Compound amplification detection kit containing forty InDel genetic polymorphic sites of human X chromosome
A genetic polymorphism and detection kit technology, applied in the field of kits for detecting human X chromosome InDel genetic polymorphism sites, can solve the problem that the number of systems and products is small, and the number of detection sites has no great advantage, etc. problems, to achieve the effect of good sample compatibility, good typing efficiency, and low mutation rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
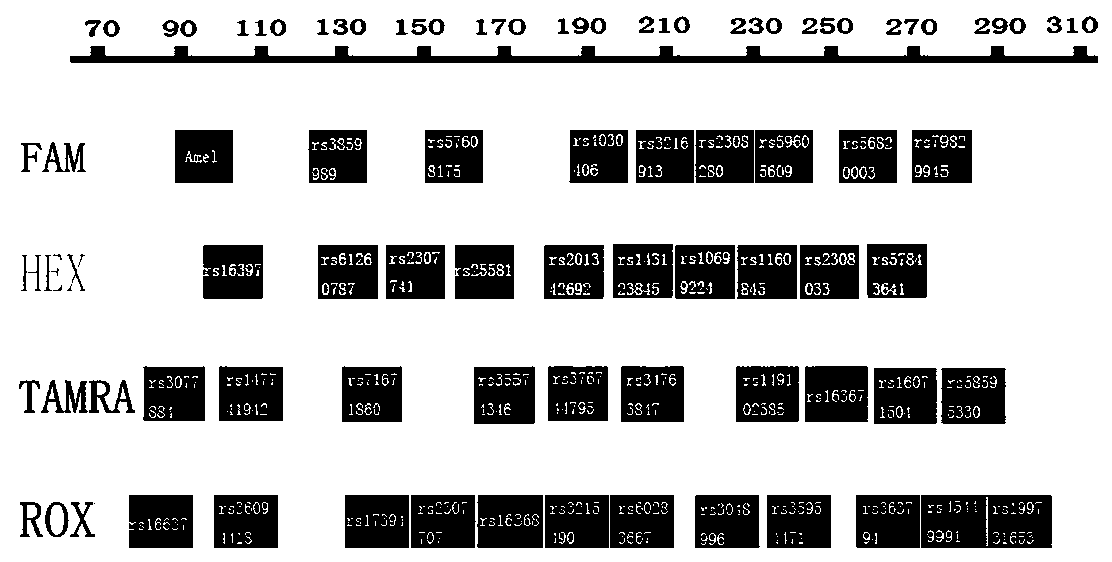
[0036] Example 1: A multiple amplification detection kit comprising 40 InDel genetic polymorphism sites of human X chromosome.
[0037] 1) Design of primers
[0038] 1-1) Screening of 40 X-InDel polymorphic sites
[0039] Based on the dbSNP database established by the National Center for Biotechnology Information (NCBI), combined with relevant literature reports, and according to the proposed screening conditions for InDel polymorphic sites, 40 polymorphic sites were screened from the X chromosome with a length of about 150 Mb. InDel polymorphic loci ensure that they are nearly evenly distributed on the X chromosome with a considerable distance between them. On this basis, Amelogenin was added as a sex identification site. The specific screening criteria for X-InDel loci are as follows: ①The length difference of X-InDel allelic fragments (the number of bases inserted or deleted) is between 3 and 30 bp; ②It is located in the intron region of X chromosome; ③dbSNP database The m...
Embodiment 2
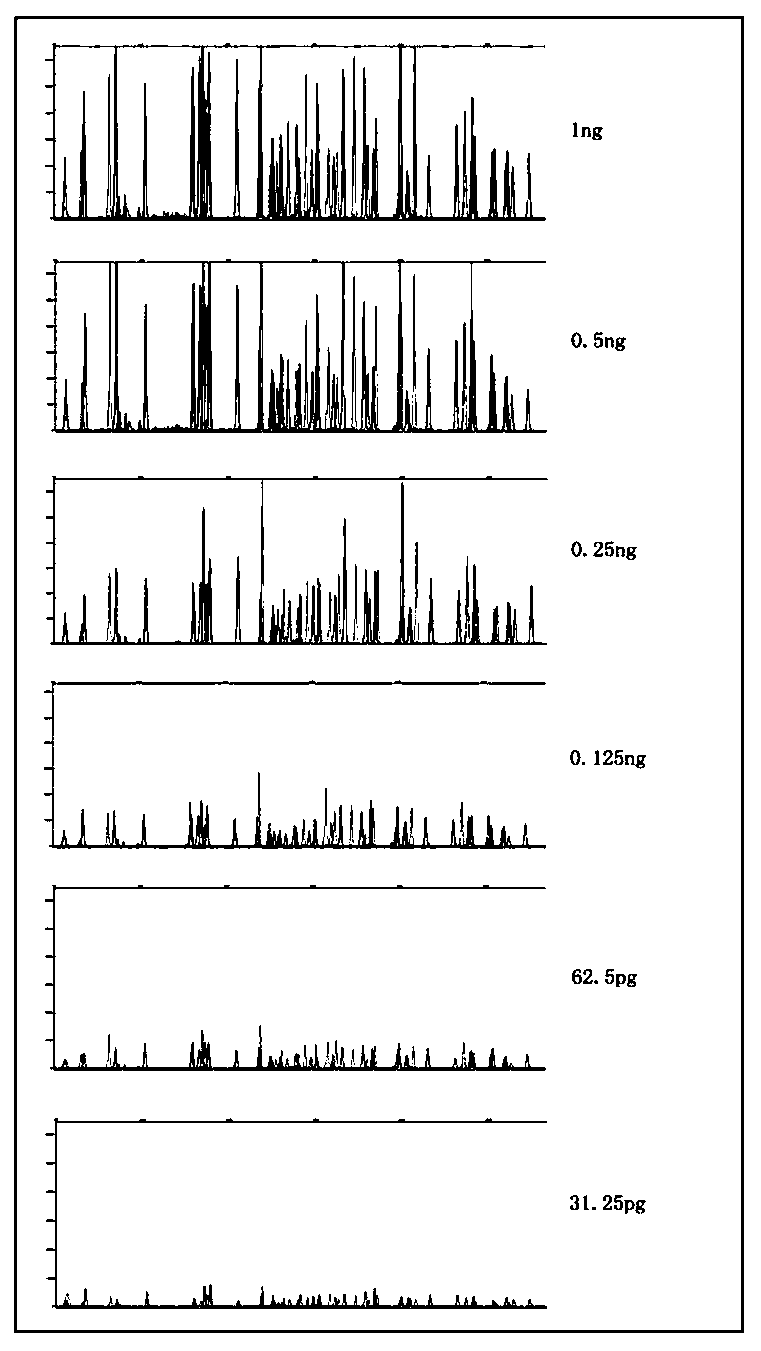
[0055] Example 2: Sensitivity test of the multiple amplification detection kit containing 40 InDel genetic polymorphism sites of human X chromosome.
[0056] The sensitivity test was performed with the multiplex amplification detection kit described in Example 1. When preparing the amplification system, the template amounts of standard genomic DNA were set to 1ng, 0.5ng, 0.25ng, 0.125ng, 62.5pg, and 31.25pg, respectively. Amplification detection was performed according to the amplification system and procedure described in Example 1. The test results obtained as figure 2 Shown: When the amount of template is higher than or equal to 62.5pg, each detection site can produce peaks normally, and the balance of peak height between colors is good; when the amount of template is further reduced to 31.25pg, the two positions of rs16637 and rs36094418 The allele peak height of the point is lower than the analysis threshold; the sensitivity of the effective detection sample of the kit ...
Embodiment 3
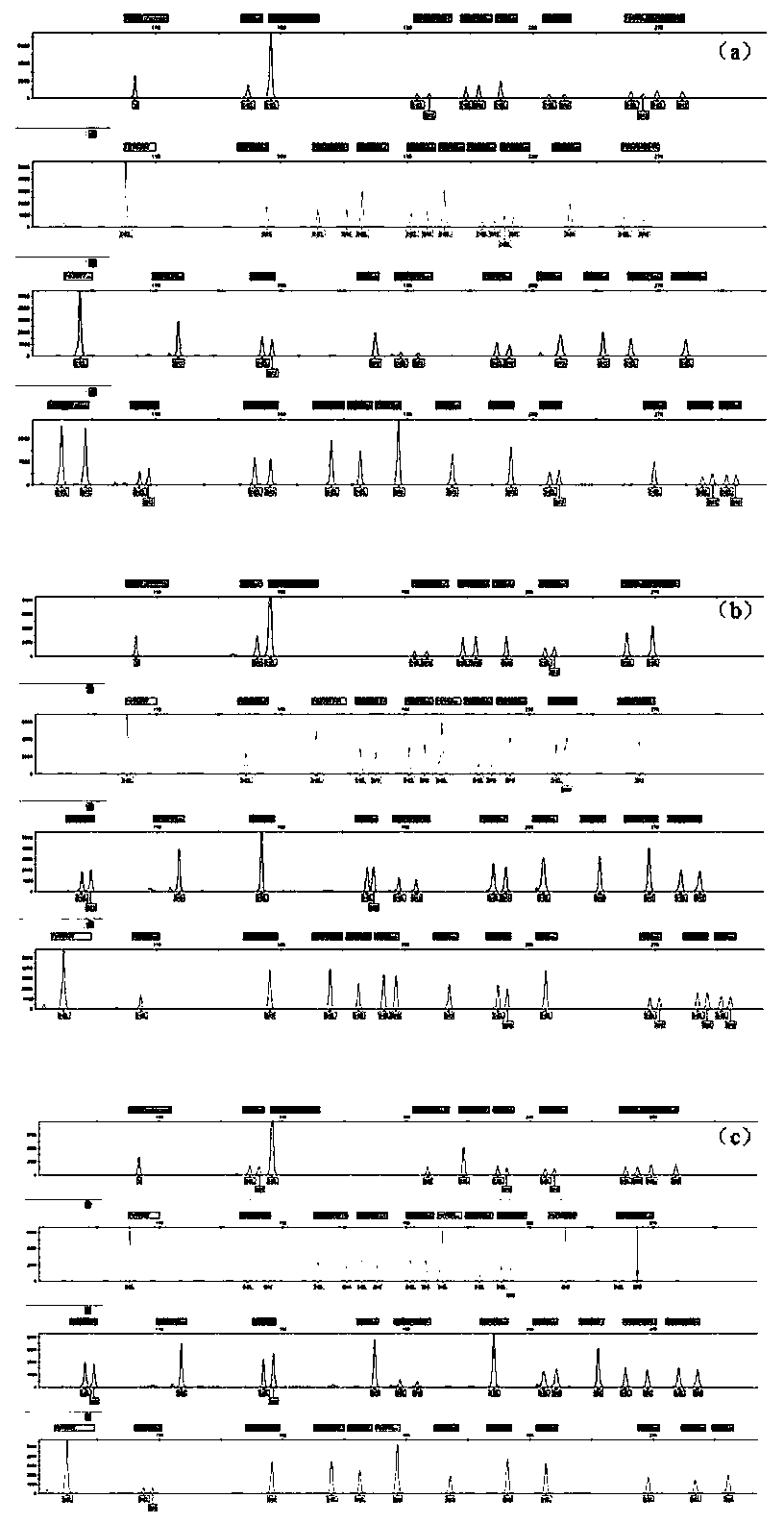
[0057] Example 3: Identification of grandmother-mother-granddaughter relationship.
[0058] The sample source was provided by volunteers, and the sample type was saliva spot; DNA was extracted by chelex-100 method. Take 2×1.2mm saliva spots, add 80μL of 5% chelex-100 solution, incubate at 56°C for 30min, shake fully, keep at 95°C for 8min, shake and mix well, and centrifuge at 12000rpm to get the supernatant.
[0059] Amplification detection: Carry out according to the amplification conditions and detection conditions described in Example 1.
[0060] The amplified products were detected by capillary electrophoresis detection and analysis system, and the results were analyzed by data analysis software GeneMapper® ID-X.
[0061] Typing map such as image 3 As shown, a, b, and c in the figure are the X-InDel of the grandmother, mother, and granddaughter respectively; the specific typing results are as follows:
[0062]
[0063]
[0064] The results showed that the grandd...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com