Methods, cells & organisms
a technology of cells and organisms, applied in the field of methods, cells & organisms, can solve the problems of low efficiency at which this occurs in both alleles, time and cost of developing such engineered proteins, and high undesirable effects, and achieve the effect of large and precise nucleotide sequence deletions or insertions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Precise DNA Modifications
[0174](a) Use of Nickase for HDR
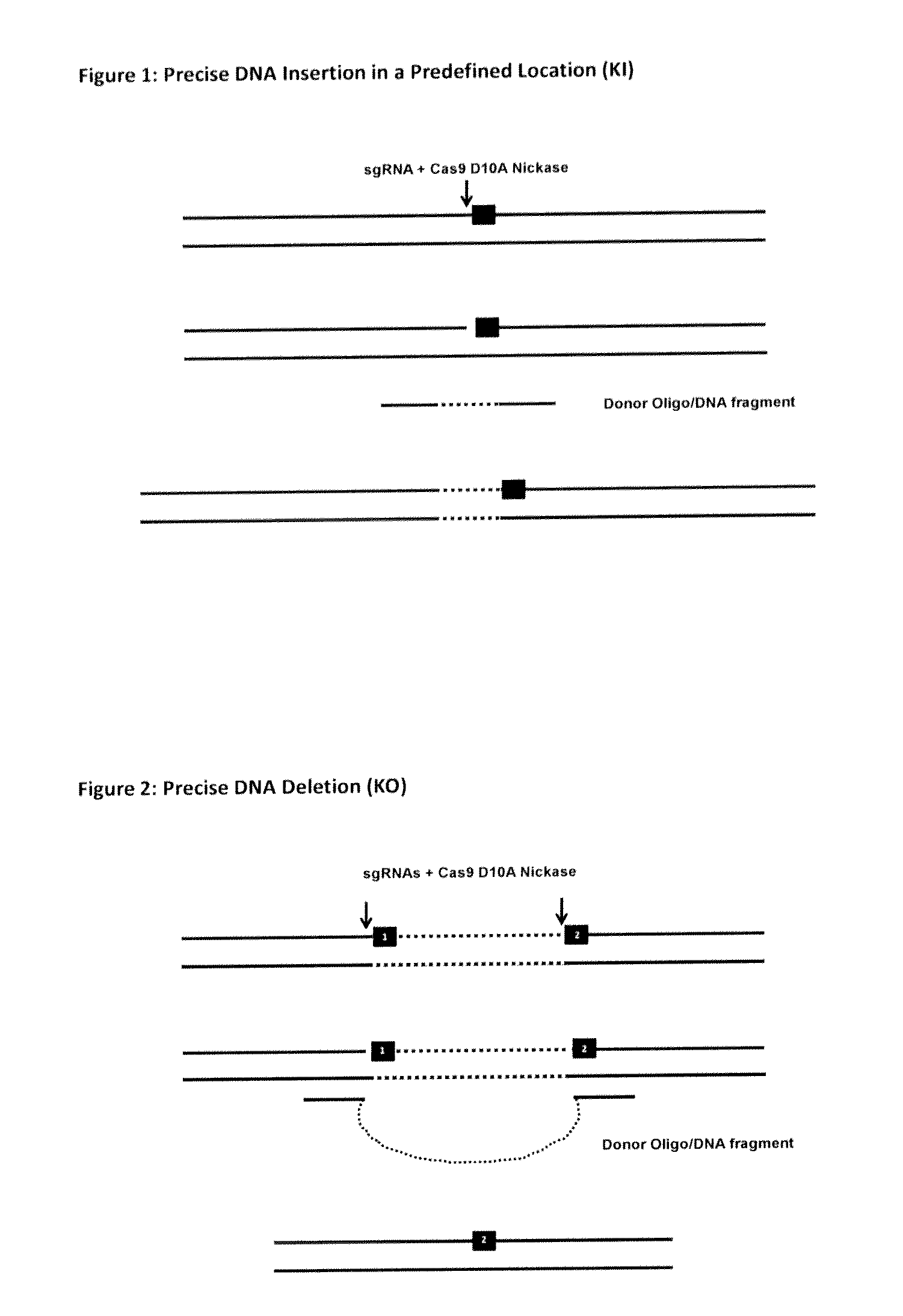
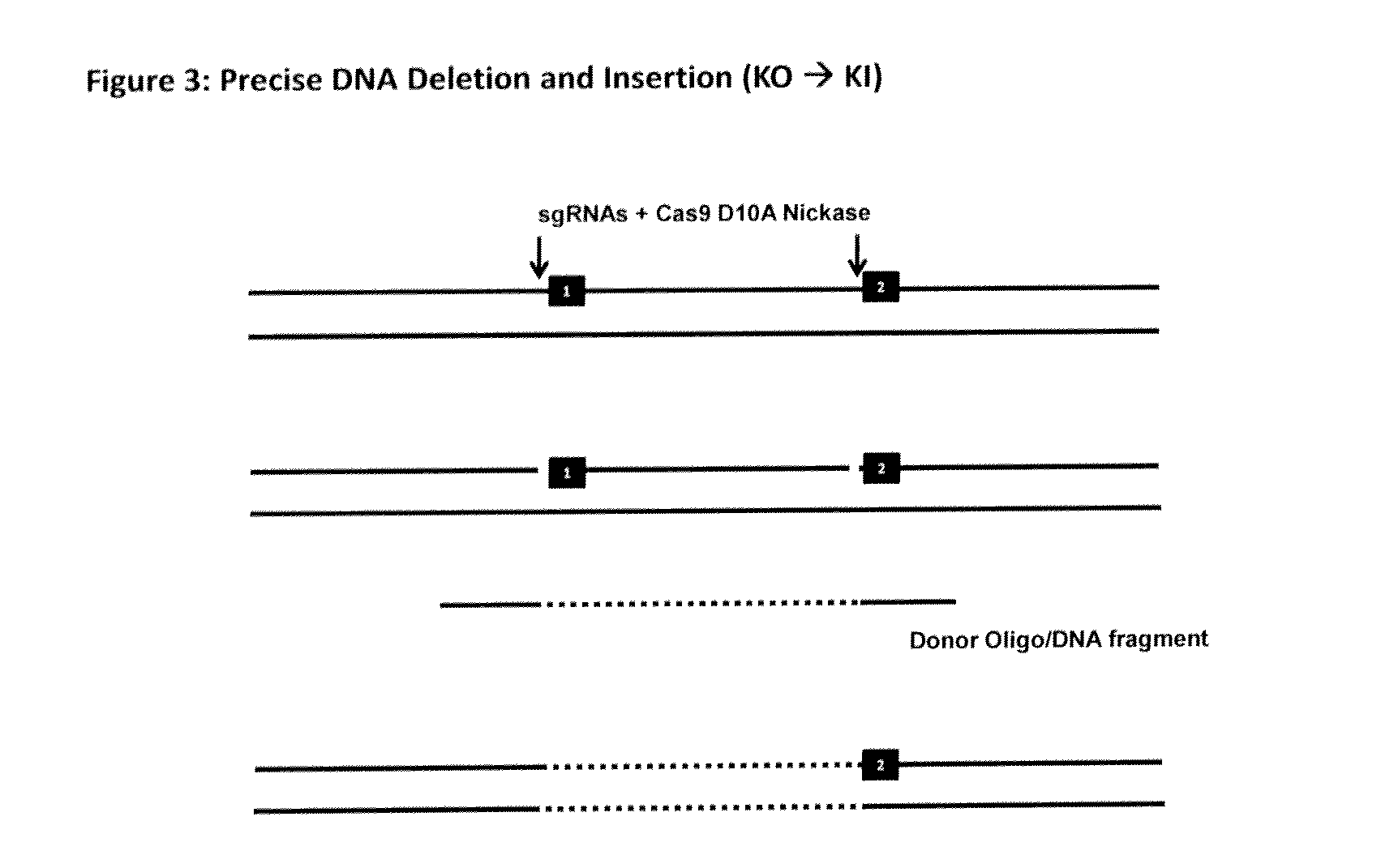
[0175]It has been reported that the Cas9 nuclease can be converted into a nickase through the substitution of an aspartate to alanine (D10A) in the RuvCl domain of SpCas9 (Cong et al). It is noteworthy that DNA single-stranded breaks are preferentially repaired through the HDR pathway. The Cas9 D10A nickase, when in a complex with mature crRNA:tracrRNA, can specifically induce DNA nicking at a precise location. With this in mind, we propose extending the application of the CRISPR / Cas system by creating a nick in a given location in a genome using Cas9 D10A nickase and then exploiting the HDR pathway for inserting a single-stranded DNA fragment (endogenous or exogenous) which will contain DNA homology flanking the nick. Typically for recombineering 50 by is enough for efficient recombination) flanking the nicked DNA junction to bring in and insert a given DNA in a precision location; similar size homology will be used with the ...
example 2
Recycling PAM for Sequential Insertions or Deletions
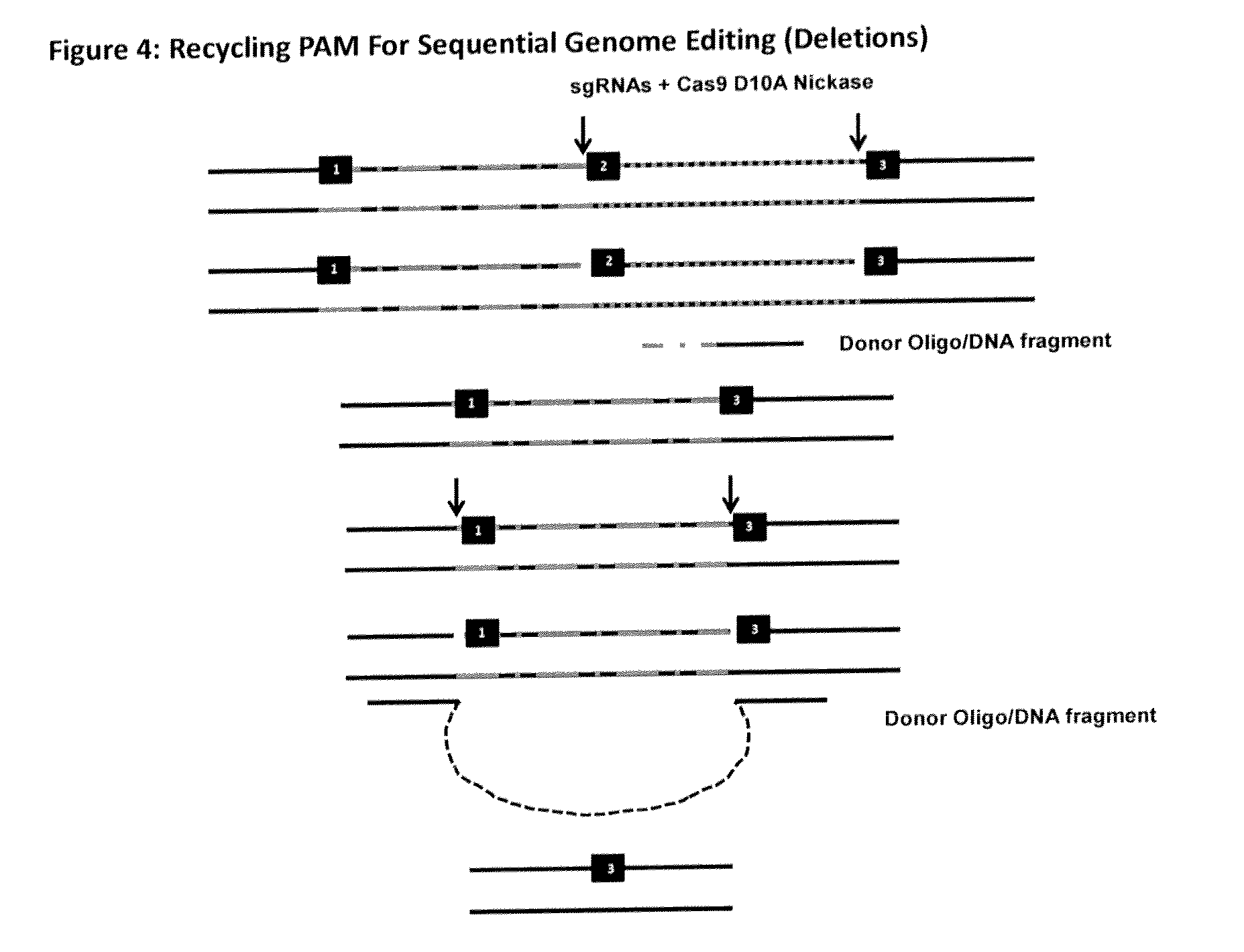
[0177]In certain settings it may be useful to edit a genome by chromosome walking. Using any of the three examples outlined above, it could be possible to carry out sequential genome editing in a stepwise fashion whereby the PAM sequence used in a previous round of CRISPR / Cas mediated genome editing, can be re-used to carry out multiple rounds of genome editing such as deletions, insertions or the simultaneous deletion and insertion. An example of sequential deletion whereby the PAM sequence from the previous genome editing step is recycled is shown in FIG. 4. Using the PAM recycling approach, it is possible to carry out sequential insertions as well as sequential simultaneous deletion and insertion.
example 3
Rapid Insertion of Lox Sites Using CRISPR / Cas System
[0178]Targeting efficiency using conventional homologous recombination methods in ES cells is low. In a different setting, the CRISPR / Cas system can be used to rapidly and efficiently introduce lox sites or other recombinase recognition sequence such as Frt in a defined location to act as a landing pad for genome editing using recombinase mediated cassette exchange (RMCE) (Qiao J, Oumard A, Wegloehner W, Bode J: Novel tag-and-exchange (RMCE) strategies generate master cell clones with predictable and stable transgene expression properties. J Mol Biol 2009, 390(4):579-594; and Oumard A, Qiao J, Jostock T, Li J, Bode J: Recommended Method for Chromosome Exploitation: RMCE-based Cassette-exchange Systems in Animal Cell Biotechnology. Cytotechnology 2006, 50(1-3):93-108). Once the lox sites are introduced into the genome, inversion, deletion or cassette exchange to delete and introduce DNA fragment varying in size at this site can be e...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com