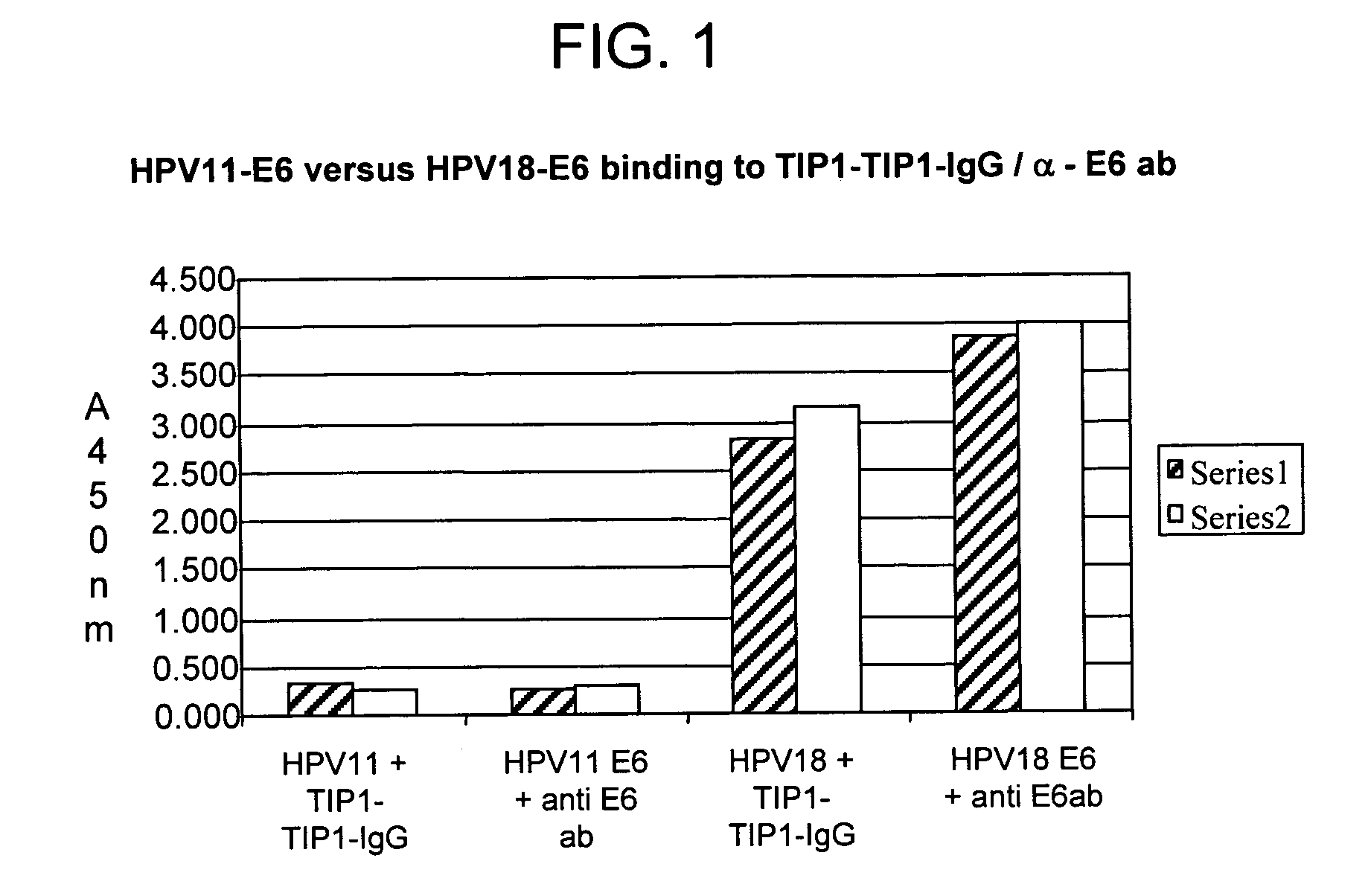
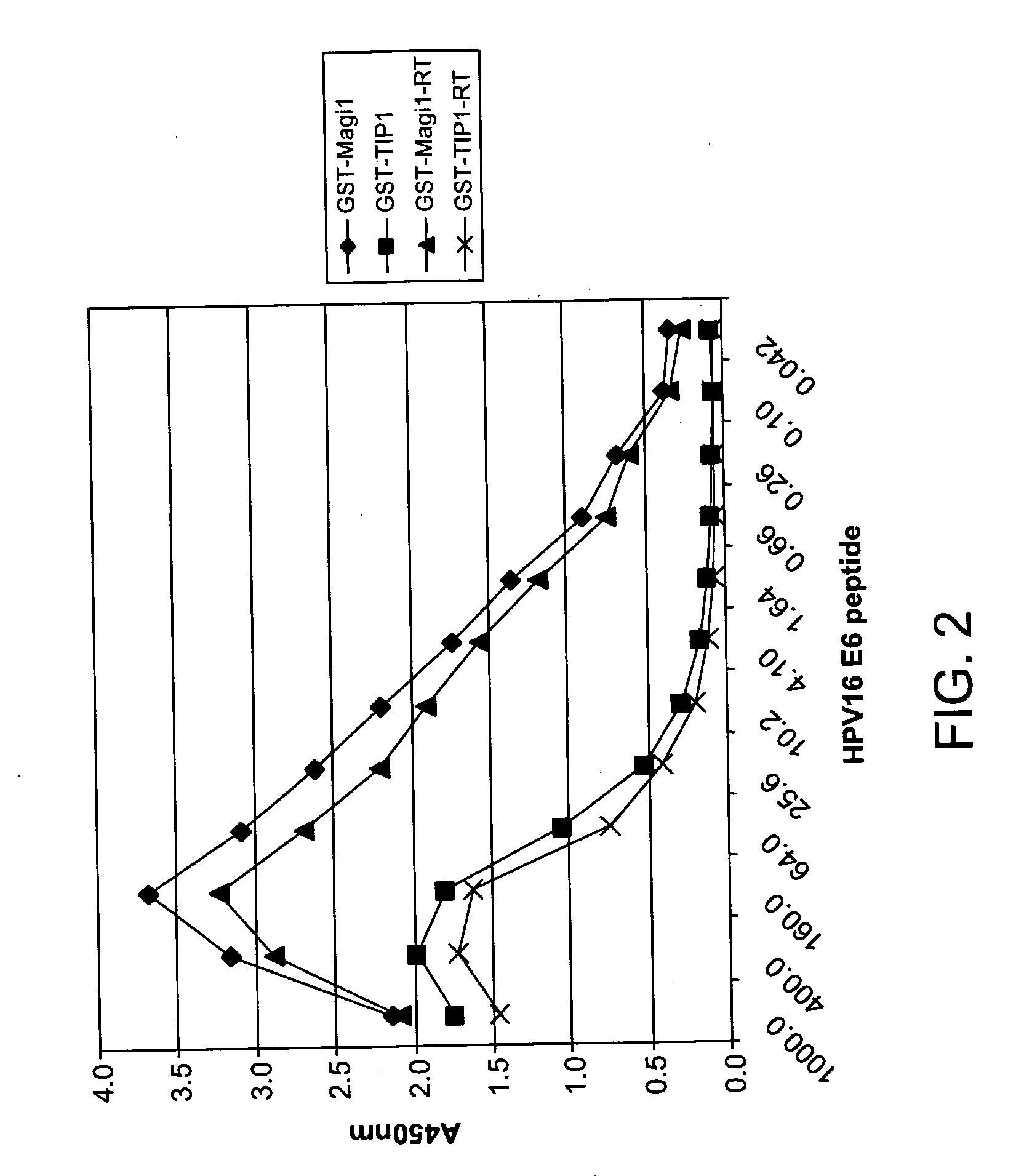
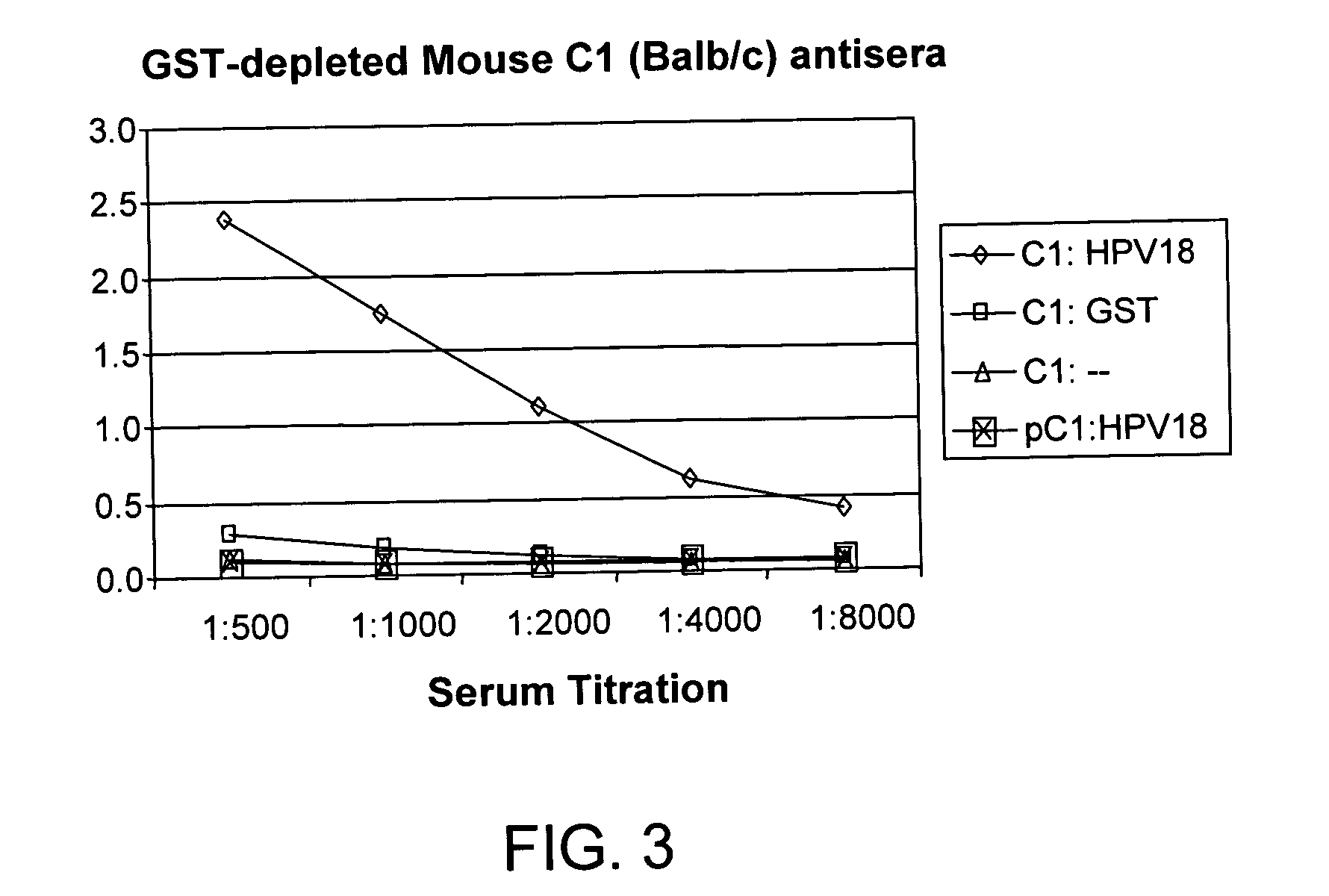
Methods of diagnosing cervical cancer
a cervical cancer and diagnostic method technology, applied in the field of detection of biological markers, can solve the problems of 5,000 deaths each year, increased treatment options, and large proportion of hpv-infected persons
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Sequence Analysis of HPV E6 Proteins to Determine Oncogenic Potential
[0309] PDZ proteins are known to bind certain carboxyl-terminal sequences of proteins (PLs). PL sequences that bind PDZ domains are predictable, and have been described in greater detail in U.S. patent application Ser. Nos. 09 / 710,059, 09 / 724,553 and 09 / 688,017. One of the major classes of PL motifs is the set of proteins terminating in the sequences -X-(S / T)-X-(V / I / L). We have examined the C-terminal sequences of E6 proteins from a number of HPV strains. All of the strains determined to be oncogenic by the National Cancer Institute exhibit a consensus PDZ binding sequence. Those E6 proteins from papillomavirus strains that are not cancerous lack a sequence that would be predicted to bind to PDZ domains, thus suggesting that interaction with PDZ proteins is a prerequisite for causing cancer in humans. This correlation between presence of a PL and ability to cause cancer is 100% in the sequences examined (Table 3A)...
example 2
[0312] Identification of PDZ Domains that Interact with the C-Termini of Oncogenic E6 Proteins
[0313] In order to determine the PDZ domains that can be used to detect oncogenic E6 proteins in a diagnostic assay, the ‘G assay’ (described supra) was used to identify interactions between E6 PLs and PDZ domains. Peptides were synthesized corresponding to the C-terminal amino acid sequences of E6 proteins from oncogenic strains of human papillomavirus. These peptides were assessed for the ability to bind PDZ domains using the G-assay described above and PDZ proteins synthesized from the expression constructs described in greater detail in U.S. patent application Ser. Nos. 09 / 710,059, 09 / 724,553 and 09 / 688,017. Results of these assays that show a high binding affinity are listed in Table 4 below.
[0314] As we can see below, there a large number of PDZ domains that bind some of the oncogenic E6 proteins. However, only the second PDZ domain from MAGI-1 seems to bind all of the oncogenic E6 ...
example 3
[0316] Generation of Eukaryotic Expression Constructs Bearing DNA Fragments that Encode HPV E6 Genes or Portions of HPV E6 Genes
[0317] This example describes the cloning of HPV E6 genes or portions of HPV E6 genes into eukaryotic expression vectors in fusion with a number of protein tags, including but not limited to Glutathione S-Transferase (GST), Enhanced Green Fluorescent Protein (EGFP), or Hemagglutinin (HA).
[0318] A. Strategy
[0319] cDNA fragments were generated by RT-PCR from HPV cell line (cervical epidermoid carcinoma, ATCC# CRL-1550 and CRL-1595 for HPV E6 16 and 18, respectively) derived RNA, using random (oligo-nucleotide) primers (Invitrogen Cat.# 48190011). DNA fragments corresponding to HPV E6 were generated by standard PCR, using above purified cDNA fragments and specific primers (see Table 5). Primers used were designed to create restriction nuclease recognition sites at the PCR fragment's ends, to allow cloning of those fragments into appropriate expression vecto...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com