Method for analyzing mixed sample DNA
An analysis method and technology of mixed samples, applied in the field of DNA typing, can solve the problems of inaccurate quantification, low polymorphism, complex calculation, etc., and achieve the effect of advancing the time limit and protecting the safety of fetuses and pregnant women
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
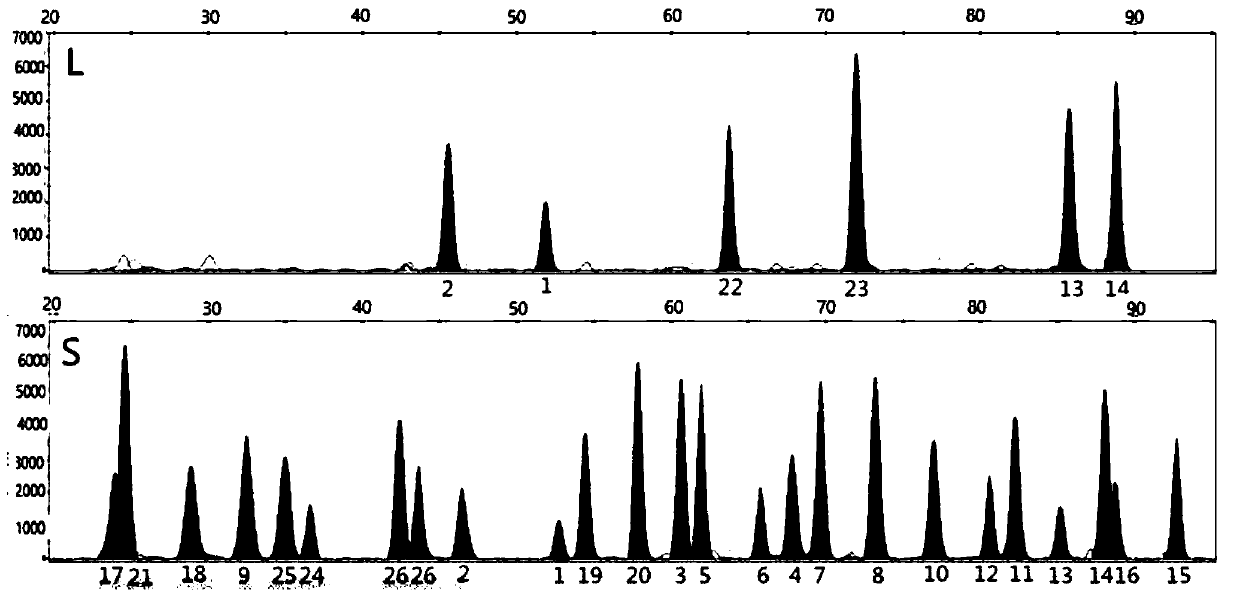
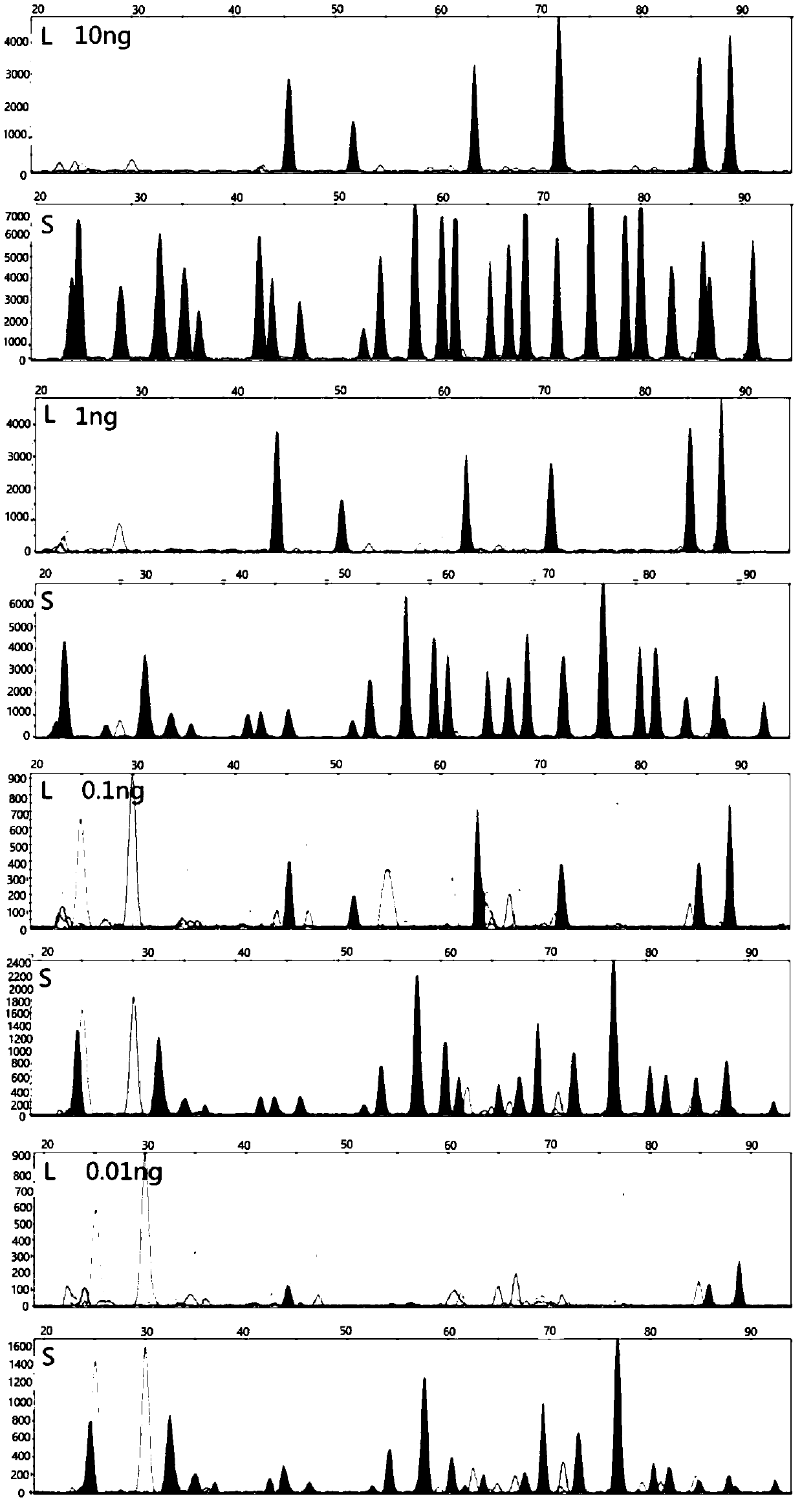
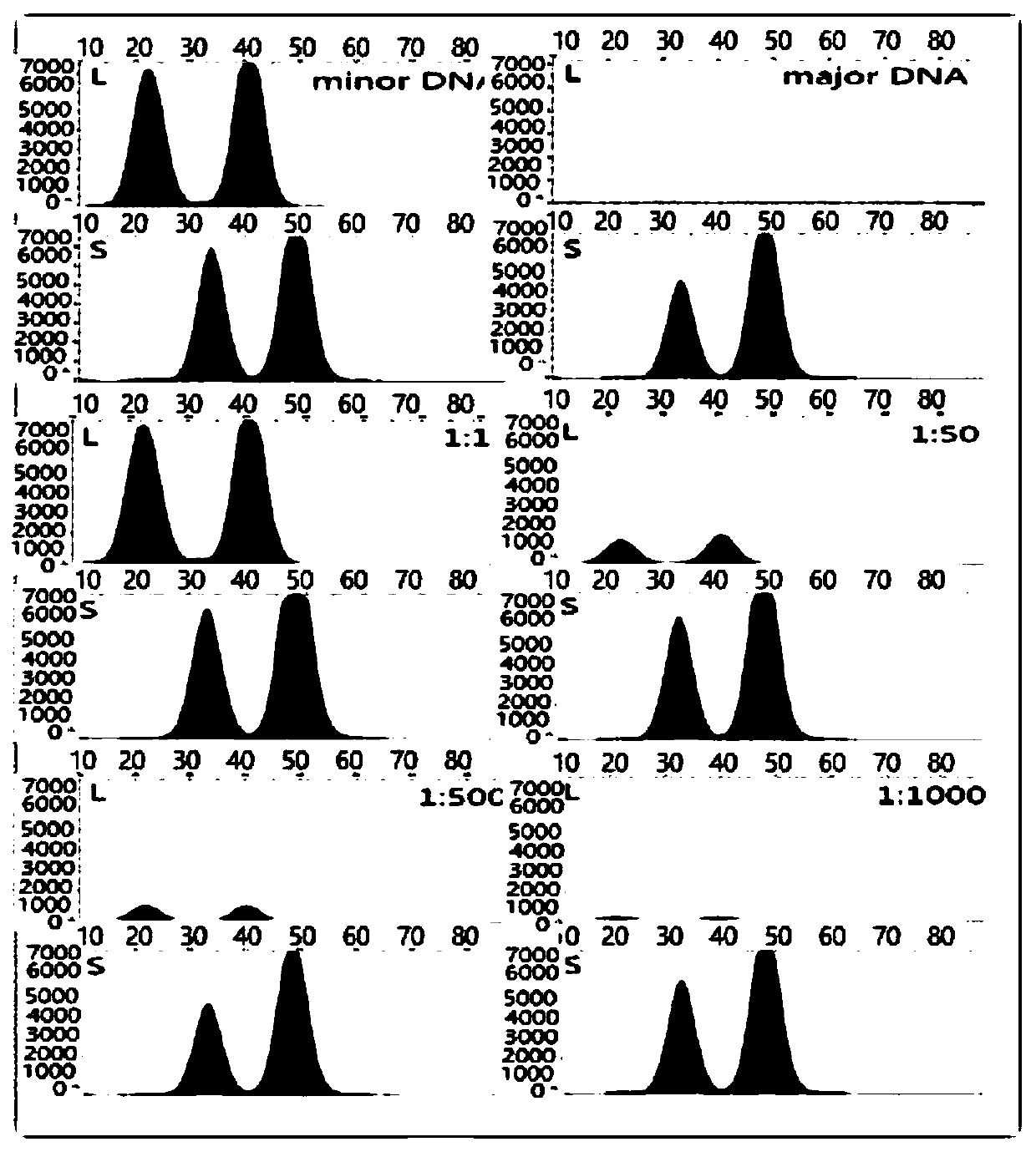
[0077] This method uses the method of INDEL+Microhaplotypes to solve the analysis problem of mixed spots, and the process is as follows:
[0078]First select the INDEL site with good frequency distribution in the Chinese population (the number of base differences in indel is 3bp-7bp, HET>0.3), and there are multiple SNP sites with good polymorphism within 200bp downstream or upstream of the INDEL site point (choose standard HET>0.1). Firstly, PCR products containing two sites of INDEL+Microhaplotypes were amplified, wherein the upstream primer was specific for the INDEL site, and the design of the downstream primer included SNPs. First, the specific amplification of INDEL is used to amplify the paternal or maternal DNA of the DNA sample, and then the polymorphism of the downstream closely linked SNP site is used to distinguish the individual. For mixed DNA samples, the typing principle combines INDEL's specific amplification technology to sort out a DNA molecule of an individ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com