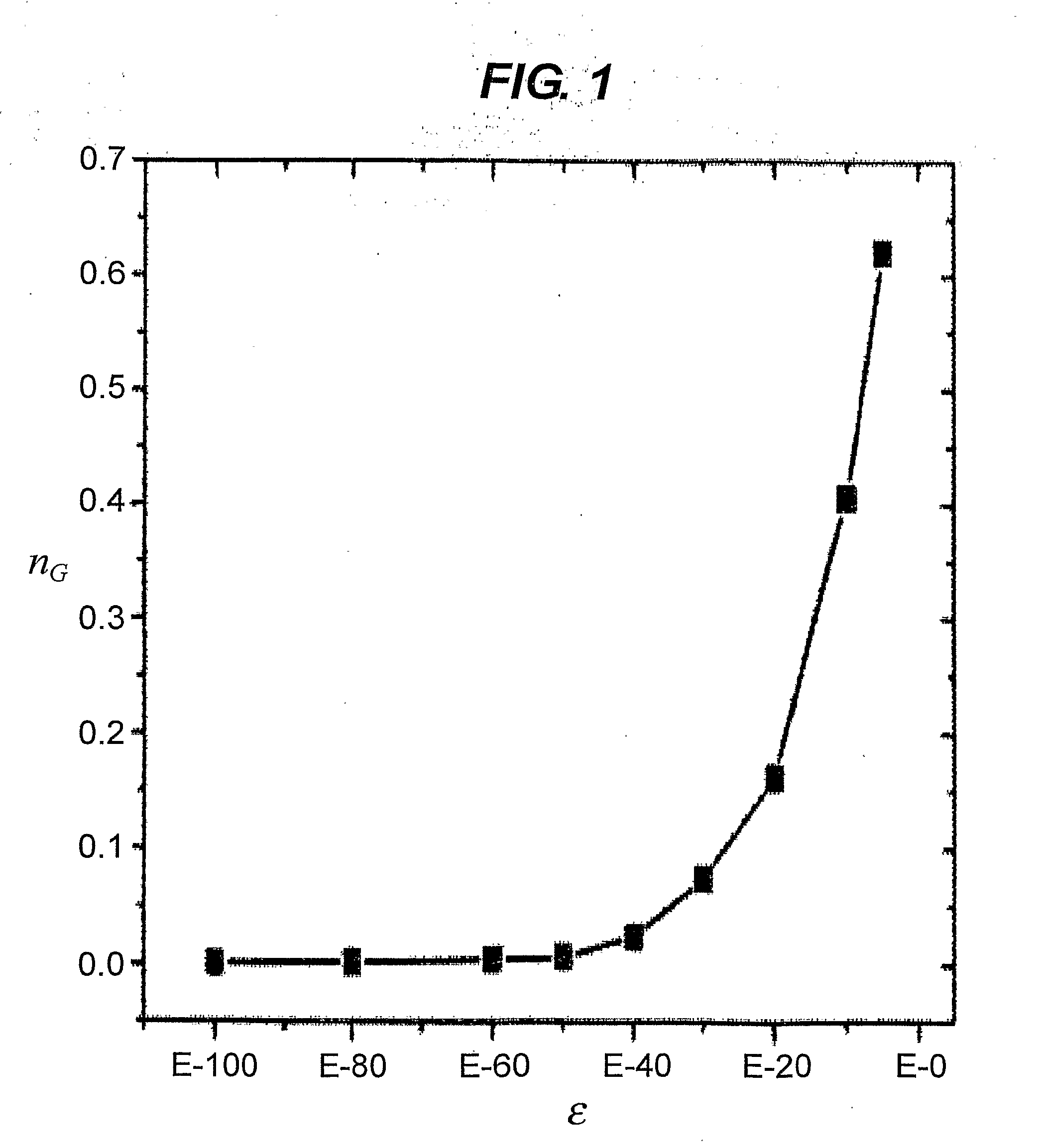
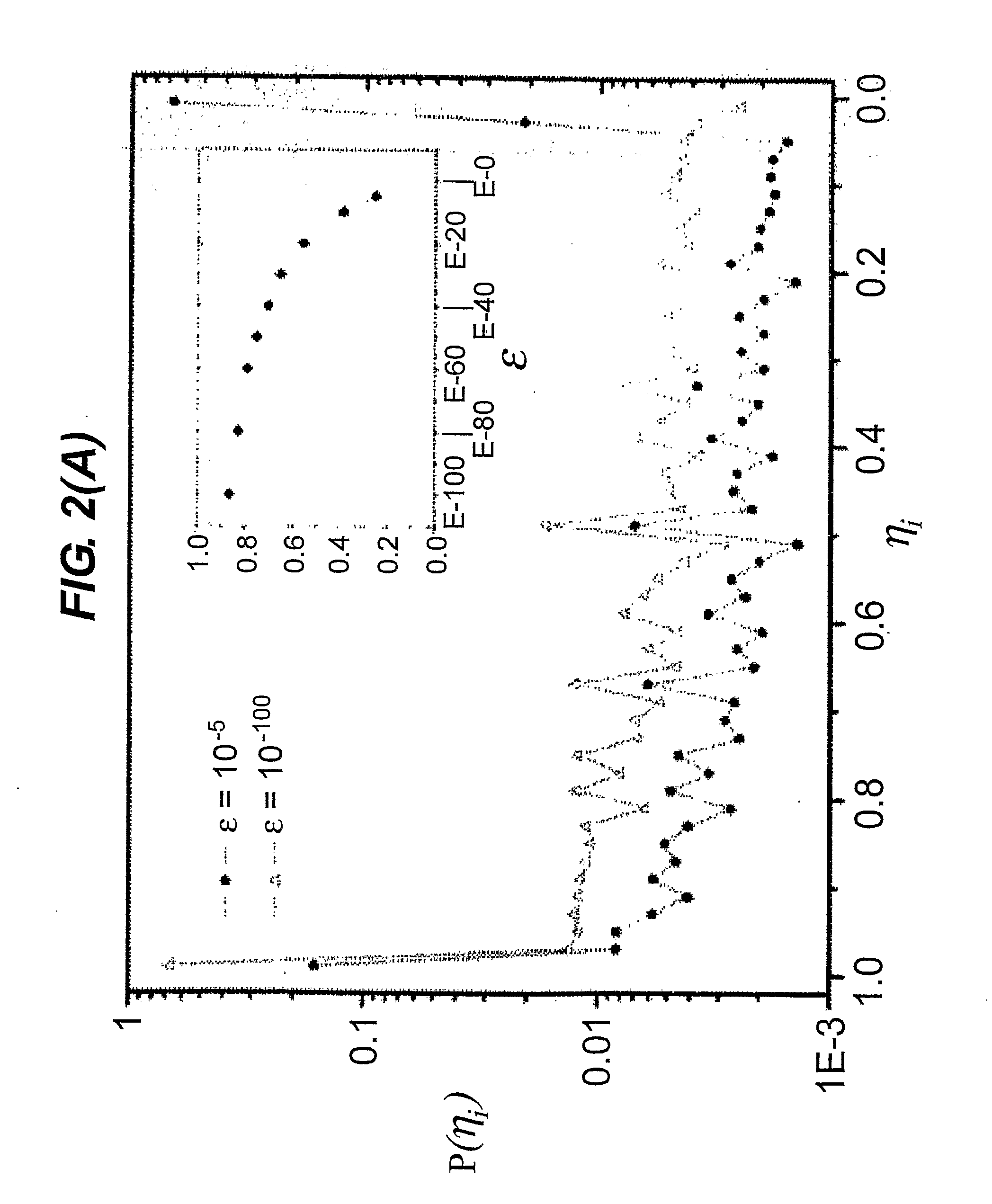
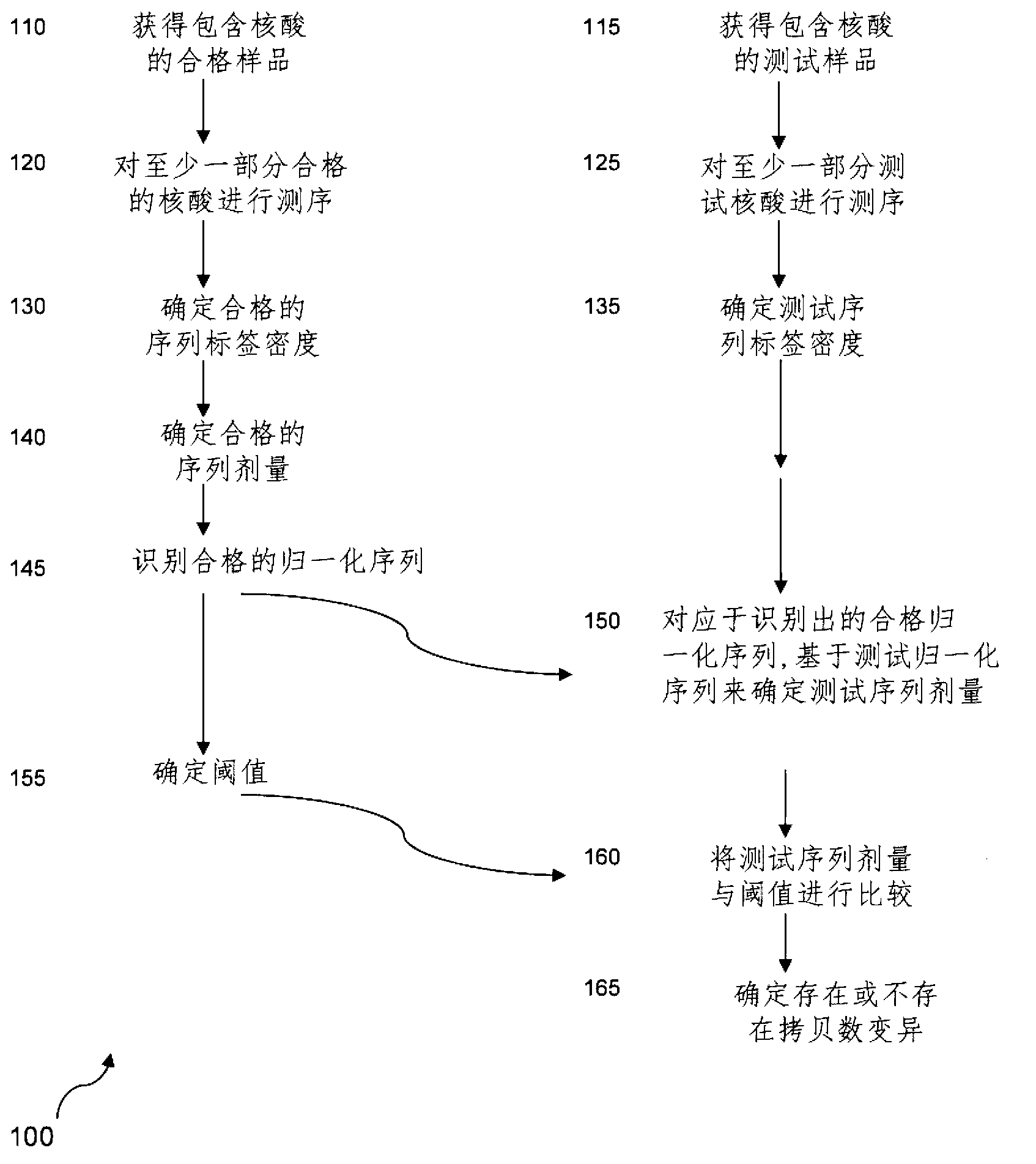
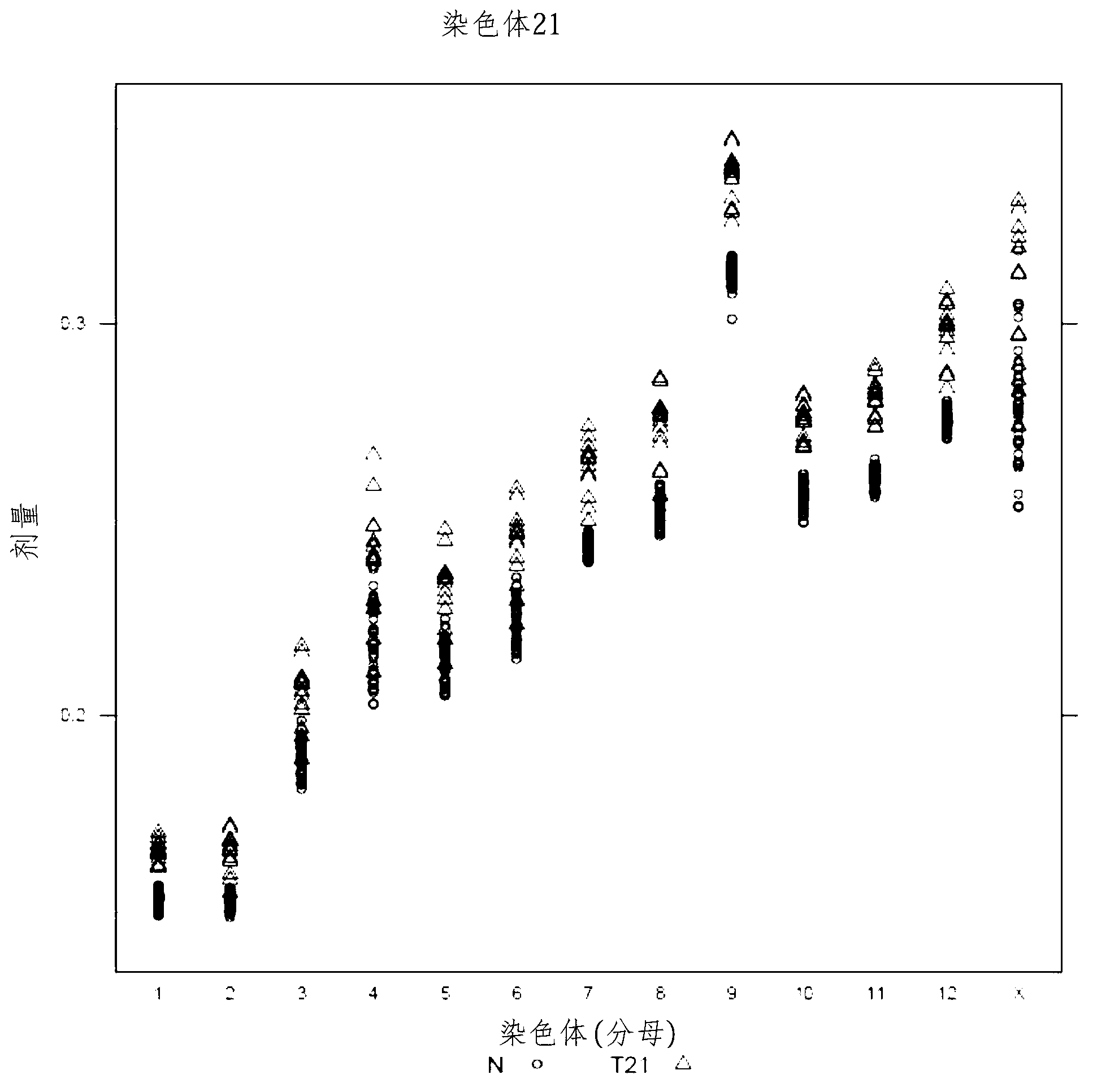
The invention provides a method for determining copy number variations (CNV) of a sequence of interest in a test sample that comprises a mixture of nucleic acids that are known or are suspected to differ in the amount of one or more sequence of interest. The method comprises a statistical approach that accounts for accrued variability stemming from process-related, interchromosomal and inter-sequencing variability. The method is applicable to determining CNV of any fetal aneuploidy, and CNVs known or suspected to be associated with a variety of medical conditions. CNV that can be determined according to the present method include trisomies and monosomies of any one or more of chromosomes 1-22, X and Y, other chromosomal polysomies, and deletions and / or duplications of segments of any one or more of the chromosomes, which can be detected by sequencing only once the nucleic acids of a test sample. Any aneuploidy can be determined from sequencing information that is obtained by sequencing only once the nucleic acids of a test sample.
Owner:VERINATA HEALTH INC
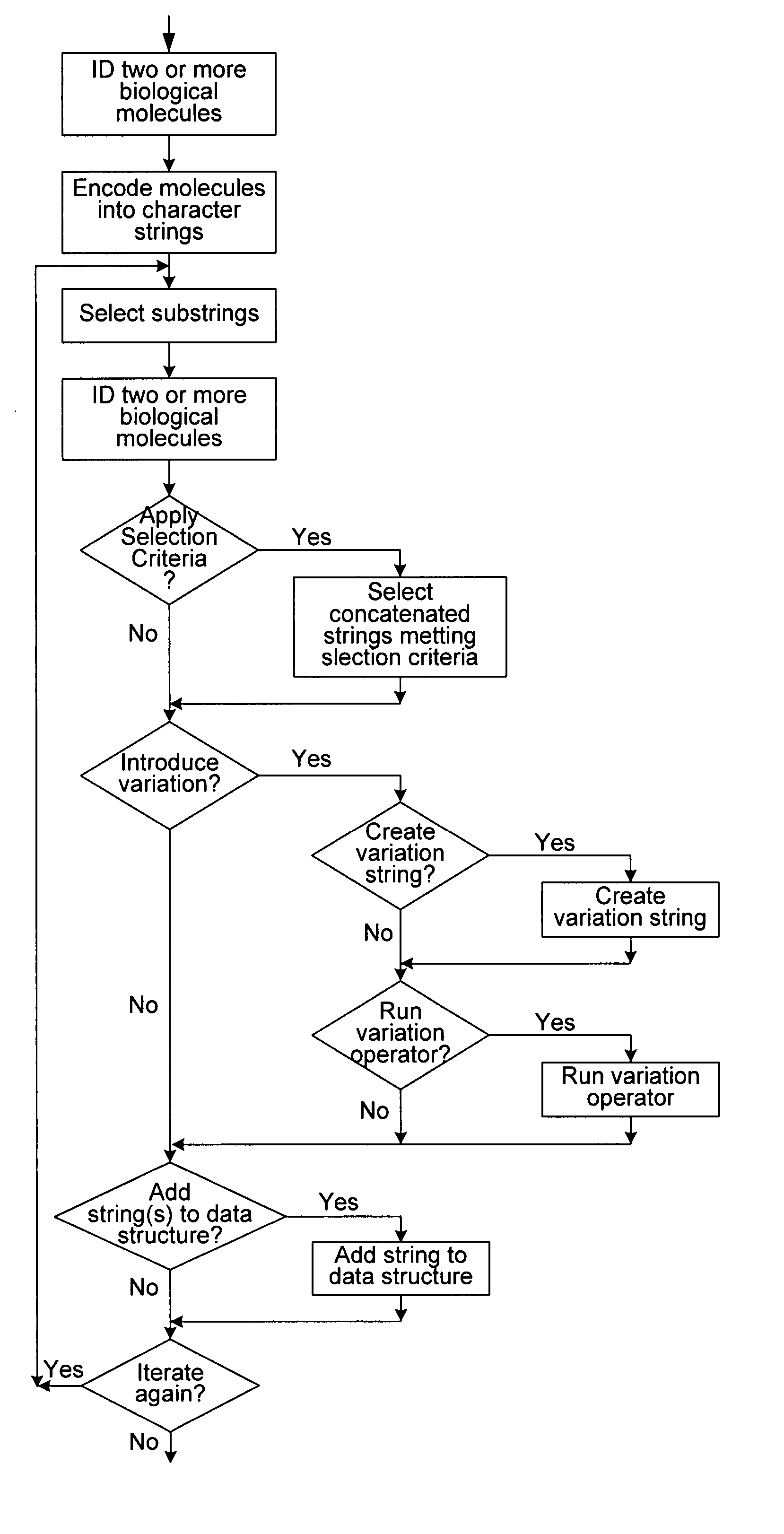
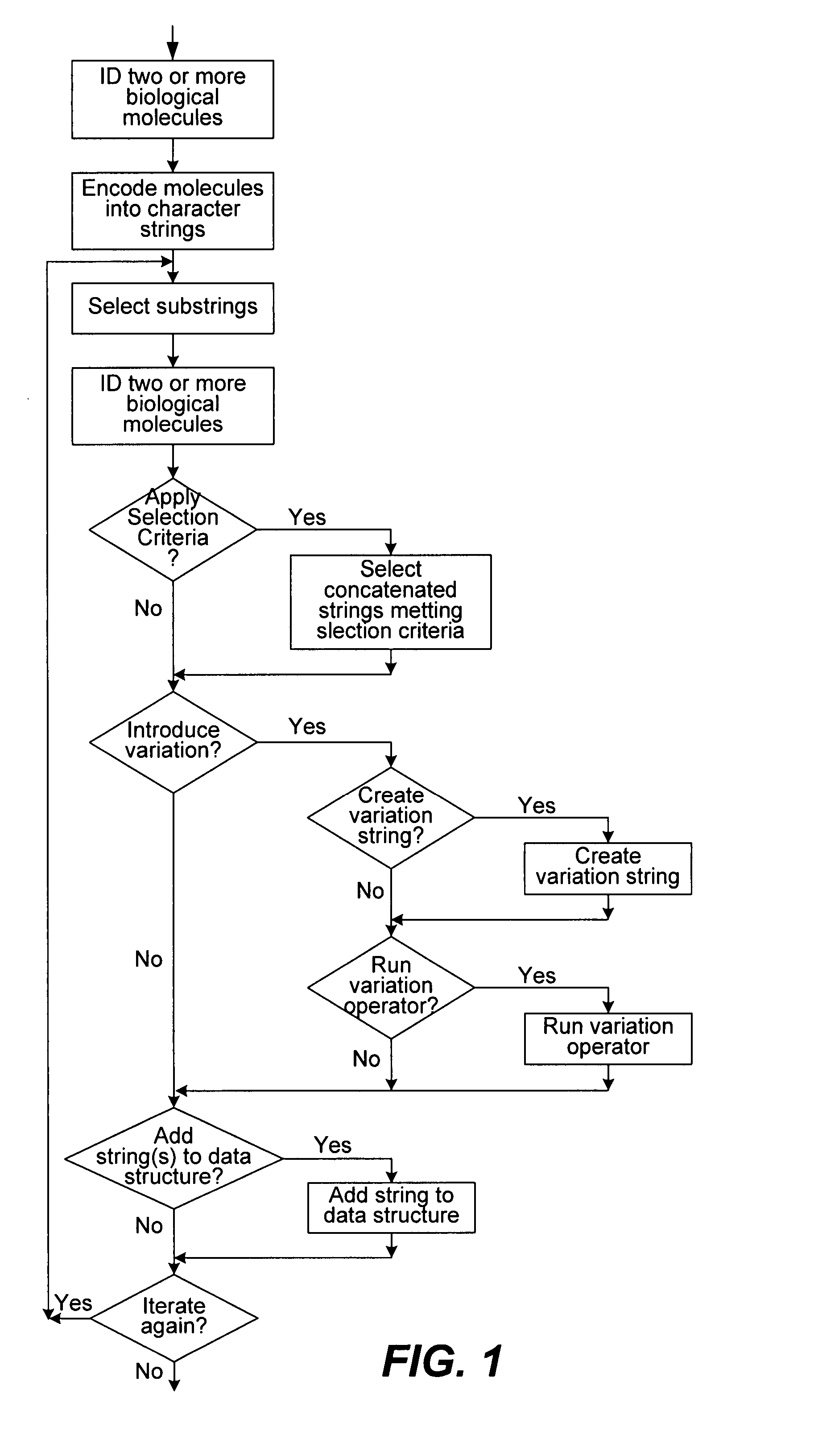
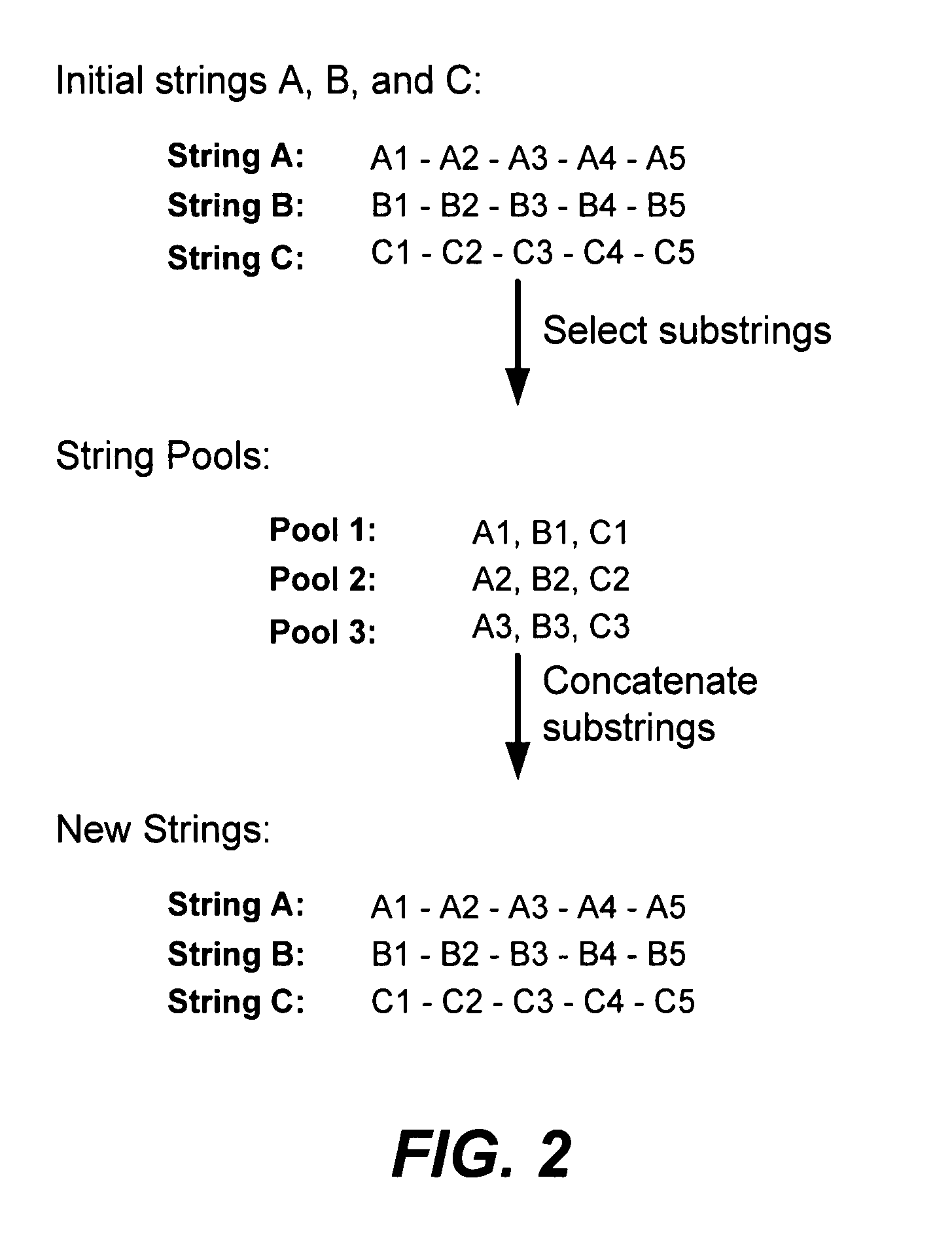
Methods of populating data structures for use in evolutionary simulations
In particular, this invention provides novel methods of populating data structures for use in evolutionary modeling. In particular, this invention provides methods of populating a data structure with a plurality of character strings. The methods involve encoding two or more a biological molecules into character strings to provide a collection of two or more different initial character strings; selecting at least two substrings from the pool of character strings; concatenating the substrings to form one or more product strings about the same length as one or more of the initial character strings; adding the product strings to a collection of strings; and optionally repeating this process using one or more of the product strings as an initial string in the collection of initial character strings.
Owner:CODEXIS MAYFLOWER HLDG LLC
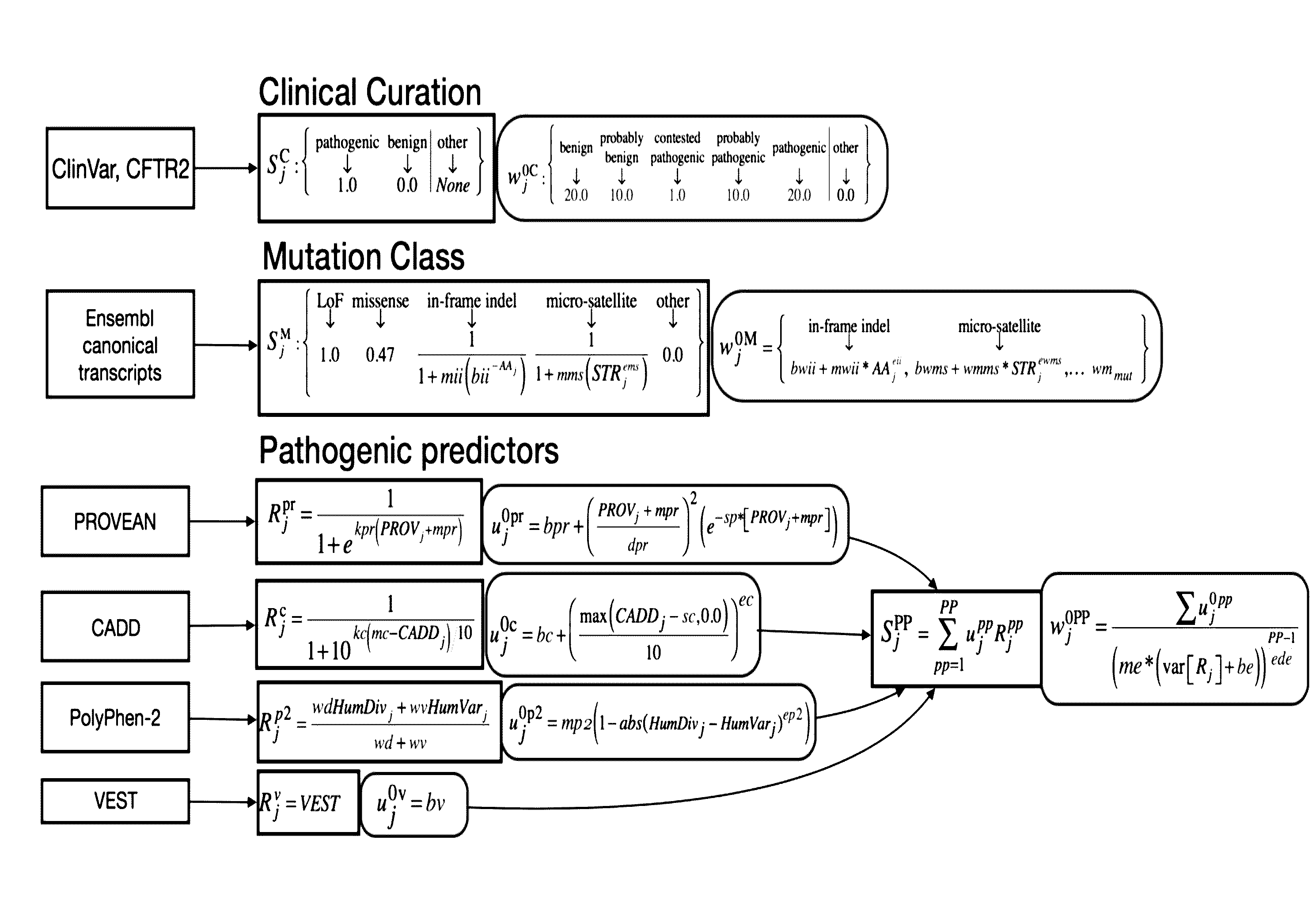
Device, system and method for assessing risk of variant-specific gene dysfunction
A device, system and method for predicting gene-dysfunction caused by a genetic mutation in the genome of an organism. A neural network may comprise multiple nodes respectively associated with multiple different gene-dysfunction metrics and multiple different confidence weights. The neural network may combine the multiple gene-dysfunction metrics according to the respective associated confidence weights to generate one or more likelihoods that a genetic mutation causes gene-dysfunction in organisms. In a training-phase, the neural network may be trained using an input data set including genetic mutations to generate new gene-dysfunction metrics and new associated confidence weights that optimize the neural network based on a cost factor. In a run-time phase, a genetic mutation may be identified and one or more likelihoods may be computed that the identified genetic mutation causes gene-dysfunction in the organism based on the new gene-dysfunction metrics and the associated new confidence weights of the neural network.
Owner:ANCESTRY COM DNA
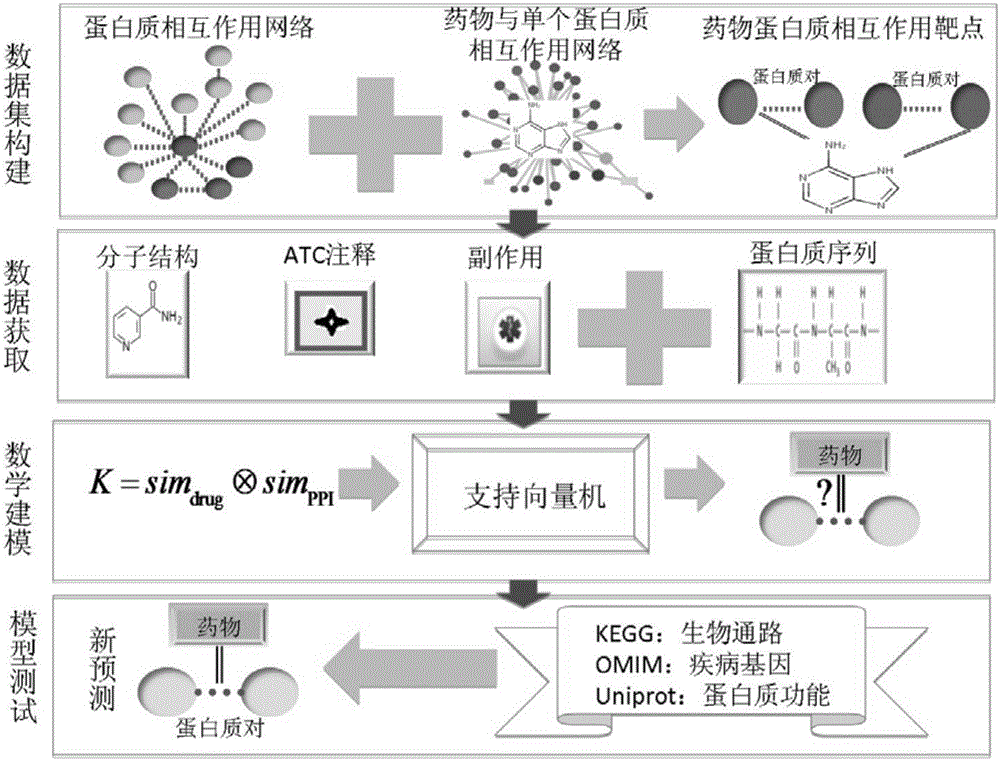
Method and system for predicting protein interaction target point of drug
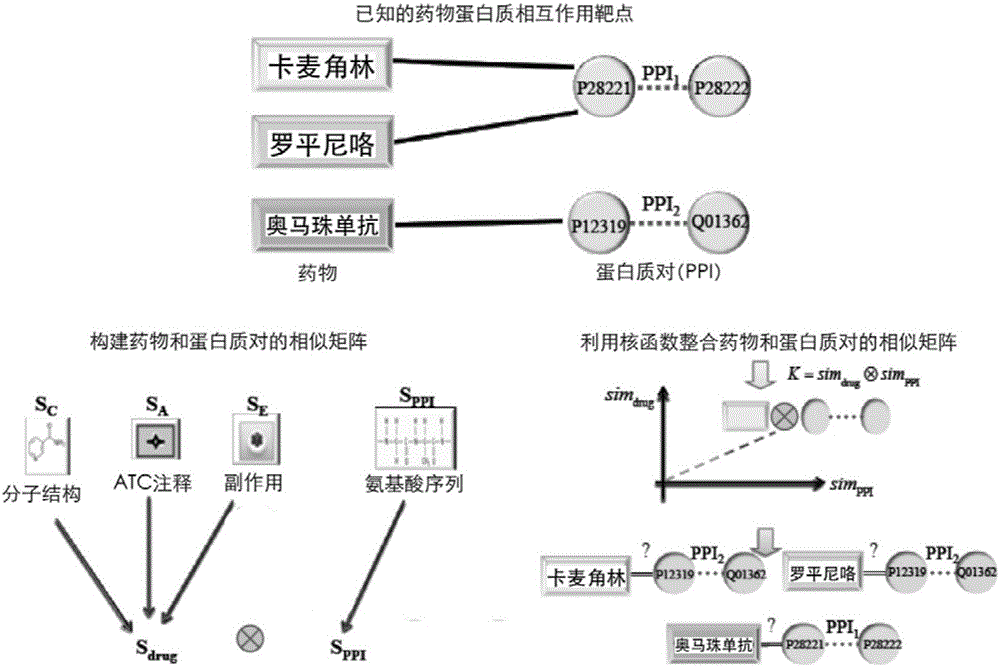
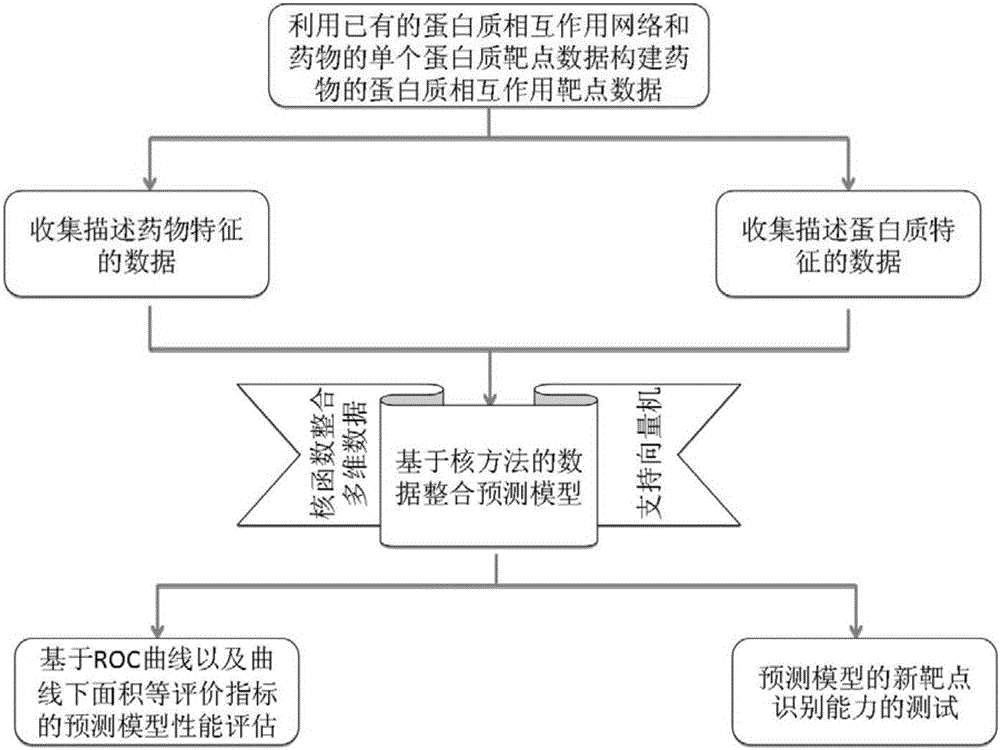
InactiveCN105160206AWiden the search spaceImprove classification effectSpecial data processing applicationsEvolutionary biologyProtein targetData set
The invention relates to a method and a system for predicting a protein interaction target point of a drug. The method comprises: 1) collecting a human protein interaction network and single protein target point data of the drug, and constructing an interactive protein target point data set of the drug; 2) obtaining description data of the drug and proteins; 3) constructing a bigraph for representing an interactive relationship between the drug and a protein pair, constructing a similar matrix for representing drug similarity and protein pair similarity, establishing a kernel function for correlating the similar matrix of the drug and the protein pair, and establishing a prediction model through a machine learning algorithm; and 4) performing independent set testing by utilizing unknown drug and interactive protein pair, and predicting a possibly existent unknown drug protein interaction target point, and verifying a prediction result through database and document retrieval. According to the method and the system, the search space of the drug target point can be expanded and the more specific drug protein interaction target point with the best classification performance can be obtained.
Owner:ACAD OF MATHEMATICS & SYSTEMS SCIENCE - CHINESE ACAD OF SCI +1
Method For Molecular Genealogical Research
InactiveUS20130149707A1Microbiological testing/measurementSequence analysisStudy methodologyOrganism
A genealogical research and record keeping system and method for identifying commonalities in haplotypes and other genetic characteristics of a biological sample of two or more individual members. Chromosomal fragments identical by descent identify family ties between siblings, parents and children and ancestors and progeny across many generations. It is particularly useful in corroborating and improving the accuracy of genealogical data and identifying previously unknown genetic relationships.
Owner:ANCESTRY COM DNA
Microbiome based identification, monitoring and enhancement of fermentation processes and products
ActiveUS20180363031A1Promote growthGood for healthMicrobiological testing/measurementBiostatisticsMicroorganismBiofuel
Monitoring, analysis and control of fermentation activities includes methods and corresponding systems directed toward agriculture, biofuels, and food production. Complex methods and corresponding systems are provided for classifying a microorganism; profiling a microbiome; sequencing multiple libraries in a single sequencing run; determining a microbiome profile in a sample; and analyzing a material from a location associated with a fermentation process. Additional implementations are directed to methods and corresponding systems for obtaining, deriving, predicting and evaluating microbiome information; control, analysis and direction of fermentation operations; and evaluating, analyzing and displaying microbiome related information in two and three dimensional plots. Yet additional methods and corresponding systems permit identification and analysis of microorganisms capable of imparting beneficial properties to phases of fermentation processes.
Owner:BIOME MAKERS INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com