Asymmetric deoxyribose nucleic acid (DNA) artificial adapters by using second-generation high-throughput sequencing technology and application thereof
A double-linked, asymmetric technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of consumables and reagents, low technical efficiency, cumbersome steps, etc., to improve stability and simplify construction Library flow, easy to save the effect for a long time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0031] The present invention will be further described below in conjunction with the drawings and embodiments.
[0032] An asymmetric DNA double link head for second-generation high-throughput sequencing, composed of two DNA oligonucleotide single strands, and its preparation method is as follows:
[0033] Dissolve the DNA oligonucleotide single-stranded in the quenching solution, adjust the final concentration to 2mM, the volume is 20 microliters, and then use the following conditions to perform double-link head annealing: 95°C for 5min; 95°C down to 12°C , 0.1°C per second; keep at 12°C, dilute the annealed double link head solution 1:4 to 500μM, and store at -20°C.
[0034] The two DNA oligonucleotide single strands can use the following marker sequences:
[0035]
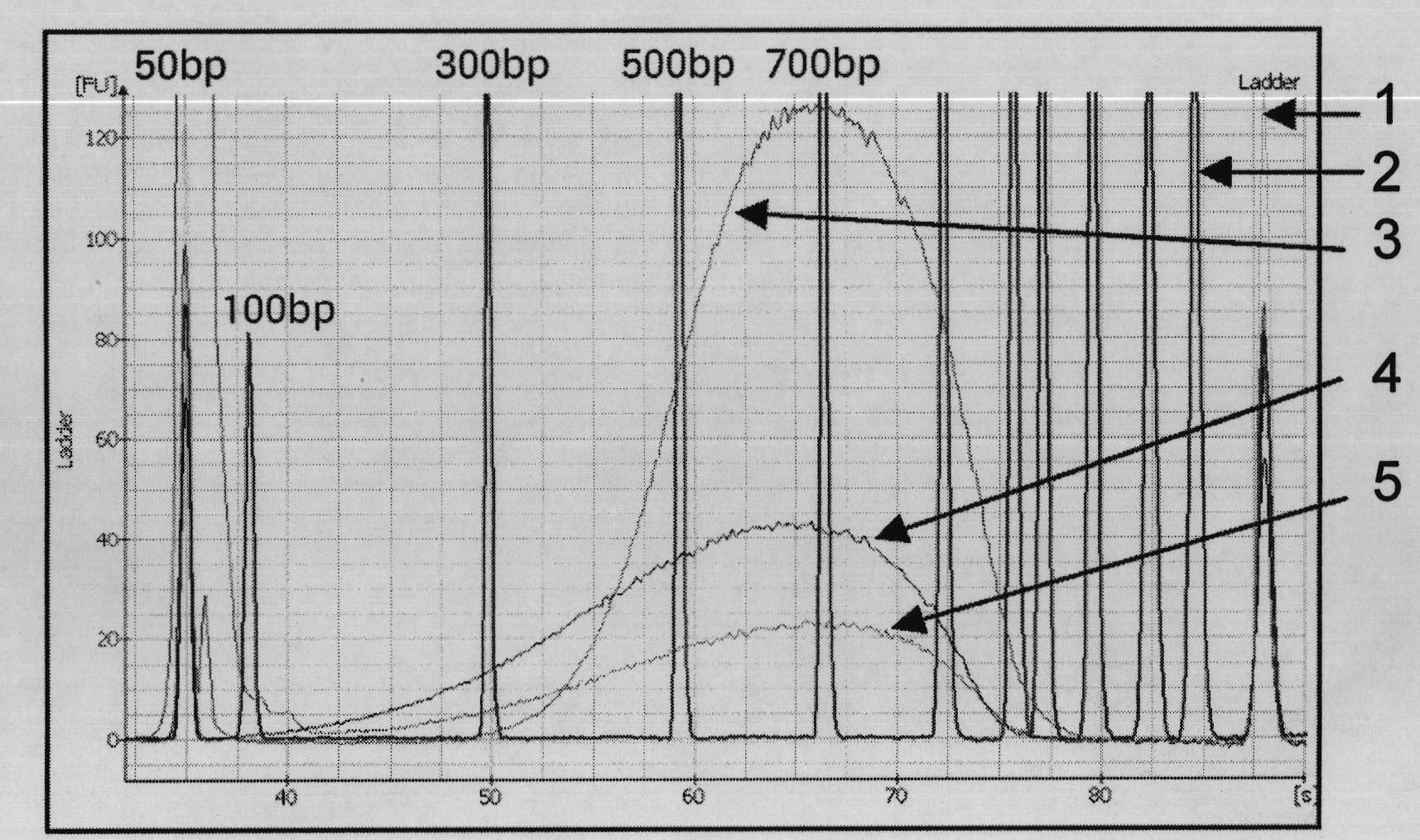
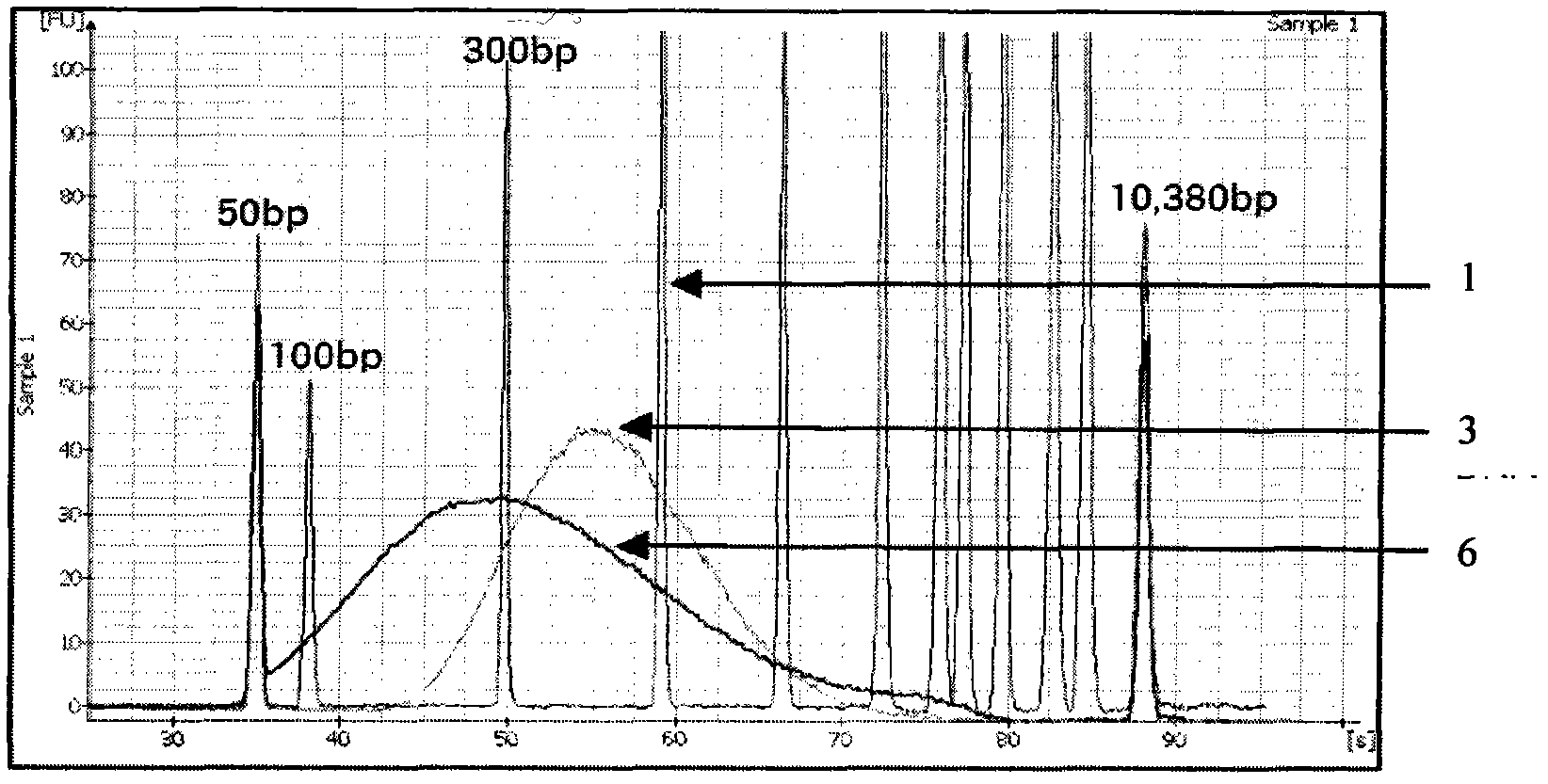
[0036] The following is the method for building a library of asymmetric DNA double link heads using this second-generation high-throughput sequencing, which uses the following process:( figure 2 Quality monitoring char...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com