A method for analyzing mixed sample DNA based on indel-snp linkage relationship
A technology of mixed samples and relationship analysis, applied in the field of forensic detection, can solve the problems of detection loss, low polymorphism, and no forensic mixed spots, etc., and achieve the effect of protecting safety, advancing time limit, and avoiding amniotic fluid or chorionic villus puncture
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] A kind of method based on INDEL-SNP linkage relationship analysis mixed sample DNA of the present invention adopts the method of INDEL+SNP to solve the analysis problem of mixed spot, and its process is as follows:
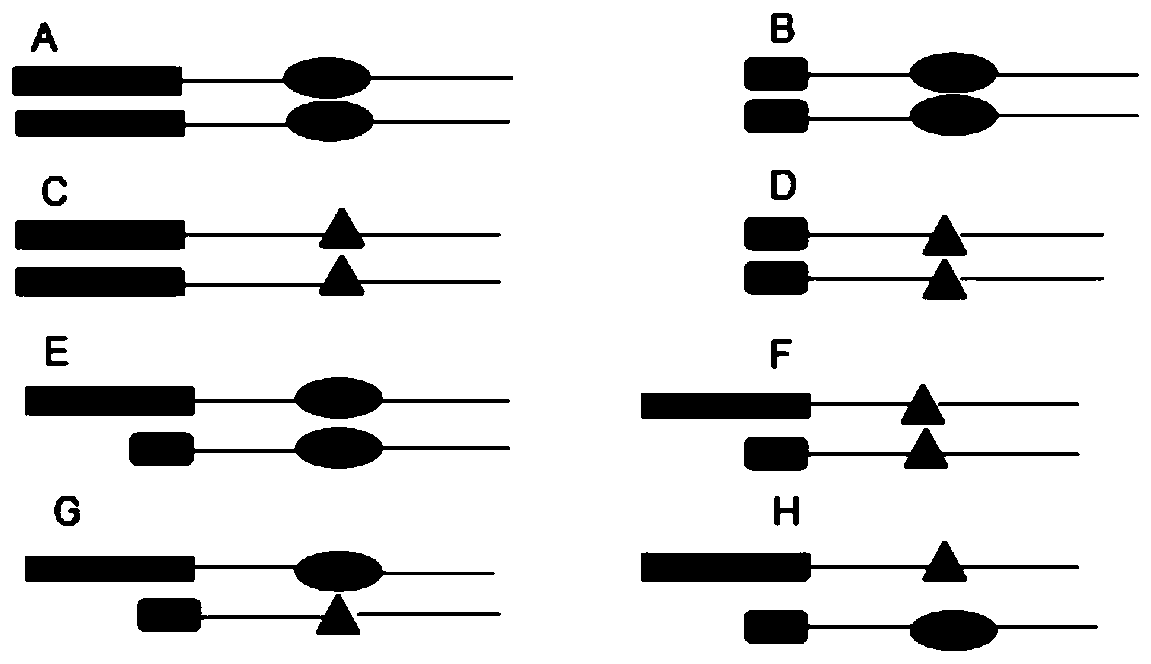
[0049] First, select an INDEL site with a good frequency distribution in the Chinese population (the base difference of INDEL is 2bp-10bp), and there is another SNP site with good polymorphism within 300bp downstream or upstream of the INDEL site (selection criteria HET>3.0). Firstly, a PCR product containing two sites of INDEL+SNP is amplified, wherein the upstream primer is specific for the INDEL site, and the downstream primer is designed to include the SNP, and a total of 8 genotypes will appear in the population such as figure 1 as shown, figure 1 are the 8 genotypes that will appear in the population. The long and short boxes represent IN-type and DEL-type INDEL polymorphisms, respectively, and ovals and triangles represent two polymorphisms with SN...
Embodiment 2
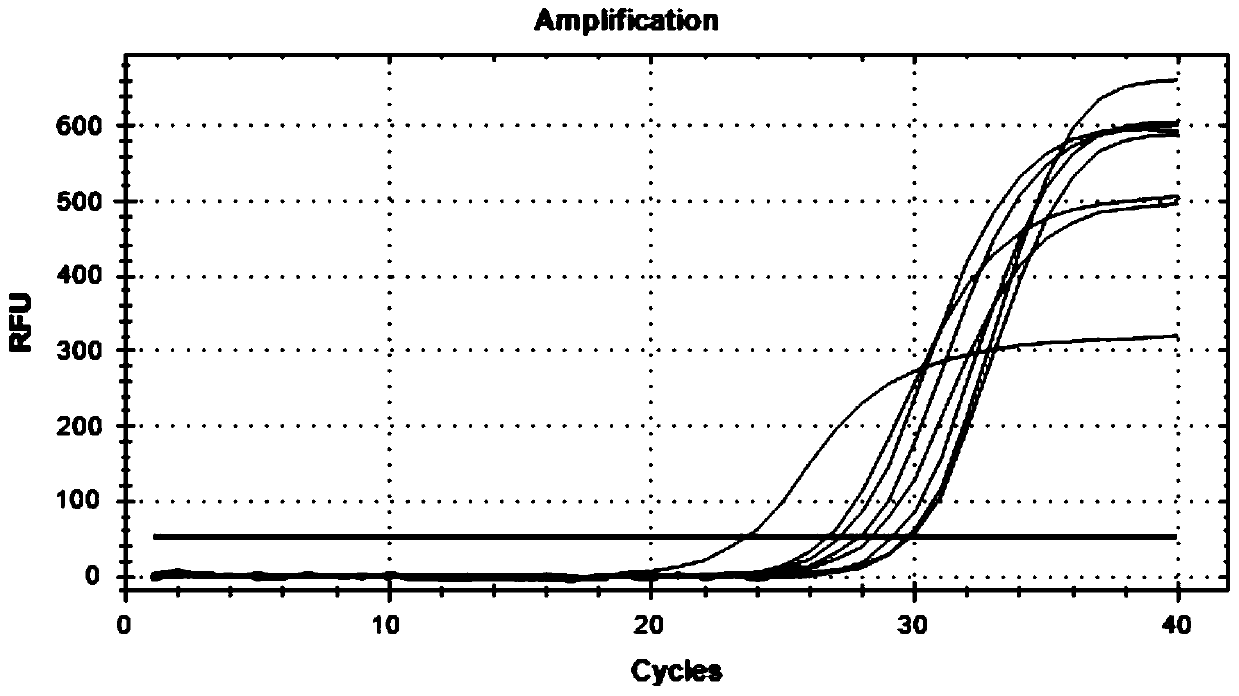
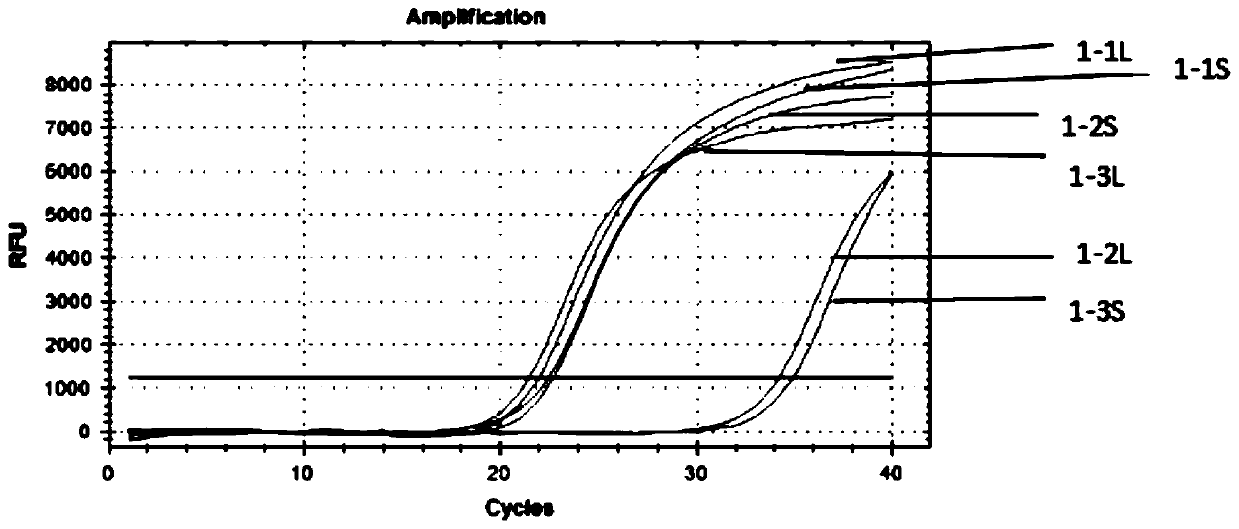
[0083] If a spot evidence is extracted from the on-site bed sheet, 11 ID-SNP types are performed on the spot. It was found that the △Cq of rs397832665 and rs397832994 loci were > 8, and the typing could be determined as LL type and SS type respectively. The △Cq of the remaining 9 sites were all <0.5, and the typing could be judged as LS type. The victim was typed as LL and SS at rs397832665 and rs397832994. There were four suspects A, B, C, and D at the scene. At the rs397832665 locus, their types were LL, LS, SS, and LL. Considering the marks and the victim's type, the suspect B and C can be ruled out. At the rs397832994 locus, the types of suspects A and D were LS and SS, respectively. Combining the spot marks and the victim's typing, suspect A could be ruled out. Among the remaining 9 loci of LS type, taking rs397844074 as an example, the victim's type is SS type, and the suspect D's type is LL type, so suspect D cannot be ruled out.
[0084] Analytical method described...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com