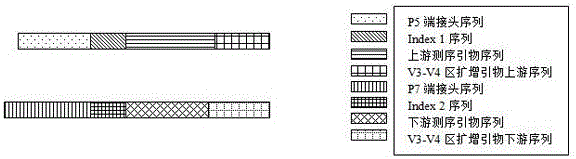
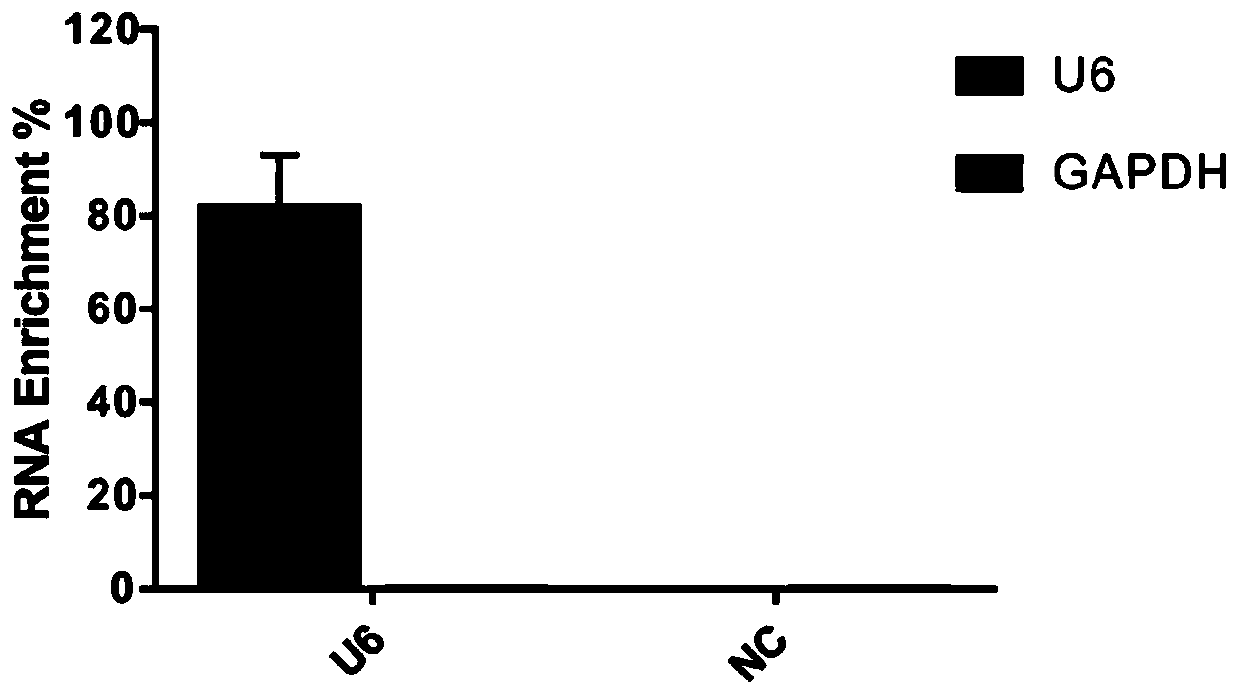
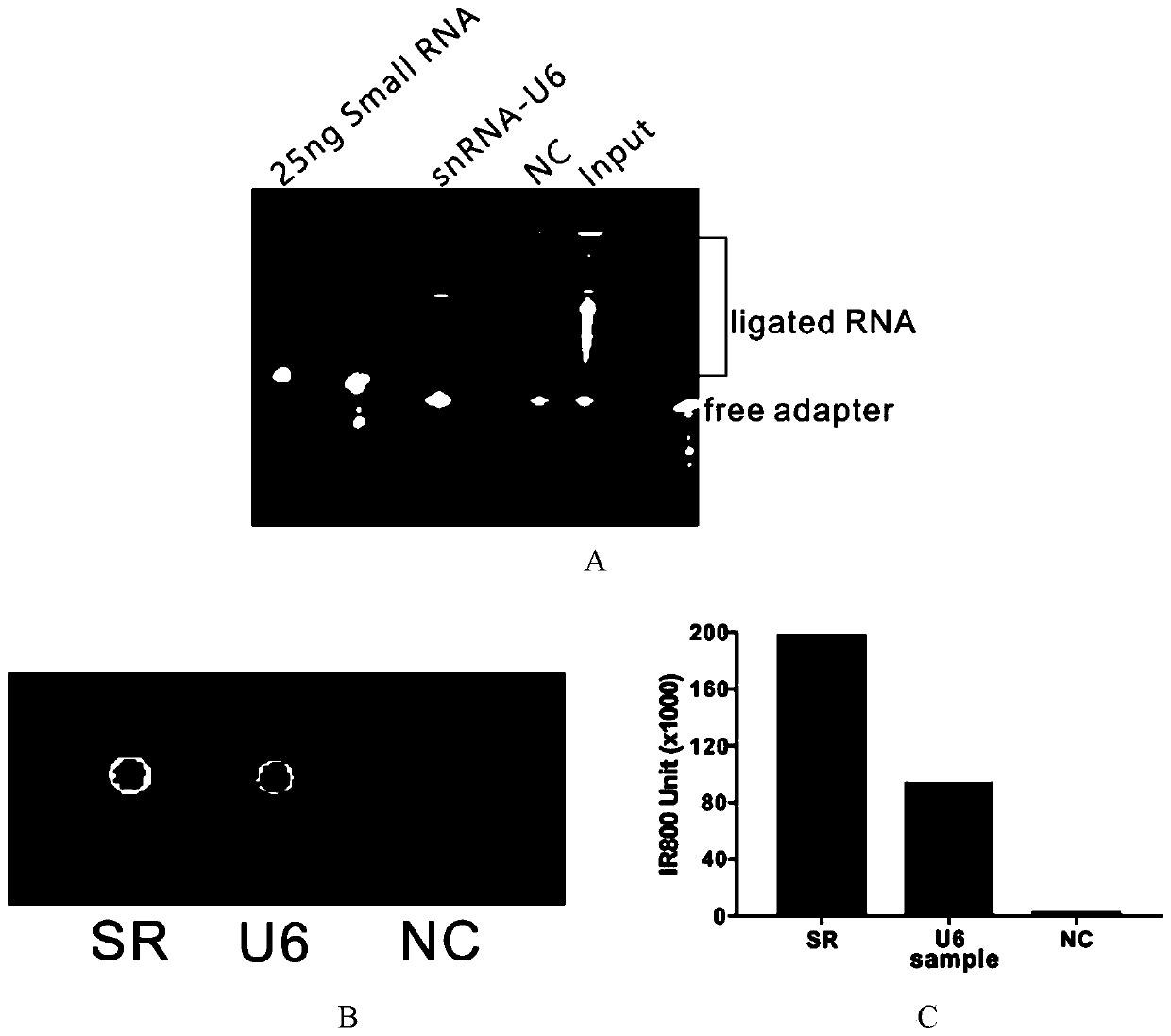
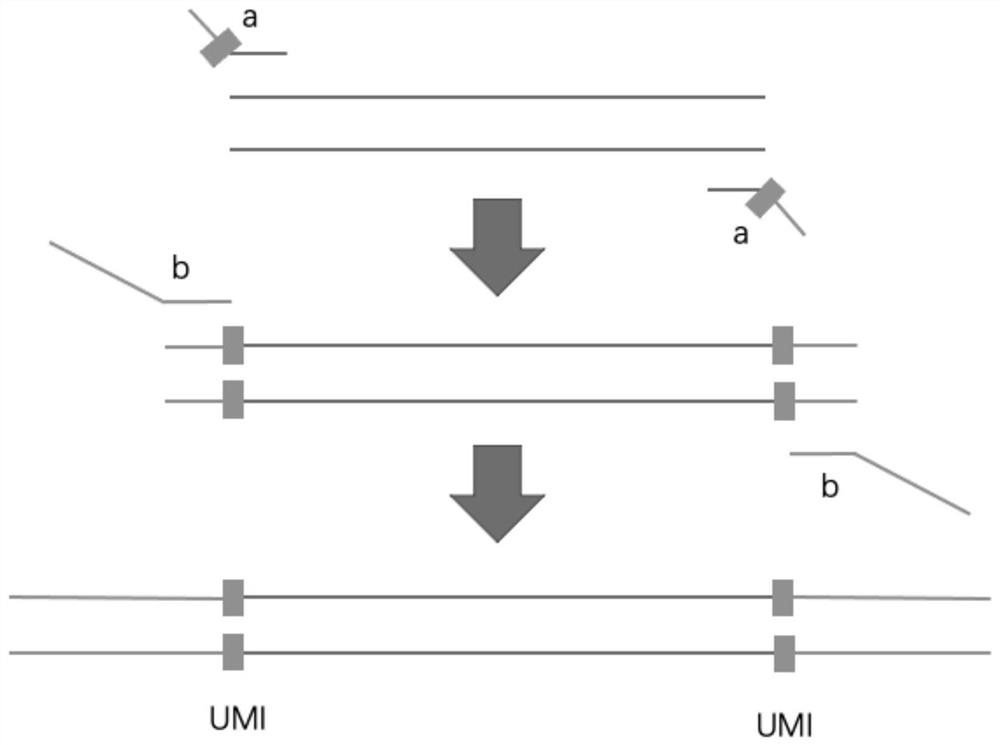
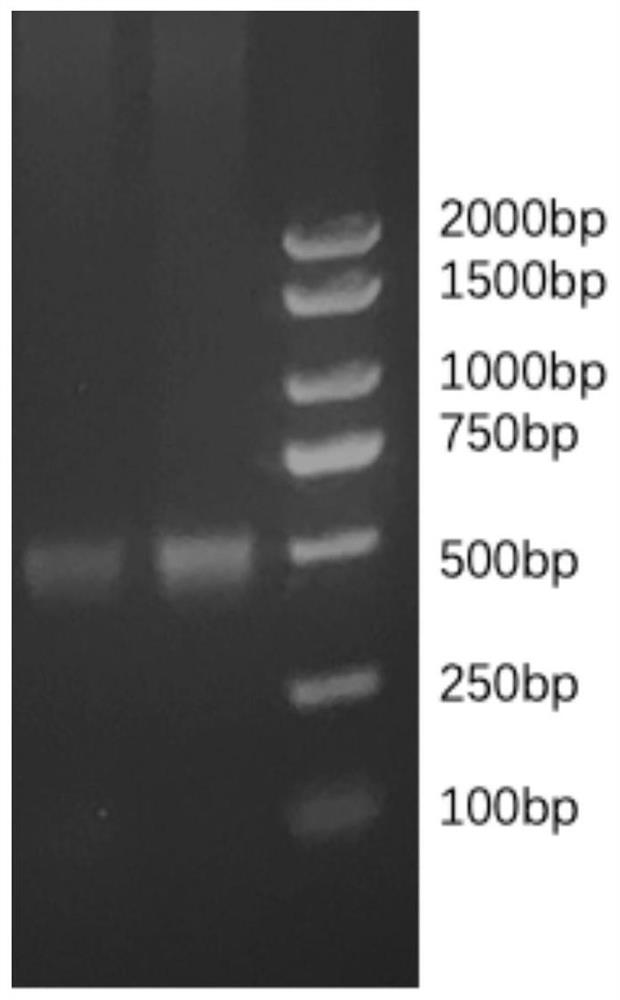
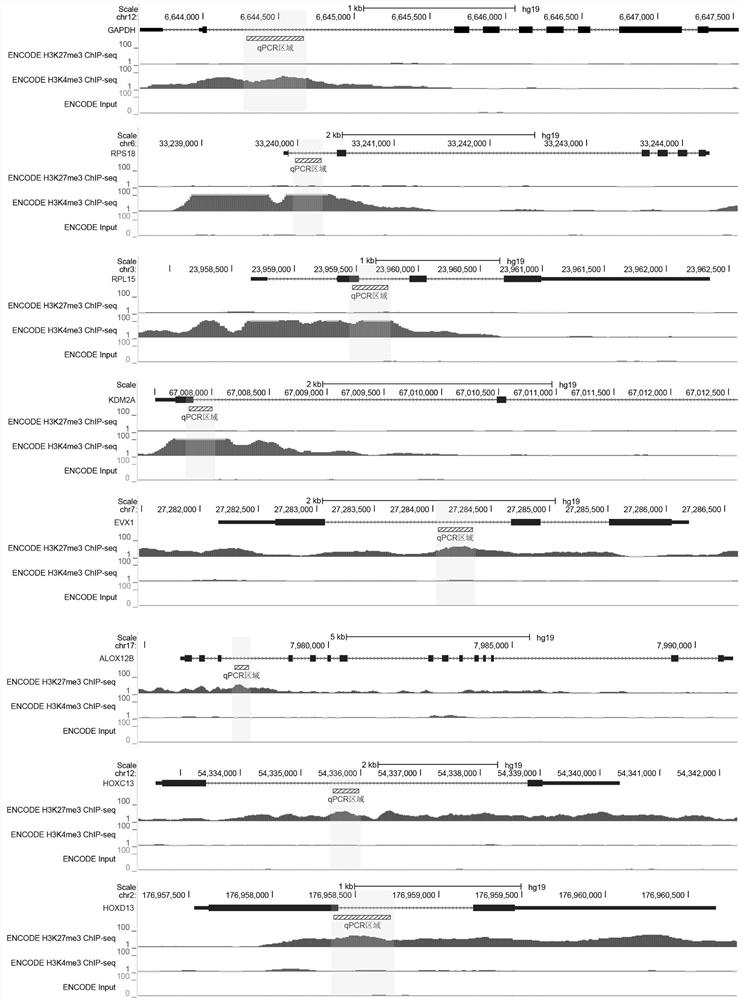
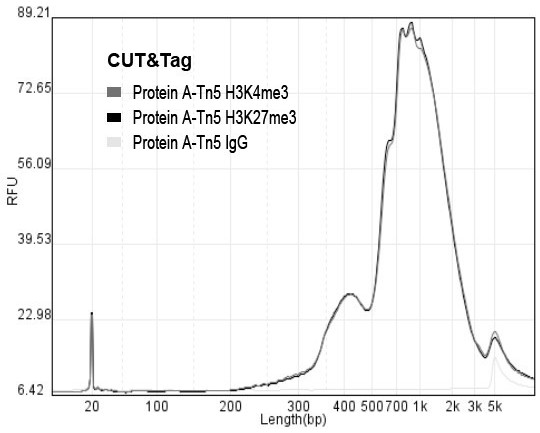
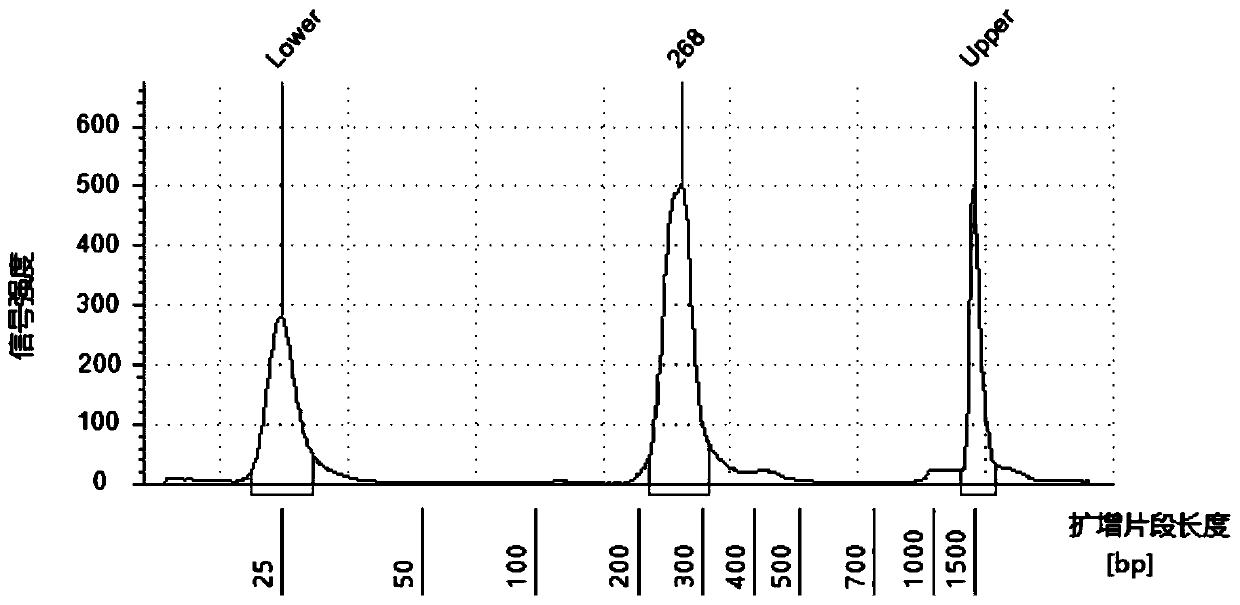
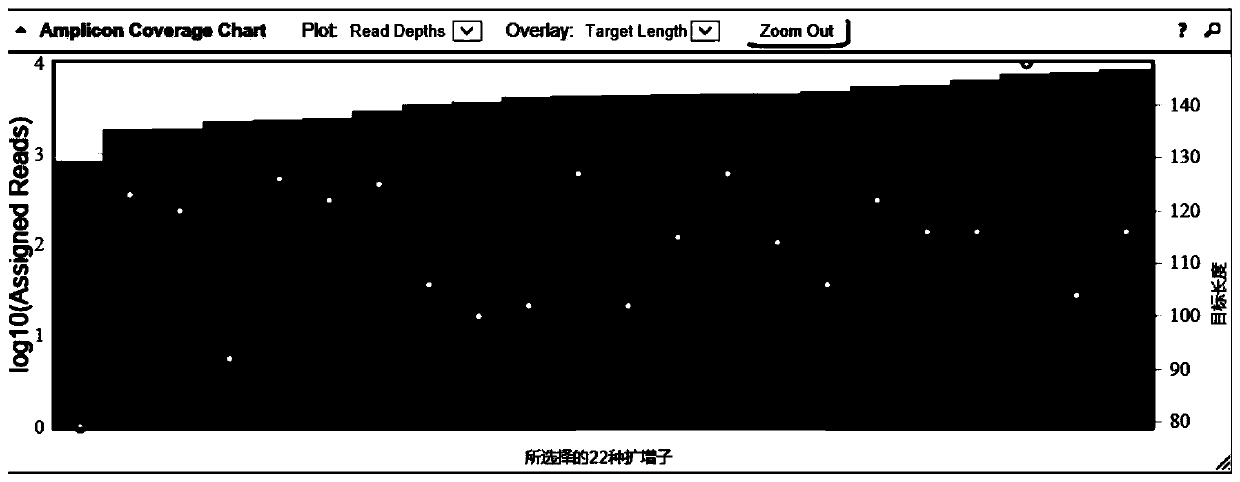
Patents
Literature
31results about How to "Shorten library building time" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
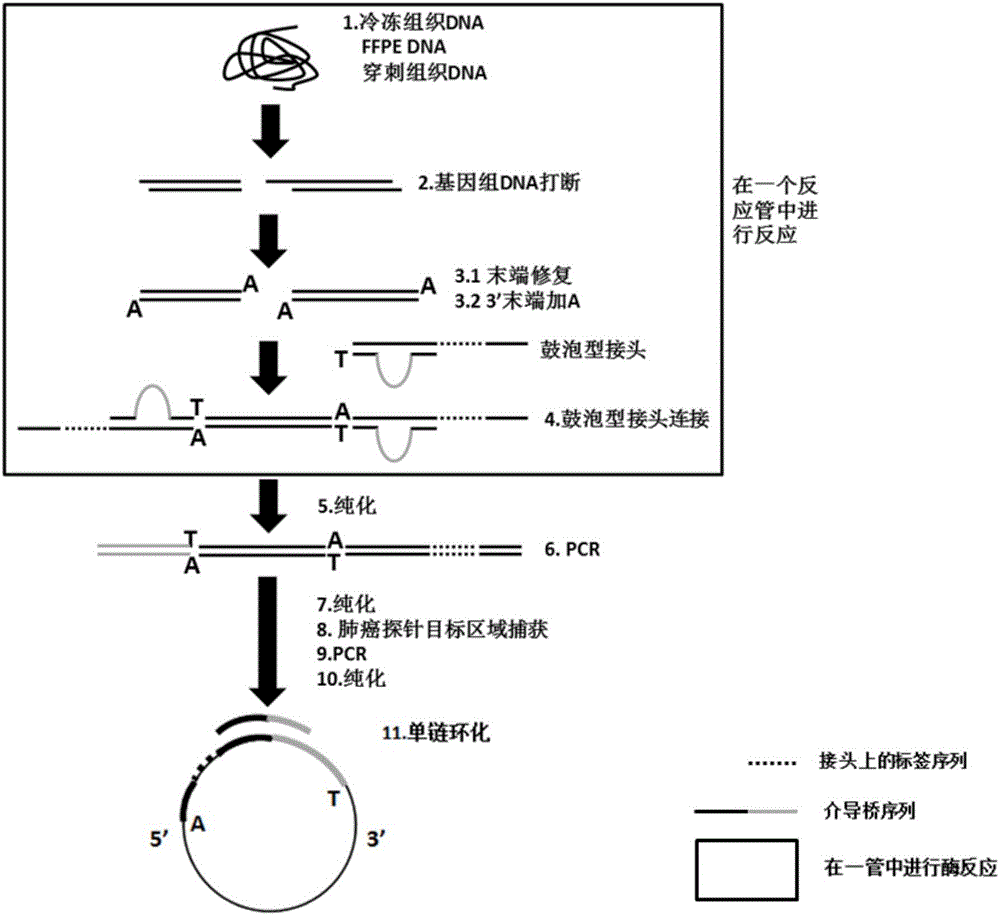
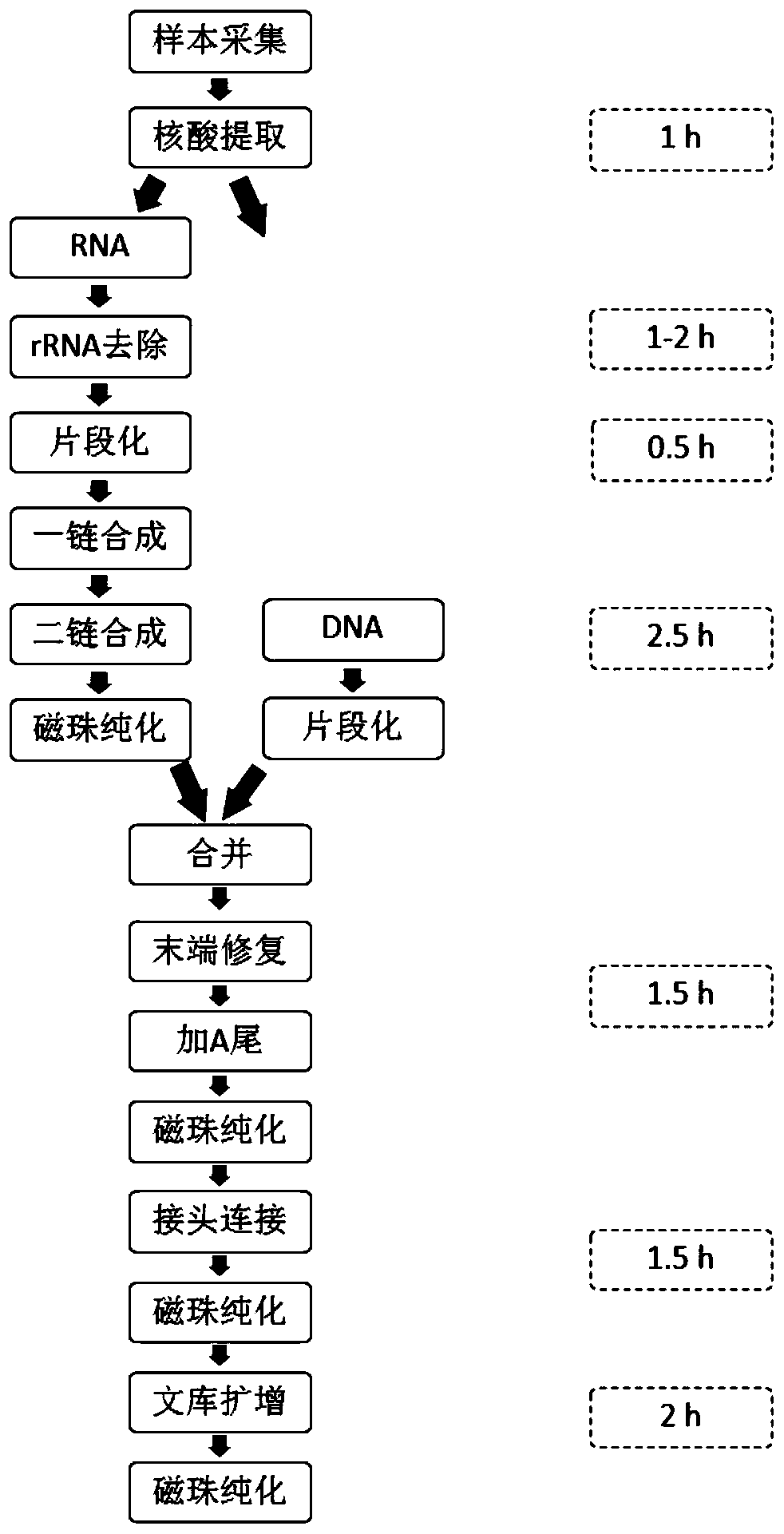
Building method of library for detecting non-small cell lung cancer gene mutation and kit
PendingCN106497920AImprove recycling efficiencyImprove utilization efficiencyMicrobiological testing/measurementLibrary creationGene targetingGene mutation
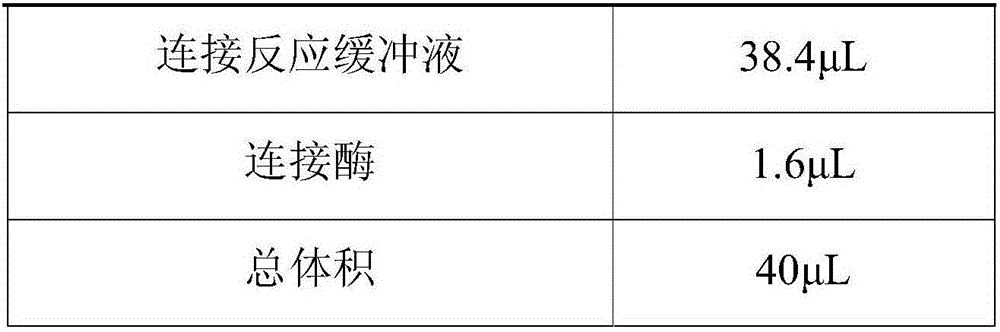
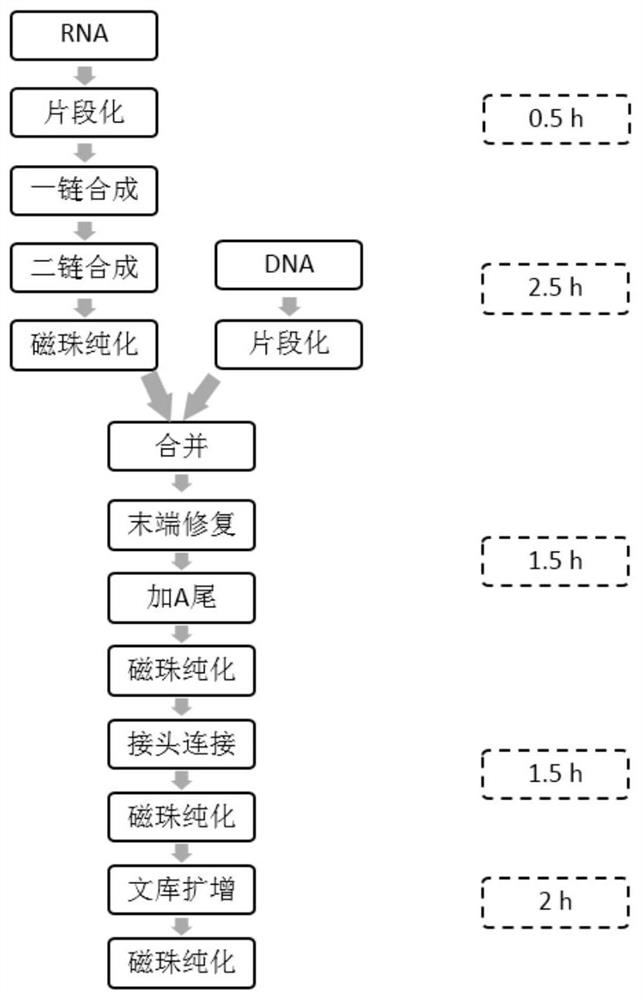
The invention discloses a building method of a library for detecting non-small cell lung cancer gene mutation and a kit. The method includes: using tubular reaction to complete genome DNA breaking and connector connection, performing hybrid capture on connection products after amplification and non-small cell lung cancer related gene target area probes, and performing BGISEQ-500 / 1000 platform sequencing and data analysis to obtain mutation conditions. The method has the advantages that the experiment flow is optimized greatly by the tubular reaction, operation complexity and time are reduced, and the requirements on clinical sample initial amount are lowered; multiple genes and multiple sites can be detected in one step, point mutation, insertion and deletion, structural variation and copy number variation are covered, the detecting result is accurate and overcomes the defect that a PCR capture method cannot detect the structural variation in one step, and the effectiveness of the high-throughput sequencing applied to the detection of the non-small cell lung cancer gene mutation; the method is wide in coverage, high in cost performance, capable of providing a reference basis for the diagnosing, treatment and drug use performed by doctors, and the method is suitable for being popularized and used in a large-scale manner.
Owner:BGI BIOTECH WUHAN CO LTD
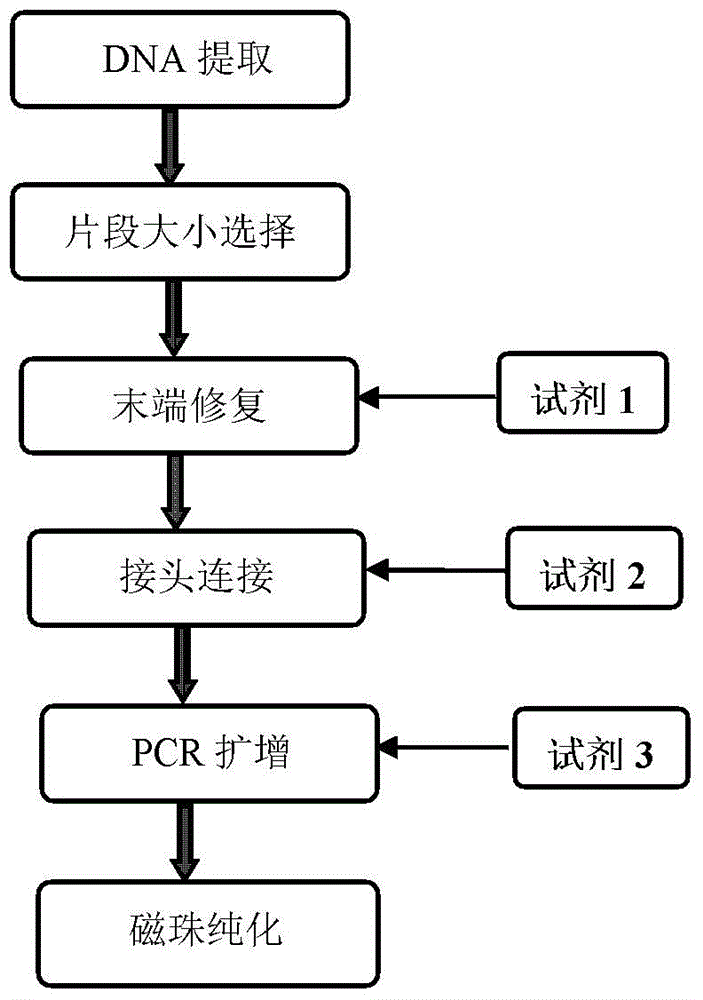
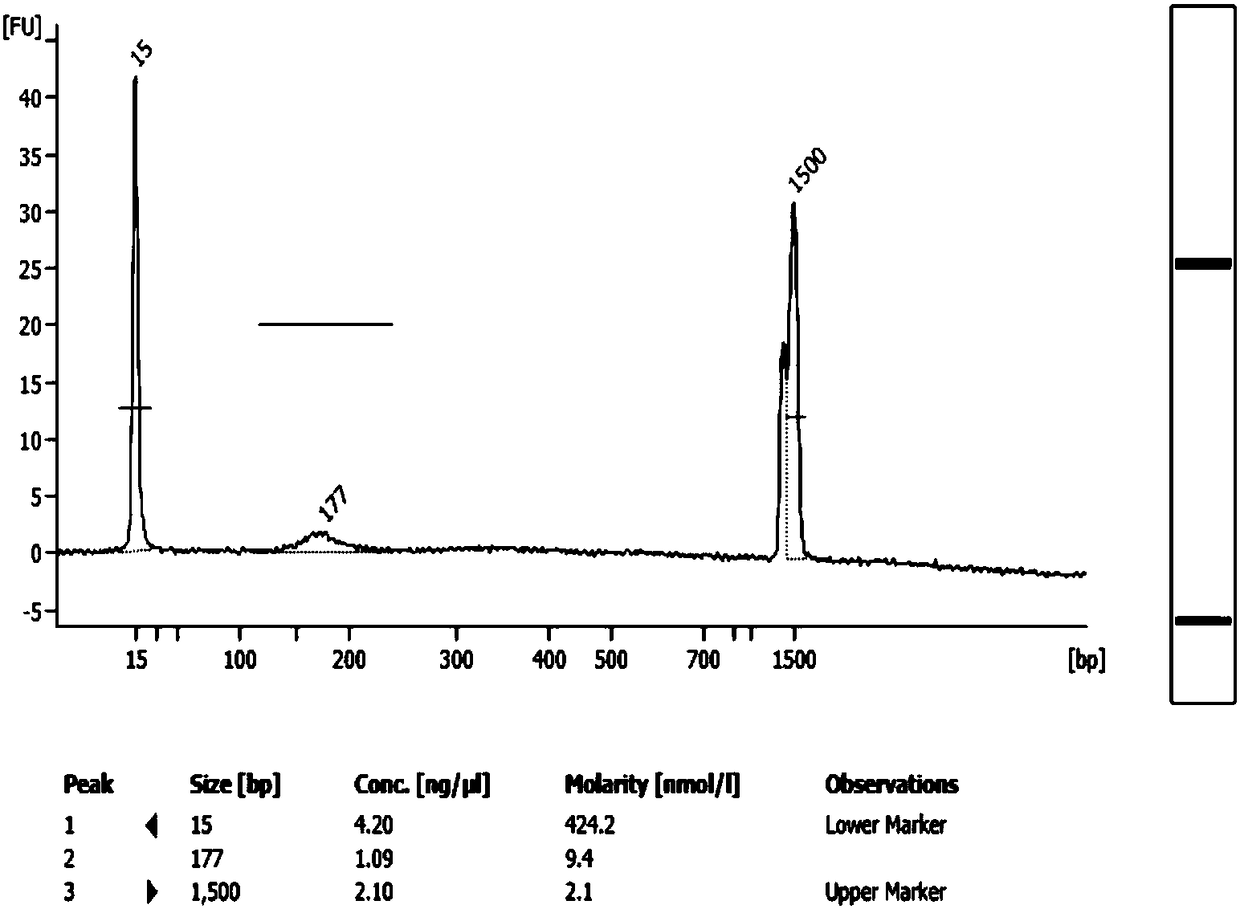
Construction method of small-fragment DNA (deoxyribonucleic acid) library based on Illumina Hiseq 2500 sequencing platform
InactiveCN104946623AReduce dosageReduce lossesMicrobiological testing/measurementLibrary creationMagnetic beadBioinformatics
The invention discloses a construction method of a small-fragment DNA (deoxyribonucleic acid) library based on an Illumina Hiseq 2500 sequencing platform, which comprises the following steps: blood free DNA extraction, fragment size screening, terminal repair, 3' terminal linker addition, PCR (polymerase chain reaction) amplification, magnetic bead purification and the like. Compared with the library construction process based on a sequencing platform in the past, the method disclosed by the invention simplifies the experiment process, shortens the library construction time, optimizes the reaction system, greatly reduces the reagent consumption, lowers the library construction cost, lowers the library loss, and is suitable for constructing an artificially-fragmented small DNA library.
Owner:浙江圣庭医学检验实验室有限公司
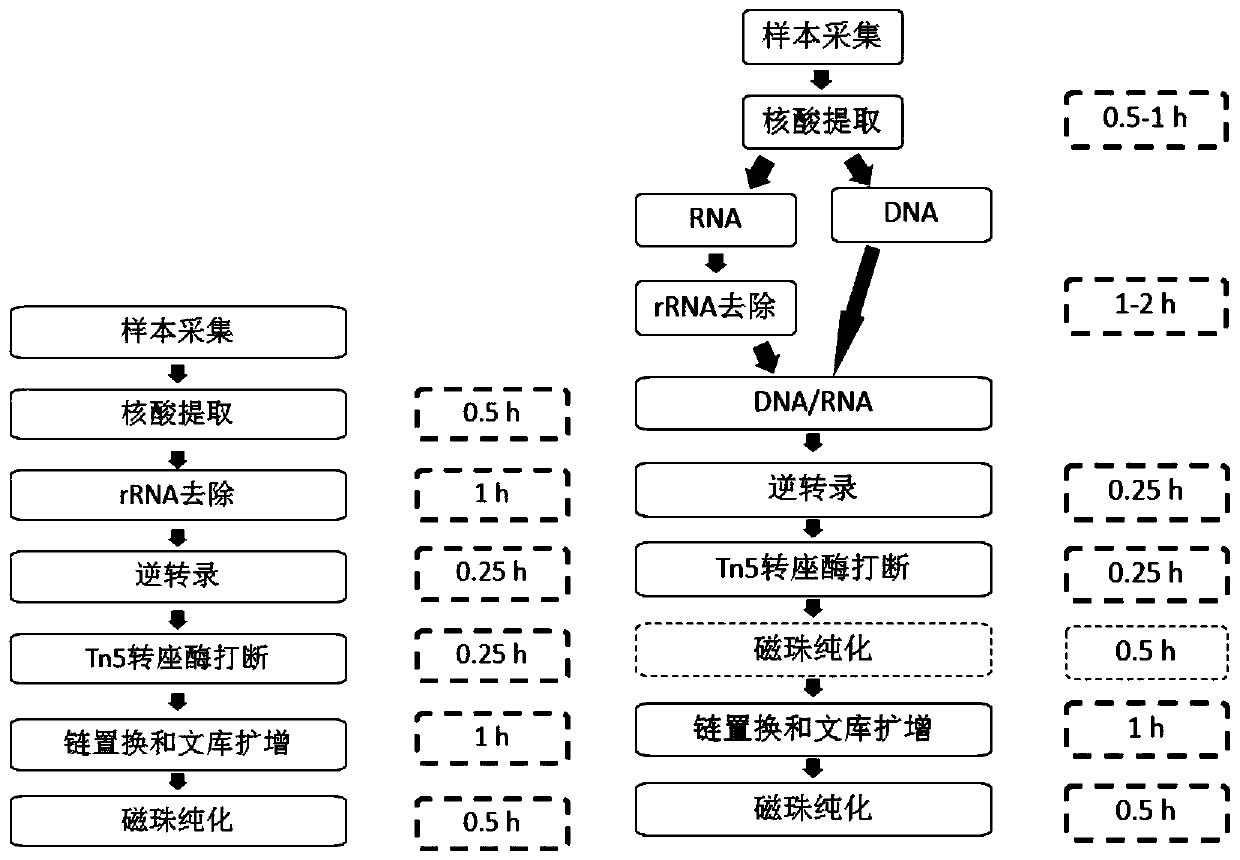
Method and kit for simultaneously constructing sequencing library by DNA and RNA
ActiveCN111139532AAvoid stickingHigh interrupt efficiencyTransferasesLibrary creationTn5 transposaseMagnetic bead
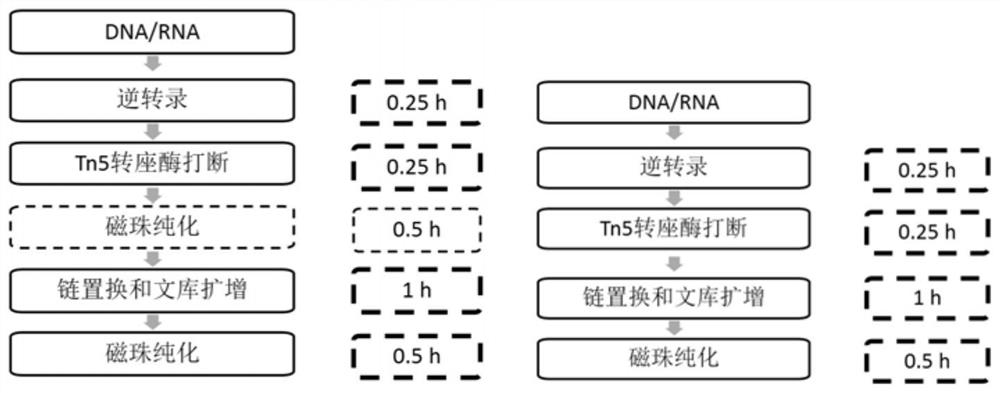
The invention discloses a method and a kit for simultaneously constructing a sequencing library by DNA and RNA. The method comprises the following steps: (1) carrying out reverse transcription on RNAin a reaction substrate to form RNA / cDNA composite double strands; (2) directly adding Tn5 transposase and a fragmentation buffer solution into a mixture of a reverse transcription product cDNA / RNA composite double strand and a DNA double strand for fragmentation, and adding a part of linker sequence to the 5' end of the double strands during fragmentation; (3) strand displacement and library amplification: adding an amplification buffer solution and a strand displacement and amplification enzyme mixture into the obtained product, and carrying out PCR amplification; and (4) carrying out magnetic bead purification to obtain a sequencing library of the original DNA / RNA. According to the method, the library building steps are greatly reduced, the library building time is shortened, the library output and offline data quality is improved, and more real information is helped to be obtained.
Owner:VAZYME BIOTECH NANJING
Sequencing library construction method and kit for pathogenic microorganism detection
ActiveCN111188094AHigh outputQuality improvementMicroorganism preservationLibrary creationReverse transcriptaseMagnetic bead
The invention discloses a sequencing library construction method and kit for pathogenic microorganism detection. The method comprises: (1) putting an extracted sample into a sample preservation solution, carrying out DNA / RNA co-extraction, and removing host rRNA; (2) adding reverse transcriptase to enable RNA in the sample to be subjected to reverse transcription to form RNA / cDNA composite doublestrands; (3) directly adding mutant Tn5 transposase and a breaking buffer solution into a mixture of the reverse transcription product cDNA / RNA composite double strand and a DNA double strand for fragmentation, and after the fragmentation is finished, carrying out a termination reaction by using a termination reaction solution 5*TS-plus; (3) adding an amplification buffer solution and a strand displacement and amplification enzyme mixture, and carrying out PCR amplification; and (4) carrying out magnetic bead purification to obtain a sequencing library of the original DNA / RNA. According to themethod, library building steps are greatly reduced, the library building time is shortened, the requirement on the initial quantity of samples is reduced, and the library output and offline data quality are improved.
Owner:VAZYME BIOTECH NANJING
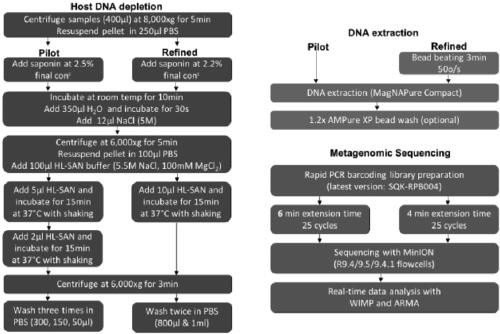
Metagenome sample library construction method and identification method based on nanopore sequencing platform and kit
ActiveCN109487345AShorten the timeTrue reflection compositionMicrobiological testing/measurementLibrary creationComputational biologyBuilding construction
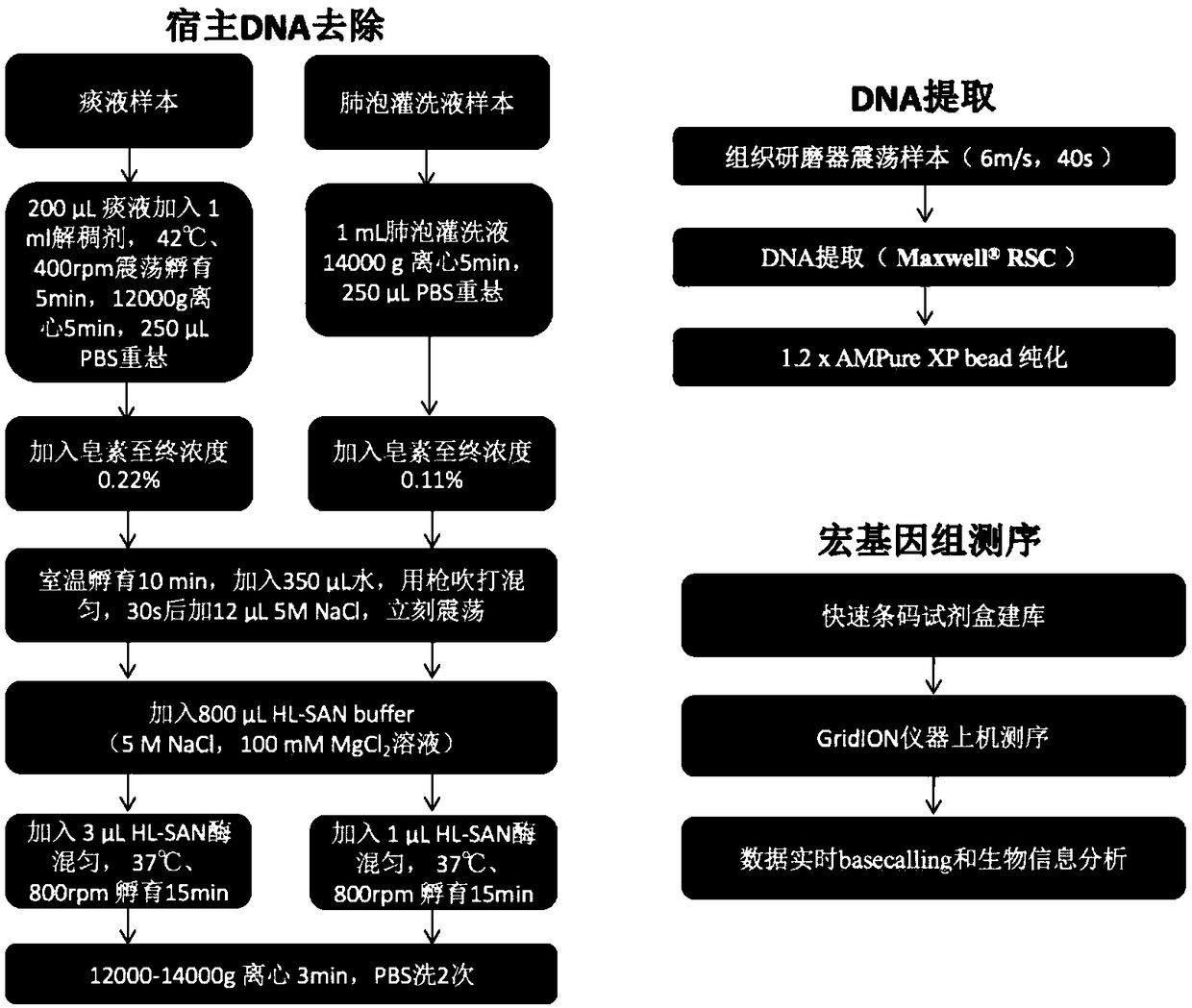
The invention relates to a metagenome sample library construction method and an identification method based on a nanopore sequencing platform and a kit. The method comprises host removal pre-treatmentand sample library construction on human and animal metagenome samples by combining incomplete host removal based on the nanopore sequencing platform so as to sequence and identify the metagenomes. Compared with a conventional process flow, the method is simpler and rapider, meets the reasonable detection demands, omits flows such as library construction PCR in the identifying process, extrudes deviation caused by PCR amplification, and can be used for identifying metagenome microorganisms more truly and accurately.
Owner:BEIJING SIMCERE MEDICAL LAB CO LTD +2
Rapid library building method for identifying target protein chromatin binding map
ActiveCN112553695ALower requirementReduce non-specificityLibrary creationProtein targetMagnetic bead
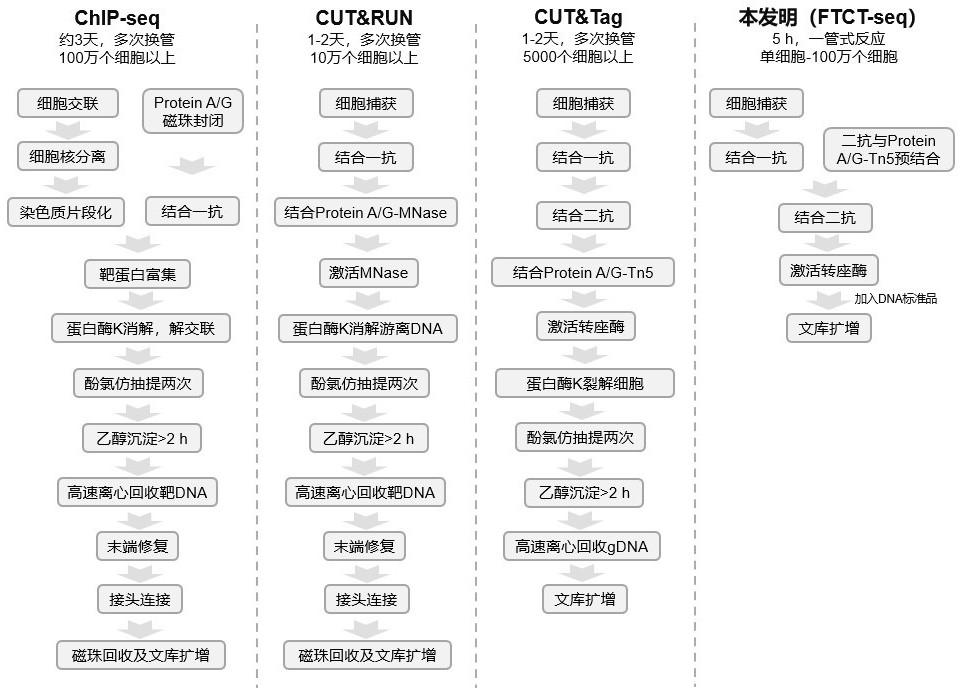
The invention discloses a rapid library building method for identifying a target protein chromatin binding map, which comprises the following steps: collecting sample cells, adding activated magneticbeads into the cells, incubating, and incubating the magnetic beads with a primary antibody; pre-binding a secondary antibody with recombinant fusion transposase; incubating the magnetic beads with the pre-combined secondary antibody and recombinant fusion transposase, adding a transposase activator and a cell perforating agent into the magnetic beads, and incubating; and terminating the transposase reaction, and carrying out library amplification. According to the method, the process of a traditional method is remarkably simplified, the library building time is shortened, the library buildingefficiency is improved, the loss is small, and the requirement for library building materials is lower. Meanwhile, three DNA standard substances with different concentrations are doped into the library building process, so that the effect of accurately quantifying the DNA copy number in the library can be achieved. The method is very suitable for researching a small amount of precious samples orsamples with different treatment states.
Owner:YEASEN BIOTECHNOLOGY (SHANGHAI) CO LTD
Construction method suitable for 16S rDNA high-throughput sequencing library
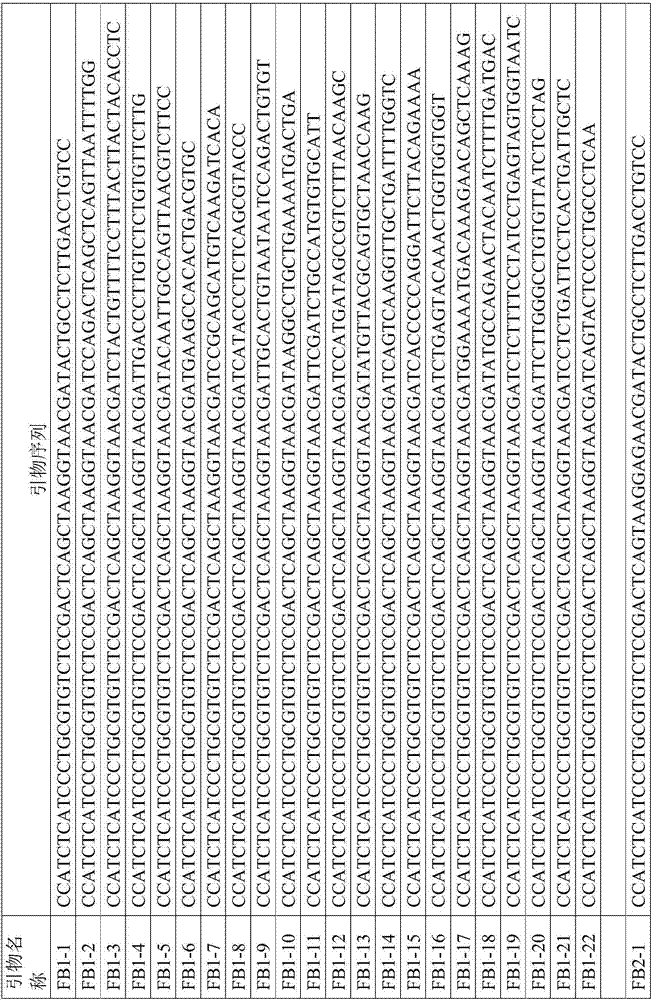
InactiveCN106282177AShorten library building timeStrong specificityMicrobiological testing/measurementLibrary creationPurification methodsBuilding construction
The invention provides a construction method suitable for a 16S rDNA high-throughput sequencing library. An upstream primer of primers is adopted as any piece in SEQ ID NO.1-8, and a downstream primer of the primers is adopted as any piece in SEQ ID NO.9-20. The provided primers include a joint sequence, an index sequence, a sequencing primer sequence and a target fragment amplification sequence at the same time, the whole library construction process can be completed only through one-step PCR, and compared with a two-step amplification method, the library construction time is obviously shortened. After the library is constructed, a beads purification method is adopted, the beads use level and the elution mode are optimized, and compared with a traditional glue recovery and purification method, time is saved, and library purification efficiency is improved.
Owner:厦门基源医疗科技有限公司
Fusion primer combination capable of rapidly constructing amplicon library
InactiveCN106906210ASimple and fast operationEasy to operateMicrobiological testing/measurementLibrary creationSequence designSingle sample
The invention discloses a fusion primer combination capable of rapidly constructing amplicon library. The fusion primer combination is capable of fusing a primer according to an upstream fusion primer designed by target amplicon and a downstream fusion primer designed by target amplicon, wherein the upstream fusion primer designed by target amplicon comprises a first linking sequence, a barcode sequence, a second linking sequence and the specific upstream primer sequence designed by target amplicon arranged according to 5' to 3' direction in order; the downstream fusion primer designed by target amplicon comprises a third linking sequence and the specific downstream primer sequence designed by target amplicon arranged according to 5' to 3' direction in order; the fusion primer combination can subjected to one-step PCR construction to obtain the amplicon library in a simple and rapid mode, barcode is introduced when PCR is initiated, possibility of intersect pollution between a sample and the library are greatly reduced; partition requirement of the experiment places can be simplified, and the cost of library establishment of a single sample can be controlled in 30 yuan / example by the fusion primer combination.
Owner:GENETRON HEALTHBEIJING LAB CO LTD
Primer combination for rapidly constructing amplicon library through one-step process
ActiveCN106834286AExperiment operation is simpleShorten library building timeMicrobiological testing/measurementLibrary creationSequence designForward primer
The invention discloses a primer combination for rapidly constructing an amplicon library through a one-step process. The primer combination comprises a forward fusion primer designed according to a target amplicon, a reverse fusion primer designed according to the target amplicon, a forward universal primer and a reverse universal primer, wherein the forward fusion primer comprises a first linker sequence and a specific forward primer sequence designed according to the target amplicon; the reverse fusion primer comprises a second linker sequence and a specific reverse primer sequence designed according to the target amplicon; the forward universal primer comprises a third linker sequence, a barcode sequence and the first linker sequence; and the reverse universal primer comprises a universal sequence and the second linker sequence. According to the fusion primer combination, the amplicon library can be simply and rapidly constructed through one step, and before the PCR is started, the barcode is introduced, so the possibility of cross contamination between the samples and the library is greatly reduced.
Owner:GENETRON HEALTH (BEIJING) CO LTD
Method for quickly building amplicon library
InactiveCN107012139ASimple and fast operationEasy to operateMicrobiological testing/measurementLibrary creationSingle sampleBarcode
The invention discloses a method for quickly building an amplicon library. The method comprises the following steps of (1) synthesizing a fusion primer combination for building the amplicon library of a DNAA sample; (2) building a PCR reaction system of the DNA sample, mixing an upstream fusion primer designed according to a target amplicon and a downstream fusion primer designed according to the target amplicon as a primer combination in the PCR reaction system; and (3) carrying out PCR. According to the method, the amplicon library can be simply and quickly built through PCR in the step (1); a barcode is introduced at the beginning of the PCR, so that the possibility of cross contamination between samples and the library is greatly reduced and the requirements of experiment site partition can be simplified. According to the method, the cost of building the library for a single sample can be controlled within 30 yuan / case.
Owner:GENETRON HEALTHBEIJING LAB CO LTD
Method for constructing DNA library and application of method
PendingCN110607352AAppropriate fragment lengthShorten library building timeMicrobiological testing/measurementLibrary creationCDNA libraryA-DNA
The invention discloses a method for constructing a DNA library and application of the method. The method for constructing the DNA library comprises the steps: providing DNA of a chimeric marker, wherein the DNA of the chimeric marker has three-dimensional structure information; carrying out transposition treatment on the DNA of the chimeric marker to obtain a transposition product; carrying out capture treatment on the transposition product to obtain captured DNA; and carrying out amplification treatment on the captured DNA so as to obtain the DNA library. The library construction method is simple in steps, short in consumed time, particularly suitable for library construction of trace DNA samples, high in effective data proportion of sequencing and low in noise single-end suspension value.
Owner:安诺优达生命科学研究院 +4
Construction method of amplicon library for detecting low-frequency mutation of target gene
PendingCN107604045AEasy to operateHigh standardNucleotide librariesMicrobiological testing/measurementA-DNAAmplicon sequencing
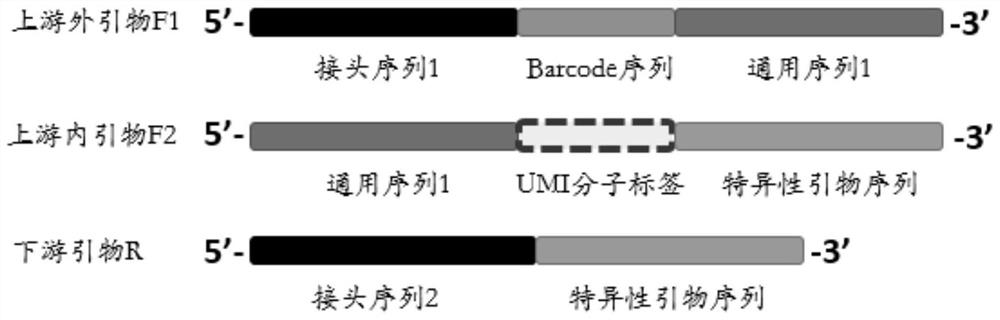
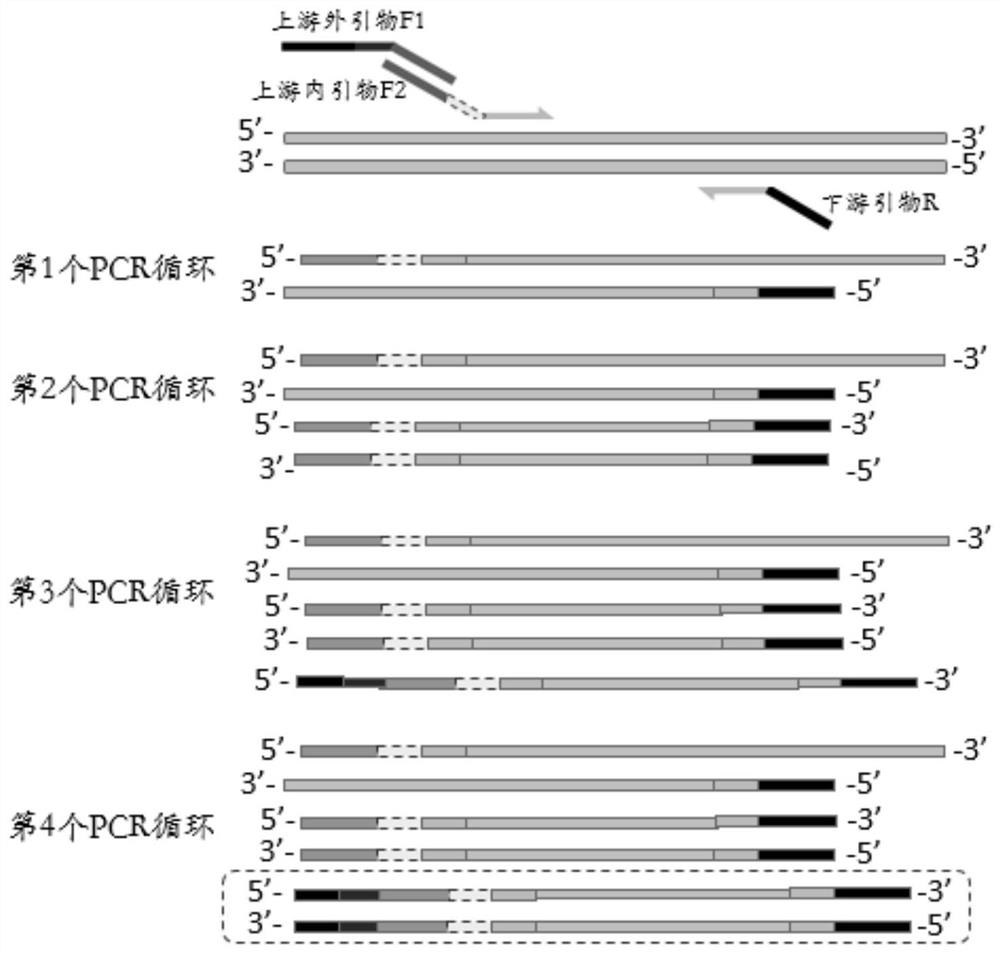
The invention discloses a construction method of an amplicon library for detecting low-frequency mutation of a target gene. The construction method of the amplicon library for detecting the low-frequency mutation of the target gene, provided by the invention, comprises the following steps: 1) designing and synthesizing a Barcode primer F1, an upstream primer F2, a downstream outer primer R1 and adownstream inner primer R2; 2) carrying out further PCR (Polymerase Chain Reaction) amplification on a cfDNA (cell-free Deoxyribonucleic Acid) of a sample to be detected by utilizing the Barcode primer F1, the upstream primer F2, the downstream outer primer R1 and the downstream inner primer R2, so as to obtain an amplified product, namely a DNA library for amplicon sequencing. By adopting the method disclosed by the invention, a tissue sample can be detected and different regions of free DNAs in samples including blood, urine, cerebrospinal fluid and the like can be rapidly, conveniently, sensitively and specifically subjected to target amplification and mutation which is as low as a 0.1 percent level is efficiently detected; the experiment operation is greatly simplified, the library loss and pollution are effectively avoided, the cost is remarkably reduced and the efficiency is improved.
Owner:GENETRON HEALTH (BEIJING) CO LTD +1
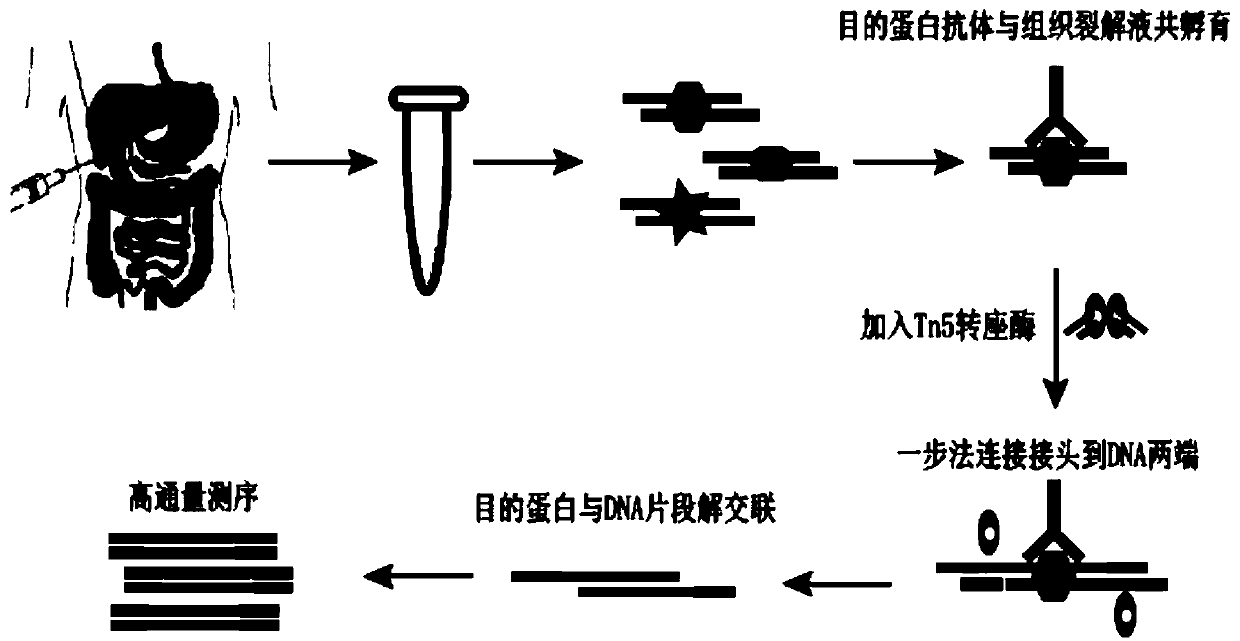
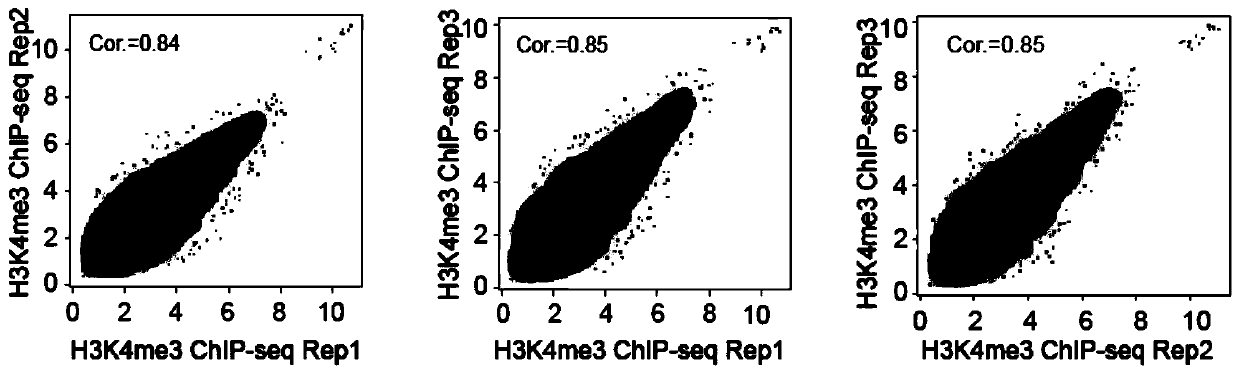
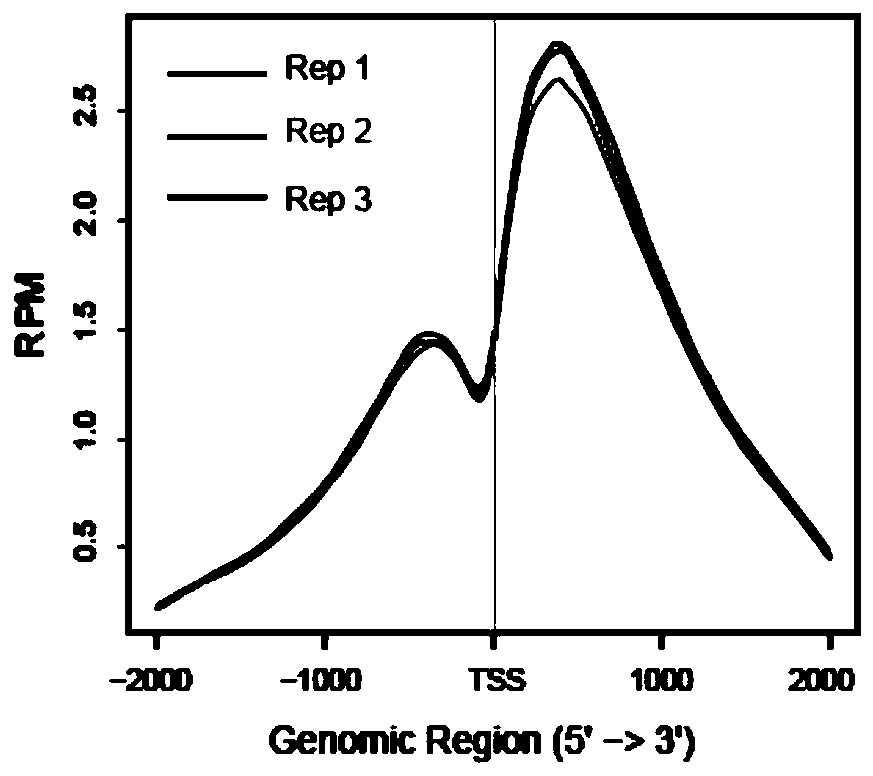
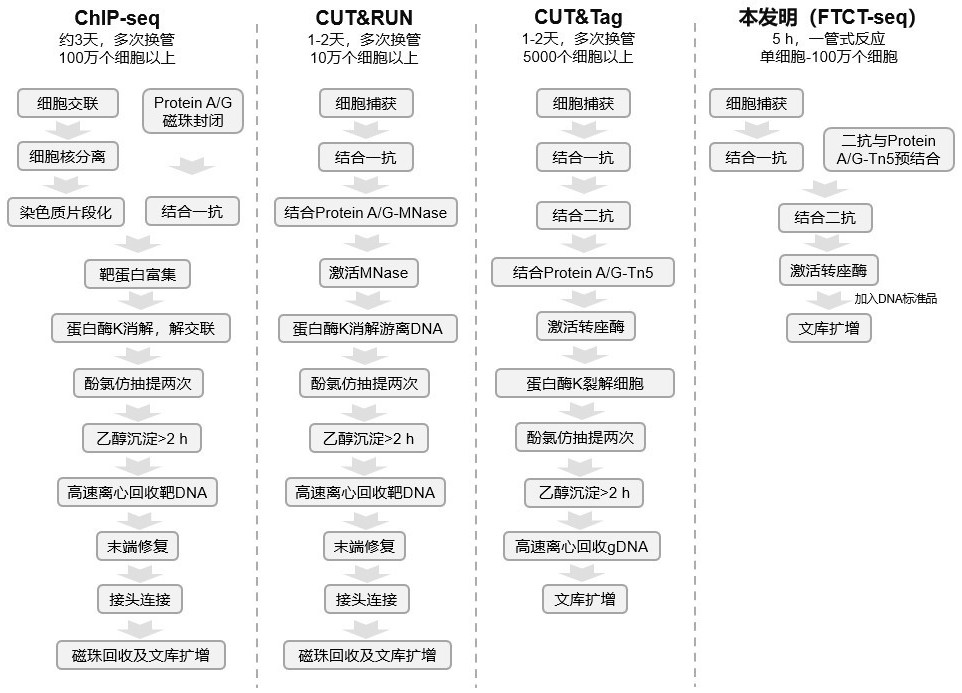
Method for preparing chromatin co-immunoprecipitation library by using trace of clinical puncture sample and application of method
InactiveCN111440843AShort cycleLow costMicrobiological testing/measurementLibrary creationTn5 transposaseTissue sample
The invention discloses a method for preparing a chromatin co-immunoprecipitation library by using a trace clinical puncture sample and an application of the method. The invention successfully develops the method for preparing the chromatin co-immunoprecipitation sequencing library based on trace of clinical puncture sample by utilizing high-sensitivity Tn5 transposase, and ChIP-seq library construction can be effectively and rapidly carried out on the trace of puncture tissue sample. Compared with a traditional ChIP-seq library building mode, the method has the advantages that the initial sample amount is greatly reduced (5-50 mg), the library building time is effectively shortened (about 1.5 days), and the requirements of the sequencing market on trace samples and large-scale sample processing speed are met.
Owner:INSITUTE OF BIOPHYSICS CHINESE ACADEMY OF SCIENCES
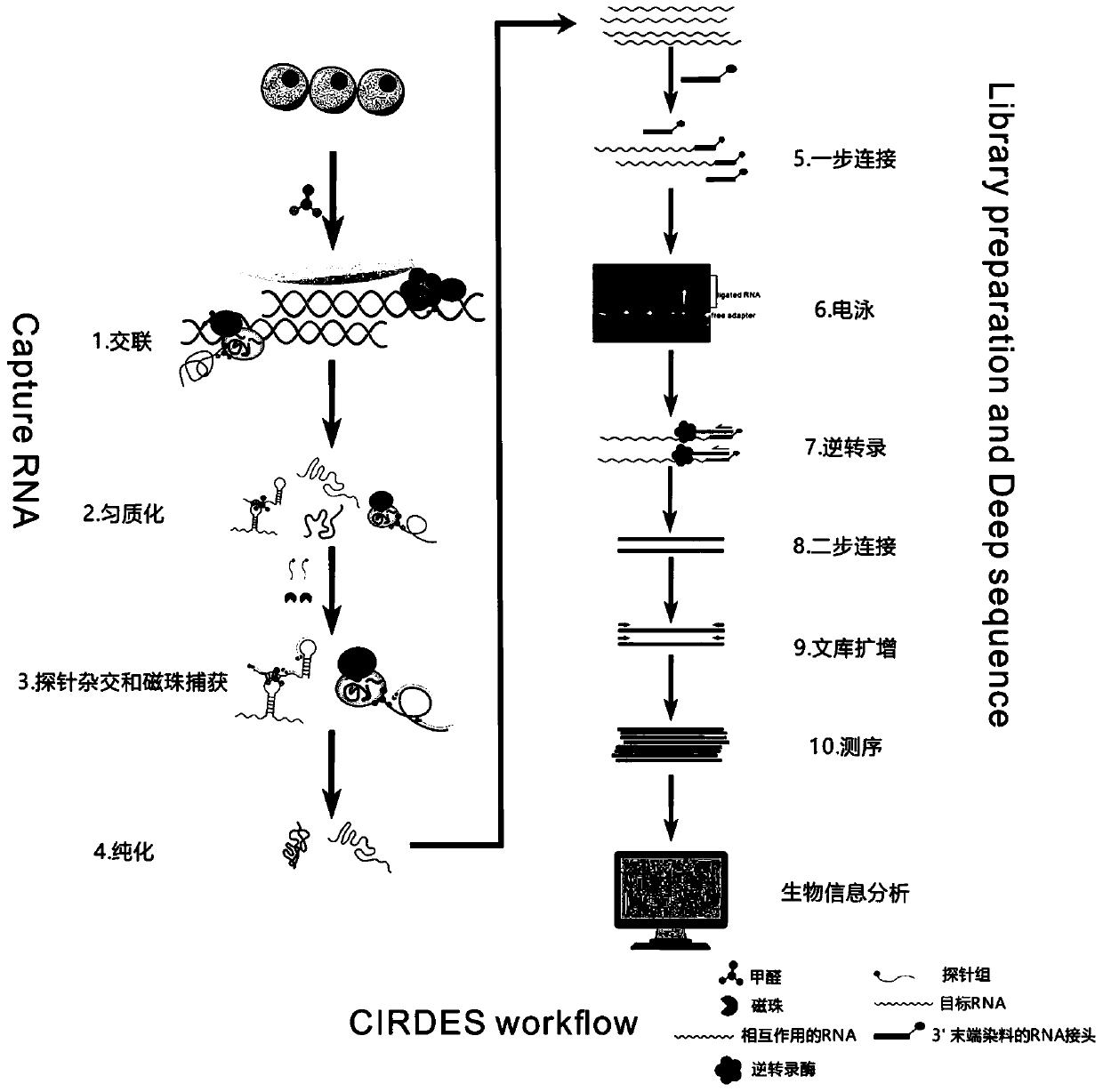
High-throughput sequencing method for efficient research of RNA interaction interactome and application of high-throughput sequencing method
ActiveCN110205365AImprove connection efficiencyReduce biasMicrobiological testing/measurementAgainst vector-borne diseasesLysisA-DNA
The invention discloses a high-throughput sequencing method for efficient research of RNA interaction interactome and application of the high-throughput sequencing method. The method includes steps: (1) arranging DNA probes at intervals of 100bp according to length of to-be-tested RNA; (2) after crosslinking fixation with 1-3% (v / v) of formaldehyde solution, subjecting cells to ultrasonication toobtain cell lysis solution in nucleic acid length range of 100-500nt; (3) adding the DNA probes for hybridization, gathering target RNA, washing, and purifying to obtain RNA fragments; (4) connecting3' terminals of the RNA fragments with RNA joints, performing product reverse transcription to obtain cDNA, connecting the 3' terminal of the cDNA with a DNA joint to obtain a library of the target RNA and RNAs in interaction with the target RAN; (5) amplifying the library, and carrying out detection analysis. The method can be used for library construction of low-content interaction RNAs.
Owner:SUN YAT SEN MEMORIAL HOSPITAL SUN YAT SEN UNIV
Library construction primer group and application thereof in high-throughput detection
InactiveCN113621609ABuild accuratelyAccurate error correctionMicrobiological testing/measurementLibrary creationAlgorithmEngineering
The invention relates to a rapid and accurate library construction primer group and application thereof in high-throughput detection, and belongs to the technical field of gene detection. The library construction primer group comprises a plurality of amplification primers for target area amplification and a sequencing linker for library construction, each amplification primer sequentially comprises a target sequence segment matched with a target area sequence, a single-molecule tag segment playing a unique recognition role and a library construction segment matched with the sequencing linker, and the multiple single-molecule tag segments comprise random basic groups with different lengths. The rapid and accurate library construction primer group can be used for quickly and accurately constructing a base balanced library, the library construction time is shortened, pollution and sequencing cost are reduced, data are more accurately corrected, and the accuracy of the technology is improved. The rapid and accurate library construction primer group can be more conveniently and efficiently used in scientific research and clinical research. In a scientific research project, the immune group can be sequenced without a sequencer with high productivity, and the base balance library is not needed, so that the rapid and accurate library construction primer group is convenient and efficient, and the application and development of the immune group are promoted and expanded.
Owner:深圳泛因医学有限公司
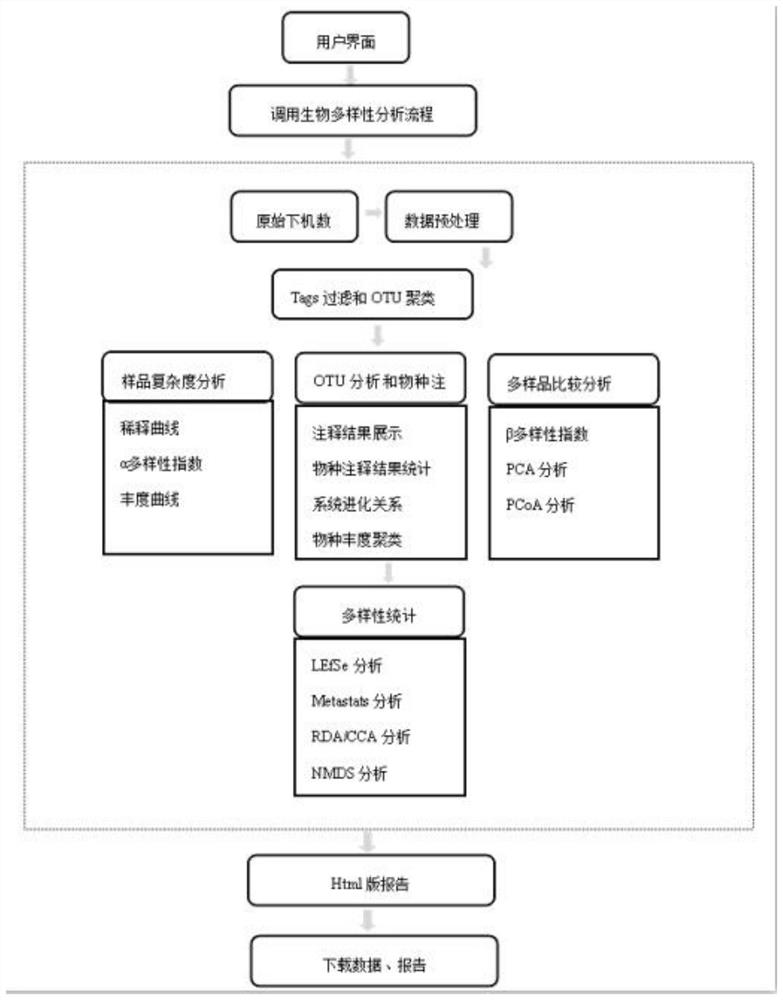
Automatic analysis method of biodiversity
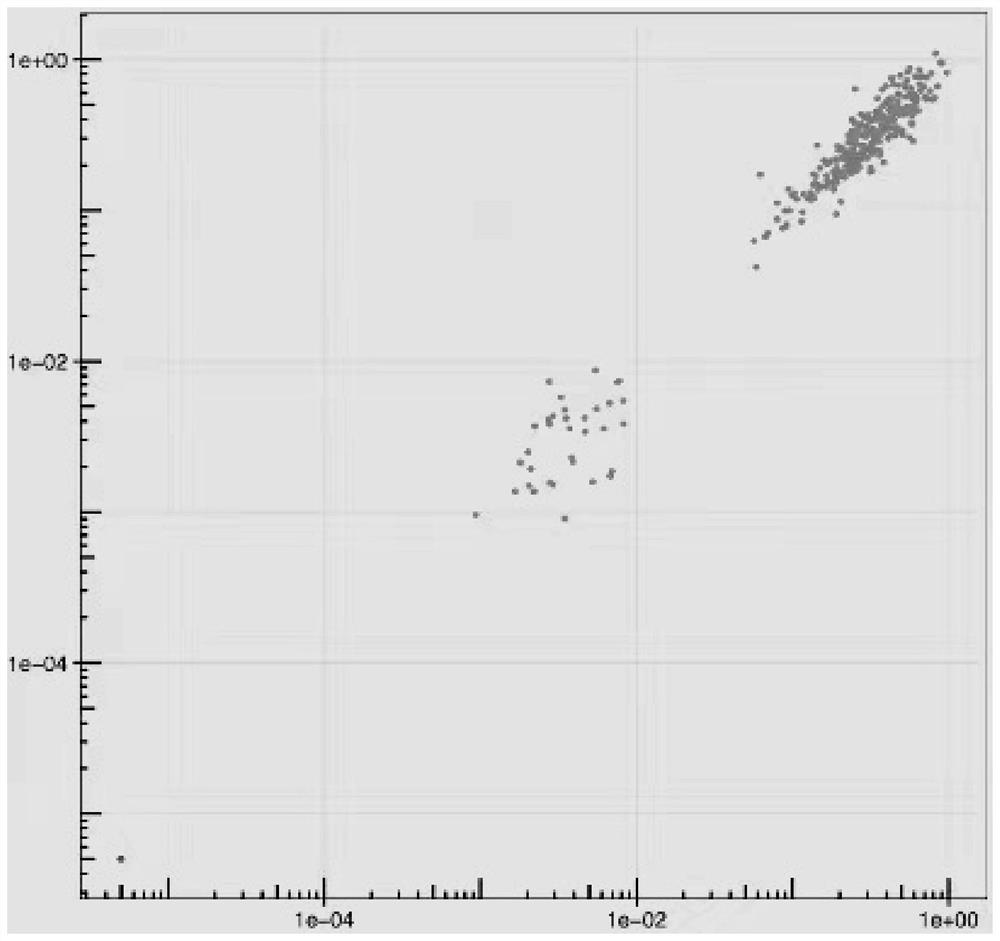
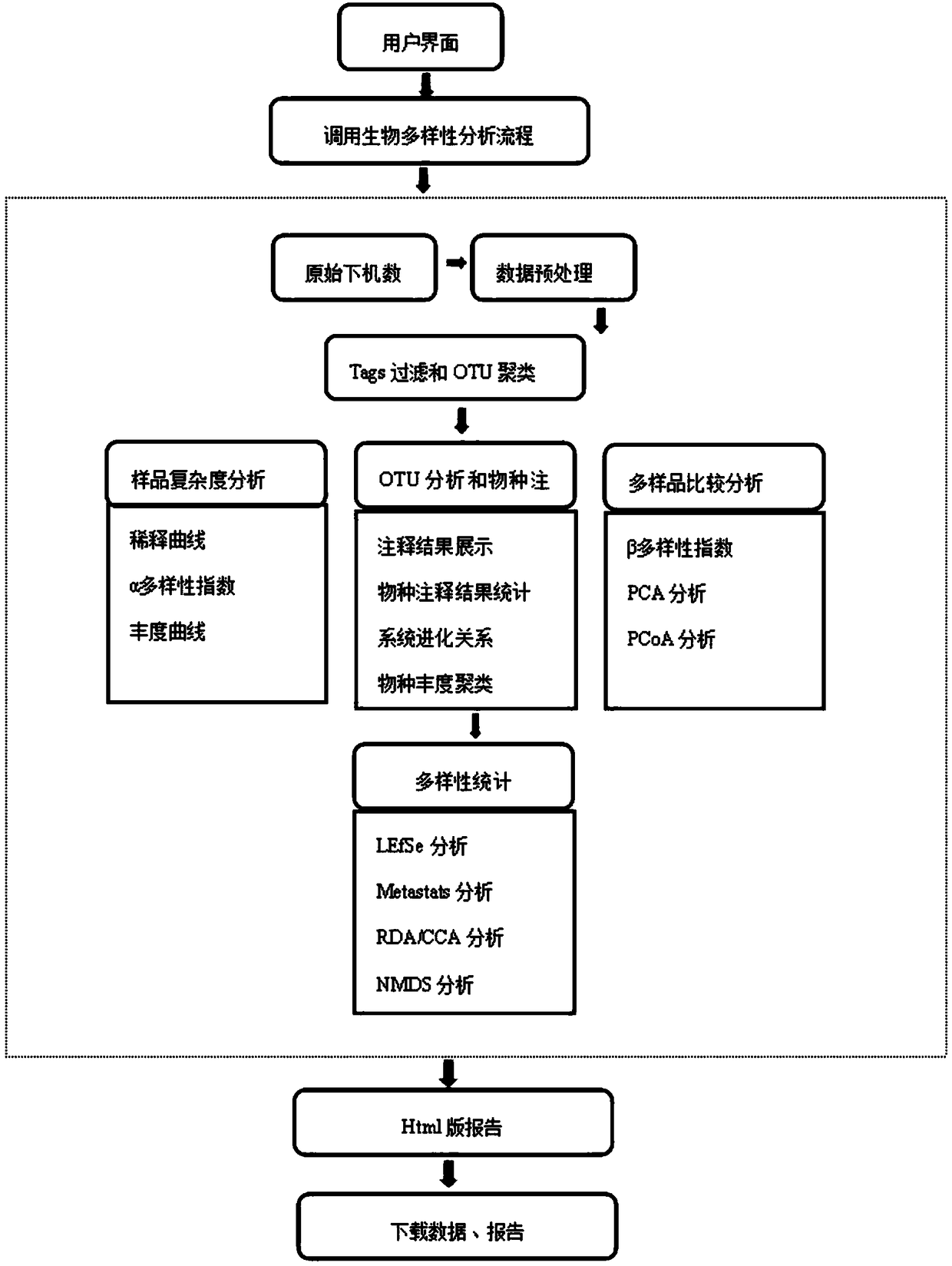
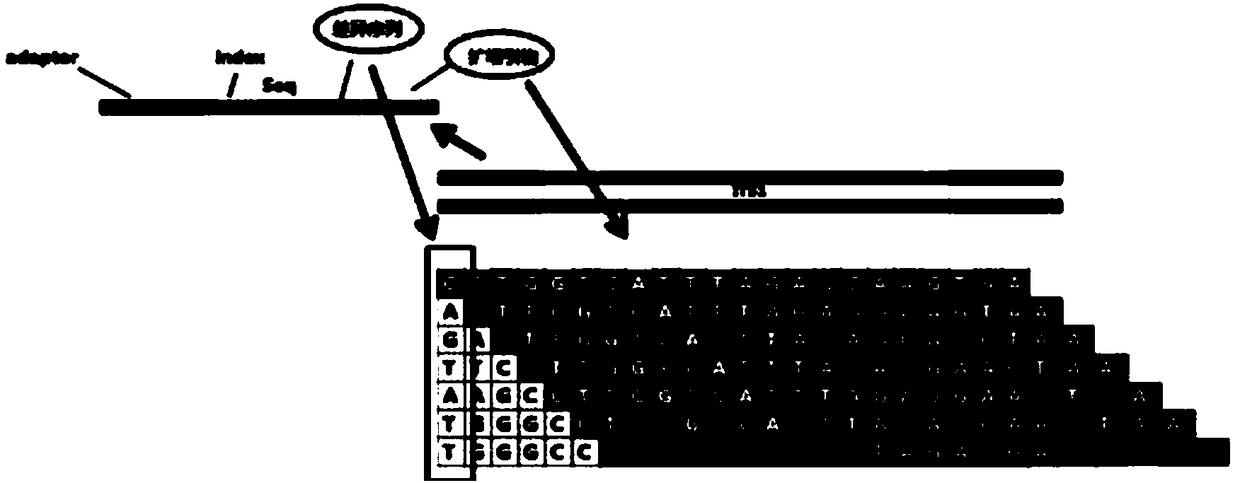
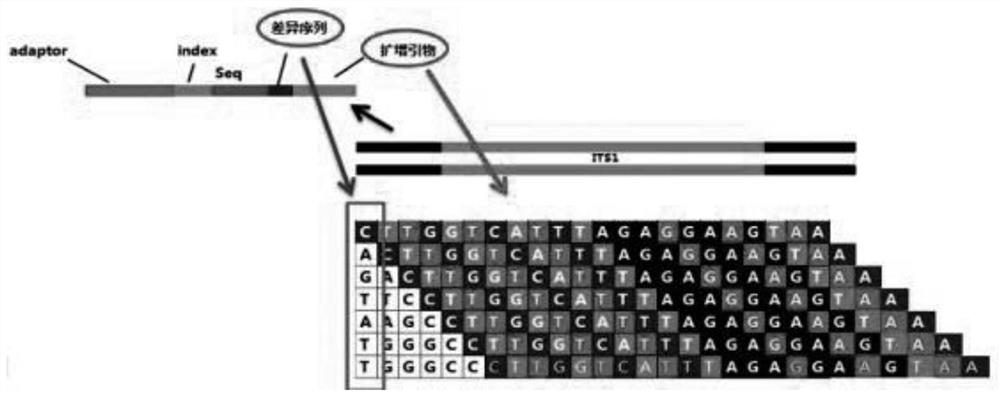
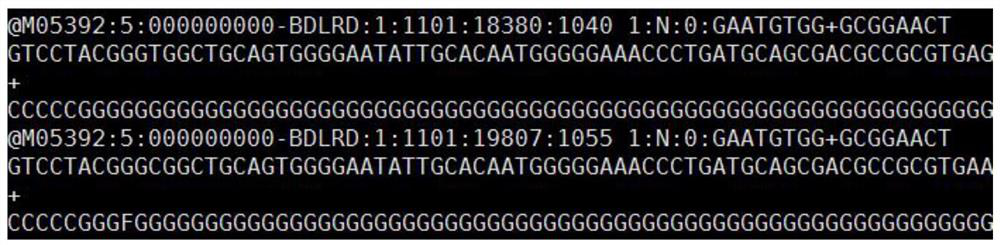
ActiveCN108388771AImproved Design PrinciplesShorten library building timeBiostatisticsSequence analysisProject cycleMicrobial diversity
The invention discloses an automatic analysis method of biodiversity. The method comprises the following specific analysis steps: step 1, using 24 pairs of primers containing sequences matched to an illumina sequencing chip, 8-base index sequences, sequencing primer sequences, difference sequences and universal amplification primer sequences, obtaining a library of on-machine sequencing through PCR reaction, and selecting a corresponding sequencing mode; step 2, converting stopping data into a fastq format by bcl2fastq software, and adding a parameter ''-create-fastq-for-index-reads''; step 3,using a Phred algorithm to intercept low-quality sequences through a filtering software BBDuk software package; and step 4, using a script in a QIIME package for secondary filtering on Tags sequencesobtained by splicing. According to the method, a project cycle is compressed, a database construction method, a data filtering method and software are improved, analysis deviations brought by chimeras and sequencing errors are effectively decreased, processes from data production to filtering and cleantags forming can be uniformity carried out without the need for distinguishing of each project even for microbial diversity projects, and processing time is saved.
Owner:安徽微分基因科技有限公司
Library construction method and application
ActiveCN112301430ALower requirementThe method of building a database is simple and fastMicrobiological testing/measurementLibrary creationDNABioinformatics
The invention discloses a library construction method and application. The invention provides an amplicon library construction method for detecting the variation condition of a target gene to-be-detected region of a to-be-detected sample. The method comprises the following steps: (1) designing and synthesizing an upstream outer primer F1, an upstream inner primer F2 and a downstream primer R according to a target region; and (2) performing one-step PCR amplification on a to-be-detected sample by using the upstream outer primer F1, the upstream inner primer F2 and the downstream inner primer Rto obtain an amplification product, namely an amplicon library of the target region. The one-step library construction technology can be applied to all second-generation platforms including IonTorrent, illumina and BGI / MGI, and on the basis of the library construction method, SNP, Ins / Del, CNV and methylation detection for DNA and detection products for gene fusion and expression of RNA samples are developed.
Owner:GENETRON HEALTH (BEIJING) CO LTD
Kit for DNA library building and application of kit
ActiveCN108660135AReduce usageReduce lossesMicrobiological testing/measurementLibrary creationBlood plasmaKlenow fragment
The invention discloses a kit for DNA library building and an application of the kit, and in particular relates to a kit for peripheral blood cfDNA library building and an application of the kit. According to the invention, the kit for DNA library building is firstly provided, wherein the kit comprises a component A and a component B; the component A consists of the following ingredients at a ratio as follows: 1-3U of T4 PNK to 1-4.5U of Klenow Fragment to 0.25-0.5U of LA Taq DNA Polymerase to 0.3-0.9U of T4 DNA Polymerase to 0.5-2.5 nmol of dNTP to 0.5-2.5nmol of dATP; and the component B consists of the following ingredients at a ratio as follows: 2.5-8nmol of ATP to 26.5 132U of T4 DNA Ligase to 0.213 0.534[mu]L of PEG 8000. According to the kit and the application method provided by the invention, DNA loss in a library building process can be reduced to the greatest extent, the usage amount of plasma can be reduced, library building cost can be reduced, library building time can beshortened, operations can be simplified and more stable and reliable results can be obtained. The kit provided by the invention is significant for improving library building efficiency and reducing labor cost.
Owner:TIANJIN MEDICAL LAB BGI +2
Sequencing library building instrument capable of continuously feeding samples and operation method thereof
ActiveCN112175943AReduce multi-sample library preparation timeImprove the adsorption rate and release rateMicrobiological testing/measurementLibrary creationReaction tubeEngineering
The invention provides a sequencing library building instrument capable of continuously feeding samples and an operation method thereof. The sequencing library building instrument at least comprises aworking space, wherein a continuous sample feeding module, a TIP head module, a reagent storage module and at least two PCR modules are arranged on the bottom side of the working space; a liquid transferring module and a magnetic rod sleeve module are movably arranged in the working space; the liquid transferring module moves in a space where the continuous sample feeding module, the TIP head module, the reagent storage module and the at least two PCR modules are located in a reciprocating manner; the magnetic rod sleeve module at least moves in a space where the PCR modules are located in areciprocating manner; the continuous sample feeding module at least comprises a plurality of rows of sample feeding reaction tubes which are arrayed at intervals; the magnetic rod sleeve module comprises a magnetic rod assembly and a magnetic rod sleeve assembly; the magnetic rod assembly and the magnetic rod sleeve assembly are matched to move to realize adsorption and release of magnetic beads;and continuous sample feeding of a plurality of samples can be realized and the utilization of consumable items is reduced.
Owner:SHAOXING INGENIGEN BIOTECH CO LTD
Plasma DNA library and construction method thereof
InactiveCN110468180AImprove liquidityEasy to operateMicrobiological testing/measurementLibrary creationDNA extractionOperability
The invention provides a plasma DNA library and a construction method thereof. The construction method comprises the following steps: adding an emulsifier into plasma to obtain emulsified plasma; performing end repair / dA on the emulsified plasma to obtain repaired plasma; performing adapter ligation on the repaired plasma to obtain a ligation product; and performing PCR amplification on the ligation product to obtain the plasma DNA library. According to the construction method, plasma is directly used as an object without the need for extracting free DNA in the plasma, and the steps of performing end repair / dA, adapter ligation and PCR amplification after the plasma is subjected to emulsifying pretreatment to improve the fluidity and operability of the plasma, and construction of a plasmaDNA library without DNA extraction can be truly implemented. The method omits the step of plasma DNA extraction, the whole experimental procedure of library construction can be completed in 3 hours, and the library construction efficiency is greatly improved. Therefore, the construction method can meet the requirement for quick batch library construction in the modern time.
Owner:BEIJING USCI MEDICAL LAB CO LTD
Method and kit for constructing whole genome high-throughput sequencing library
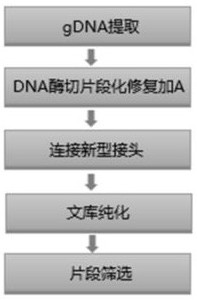
ActiveCN112301432ASimple and fast operationShort timeMicrobiological testing/measurementLibrary creationEnzyme digestionGenome
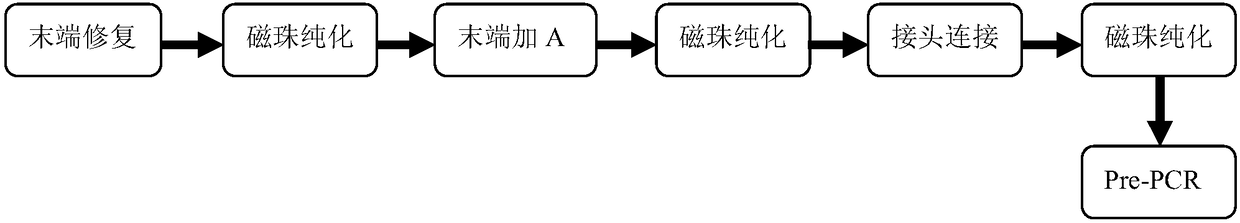
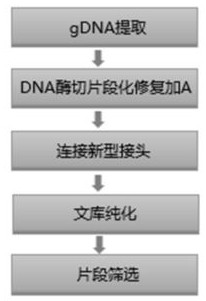
The invention discloses a method for constructing a whole genome high-throughput sequencing library. The method comprises the following steps: 1) extracting a sample gDNA; 2) carrying out enzyme digestion fragmentation on the sample gDNA, filling in the tail end, and adding A to obtain gDNA added with A; 3) connecting the gDNA added with A with a linker combination to obtain a connection product,the linker combination comprising two parts: a Y-shaped reverse linker and a high GC splint linker; 4) purifying the connection product to obtain a purified product, and 5) performing fragment screening on the purified product to obtain the sequencing library. The invention also discloses a kit for constructing the whole genome high-throughput sequencing library.
Owner:BERRYGENOMICS CO LTD
A Rapid Library Construction Method for Identifying Chromatin Binding Profiles of Target Proteins
ActiveCN112553695BLower requirementReduce non-specificityLibrary creationProtein targetEnzyme activator
The invention discloses a method for quickly building a library for identifying the chromatin binding map of a target protein. The steps include: collecting sample cells, adding activated magnetic beads into the cells for incubation, and incubating the magnetic beads with a primary antibody; secondary antibody and recombinant Pre-binding of fusion transposase; incubation of magnetic beads with pre-bound secondary antibody and recombinant fusion transposase. Add transposase activator and cell perforation agent to the magnetic beads and incubate; terminate the transposase reaction and perform library amplification. The invention significantly simplifies the flow of the traditional method, shortens the time for building a library, improves the efficiency of building a library, has small losses, and has lower requirements on materials for building a library. At the same time, by incorporating three DNA standards of different concentrations into the library construction process, the effect of accurately quantifying the DNA copy number in the library can be achieved. This method is very suitable for the study of small, precious samples or samples in different processing states.
Owner:YEASEN BIOTECHNOLOGY (SHANGHAI) CO LTD
One-step rapid construction of primer sets for amplicon libraries
ActiveCN106834286BExperiment operation is simpleShorten library building timeMicrobiological testing/measurementLibrary creationForward primerSequence design
The invention discloses a primer combination for rapidly constructing an amplicon library through a one-step process. The primer combination comprises a forward fusion primer designed according to a target amplicon, a reverse fusion primer designed according to the target amplicon, a forward universal primer and a reverse universal primer, wherein the forward fusion primer comprises a first linker sequence and a specific forward primer sequence designed according to the target amplicon; the reverse fusion primer comprises a second linker sequence and a specific reverse primer sequence designed according to the target amplicon; the forward universal primer comprises a third linker sequence, a barcode sequence and the first linker sequence; and the reverse universal primer comprises a universal sequence and the second linker sequence. According to the fusion primer combination, the amplicon library can be simply and rapidly constructed through one step, and before the PCR is started, the barcode is introduced, so the possibility of cross contamination between the samples and the library is greatly reduced.
Owner:GENETRON HEALTH (BEIJING) CO LTD
Reagent for constructing free DNA library and application thereof
ActiveCN114032619AShorten library building timeReduce the cost of building a libraryMicrobiological testing/measurementLibrary creationMethylation SiteWhole Genome Amplification
The invention discloses a reagent for constructing a free DNA library and application of the reagent. The reagent comprises carrier DNA, heat-sensitive protease and a linker connecting reagent, wherein the linker connecting reagent comprises ligase, a connecting buffer solution, propylene glycol and polyethylene glycol. By utilizing the reagent, the problem that high-throughput sequencing library construction can be carried out only after single-cell whole genome amplification in the prior art can be solved, and the problems of polluted DNA signal amplification and covering of information such as cfDNA methylation sites, cfDNA length and cfDNA sequence characteristics caused by single-cell whole genome amplification are avoided; the library building time is obviously shortened; and the library building cost is reduced.
Owner:SUZHOU BASECARE MEDICAL DEVICE CO LTD
A Biodiversity Automatic Analysis Method
ActiveCN108388771BImproved Design PrinciplesShorten library building timeBiostatisticsSequence analysisMicroorganismEngineering
The invention discloses an automatic analysis method for biological diversity. The specific analysis steps are as follows: Step 1: 24 pairs of primers, including the sequence matched with the illumina sequencing chip, 8-base index sequence, sequencing primer sequence, difference sequence, universal amplification Increase the primer sequence, obtain the library for sequencing on the machine through PCR reaction, and select the corresponding sequencing mode; Step 2: convert the off-machine data into fastq format by bcl2fastq software, and add the parameter "‑create‑fastq‑for‑index‑reads"; Step 3: Use the Phred algorithm to intercept low-quality sequences through the filtering software BBDuk package; Step 4: Use the script in the QIIME package to perform secondary filtering on the spliced Tags sequences. The invention compresses the project cycle, improves the database building method, data filtering method and software, effectively reduces the analysis deviation caused by chimeras and sequencing errors, and, even due to the microbial diversity project, can also be filtered into cleantags from data production The process is done in a unified manner, without distinguishing each item, which saves processing time.
Owner:安徽微分基因科技有限公司
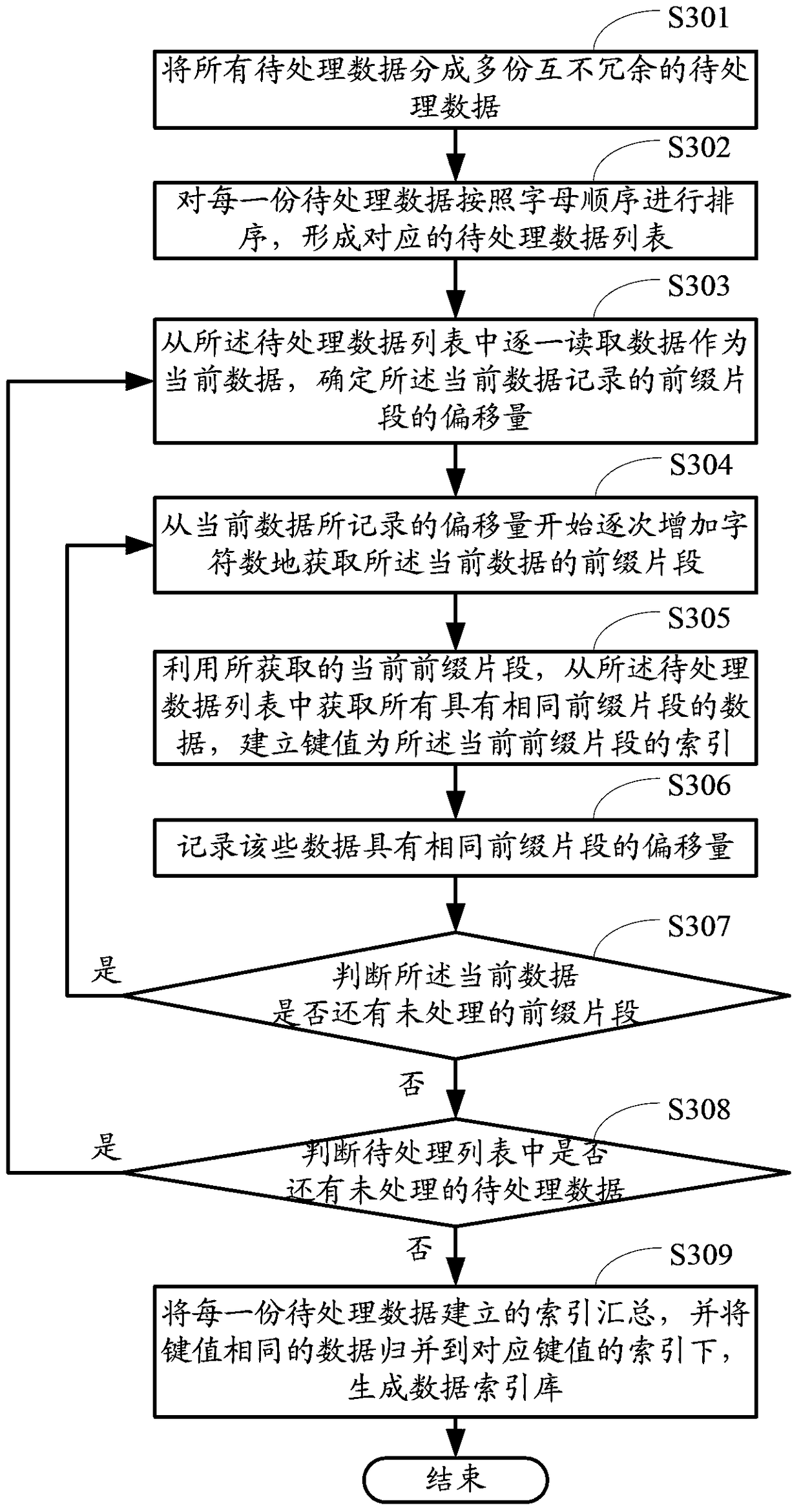
Method for establishing a data index database, method and device for generating search suggestions
ActiveCN102831224BImprove efficiencyImprove performanceSpecial data processing applicationsInternal memoryTheoretical computer science
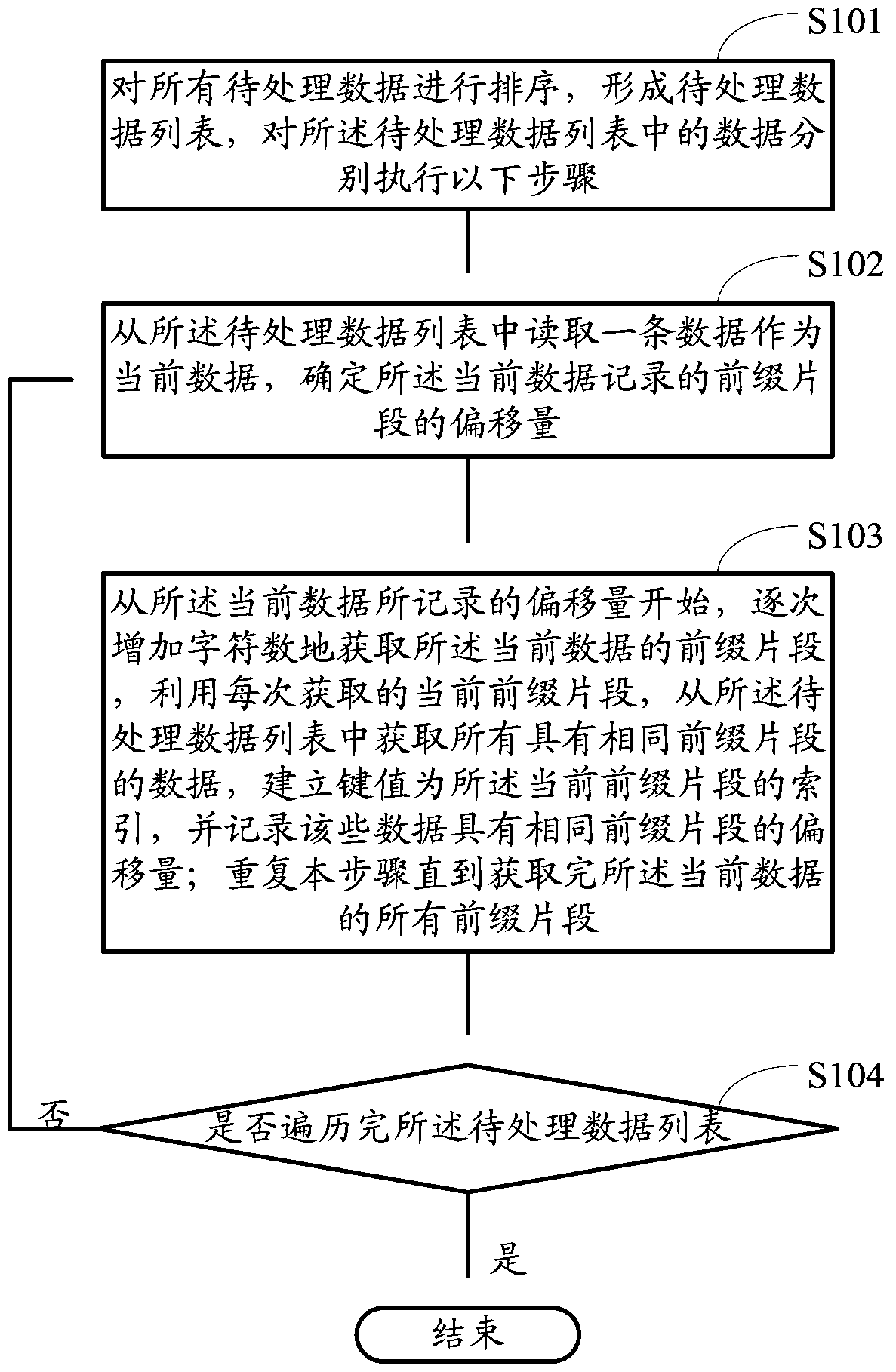
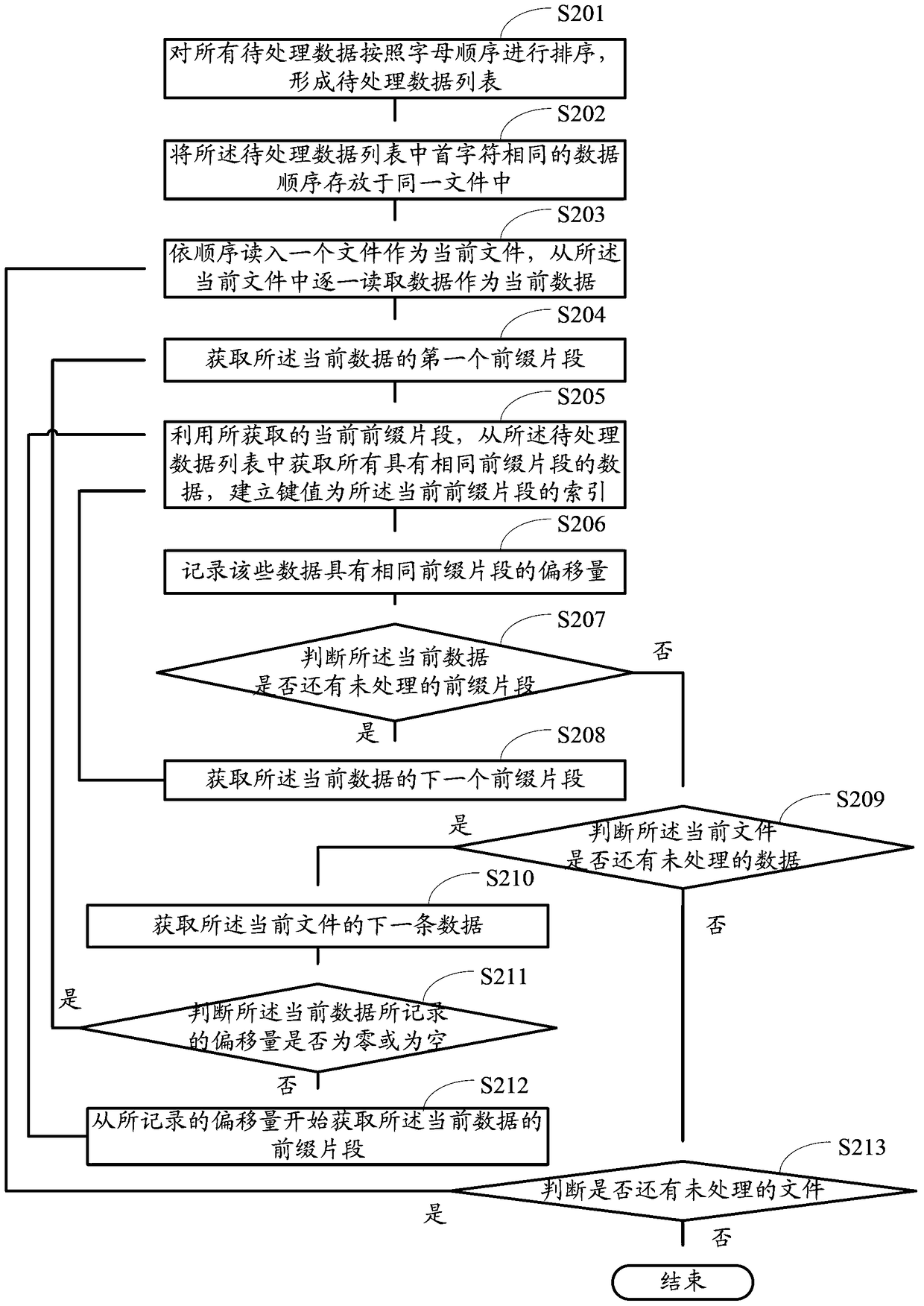
The invention provides a creating method for a data index base and searching suggest generation method and device. The creating method comprises the following steps of: sequencing all data to be processed to form a sequence of data to be processed; and reading data from the sequence of data to be processed one by one as current data, determining the offset of prefix fragments recorded by the current data, acquiring the prefix fragments of the current data by gradually increasing characters starting from the offset recorded by the current data, acquiring all data containing the same prefix fragments from the sequence of data to be processed by using the current prefix fragment acquired at each time, creating a key value as an index of the current prefix fragment until all the prefix fragments of the current data are completely acquired, and creating key values as indexes of corresponding prefix fragments. Compared with the prior art, the creating method can lower the internal memory usage amount during base creation, reduce I / O read-write operations, reduce the base creation time of the data index base and increase the timeliness of searching suggest service.
Owner:BEIJING BAIDU NETCOM SCI & TECH CO LTD
A kind of reagent and its application for constructing free dna library
ActiveCN114032619BShorten library building timeReduce the cost of building a libraryMicrobiological testing/measurementLibrary creationPolyethylene glycolLigation
The invention discloses a reagent for constructing a free DNA library and its application. The reagents include carrier DNA, heat-sensitive protease, and linker ligation reagents including ligase, ligation buffer, propylene glycol and polyethylene glycol. The use of the reagent can overcome the problem that high-throughput sequencing library construction can only be performed after single-cell whole-genome amplification in the prior art, and avoid contaminating DNA signal amplification and masking of cfDNA methylation sites caused by single-cell whole-genome amplification It can significantly shorten the construction time and reduce the cost of library construction.
Owner:SUZHOU BASECARE MEDICAL DEVICE CO LTD
A method and kit for simultaneously constructing sequencing libraries for dna and rna
ActiveCN111139532BAvoid stickingHigh interrupt efficiencyTransferasesLibrary creationTn5 transposaseMagnetic bead
The invention discloses a method and a kit for simultaneously constructing a sequencing library by DNA and RNA. The method comprises the following steps: (1) carrying out reverse transcription on RNAin a reaction substrate to form RNA / cDNA composite double strands; (2) directly adding Tn5 transposase and a fragmentation buffer solution into a mixture of a reverse transcription product cDNA / RNA composite double strand and a DNA double strand for fragmentation, and adding a part of linker sequence to the 5' end of the double strands during fragmentation; (3) strand displacement and library amplification: adding an amplification buffer solution and a strand displacement and amplification enzyme mixture into the obtained product, and carrying out PCR amplification; and (4) carrying out magnetic bead purification to obtain a sequencing library of the original DNA / RNA. According to the method, the library building steps are greatly reduced, the library building time is shortened, the library output and offline data quality is improved, and more real information is helped to be obtained.
Owner:VAZYME BIOTECH NANJING
A method and kit for constructing a library for whole-genome high-throughput sequencing
ActiveCN112301432BReduce crosstalkReduce redundancyMicrobiological testing/measurementLibrary creationHigh fluxGenetics
The invention discloses a method for constructing a whole-genome high-throughput sequencing library, comprising the following steps: 1) extracting sample gDNA; 2) enzymatically cleaving the sample gDNA into fragments, filling in the ends, and adding A to obtain the added A 3) Ligate the A-added gDNA with an adapter combination to obtain a ligation product, the adapter combination includes two parts: a Y-shaped reverse adapter and a high GC splint adapter; 4) Purify the ligation product, obtaining a purified product; and 5) performing fragment screening on the purified product to obtain a sequencing library. The invention also discloses a kit for constructing a whole genome high-throughput sequencing library.
Owner:BERRYGENOMICS CO LTD
Asymmetric deoxyribose nucleic acid (DNA) artificial adapters by using second-generation high-throughput sequencing technology and application thereof
InactiveCN102061335BImprove connection efficiencyEasy to storeMicrobiological testing/measurement3-deoxyriboseElectrophoresis
The invention provides asymmetric deoxyribose nucleic acid (DNA) artificial adapters by using a second-generation high-throughput sequencing technology, which is composed of two DNA oligonucleotide single strands. The preparation method comprises the following steps: dissolving the two DNA oligonucleotide single strands into a quenching solution; regulating the final concentration to 2mM and volume to 20microlitre; carrying out a quenching reaction to form the DNA artificial adapters which are locally and mutually complemented at a temperature of 95 DEG C for 5 minutes; reducing the temperature of 95 DEG C to 12 DEG C at the speed of 0.1 DEG C per second; keeping the temperature of 12 DEG C, and diluting an adapter solution after quenching to 500 mu M at a ratio of 1:4; and storing at the temperature of minus 20 DEG C. An application of the asymmetric DNA artificial adapters by using the second-generation high-throughput sequencing technology is as follows: A, asymmetric DNA adapters are in ligation with DNA samples; B, non-connected DNA artificial adapters are purified and isolated by using electrophoresis tapping; C, judgment is carried out; and D, a polymerase chain reaction (PCR) amplification reaction is performed based on an asymmetric sequence. The asymmetric DNA artificial adapters by using the second-generation high-throughput sequencing technology has the obvious advantages that: 1) the usage of original DNA samples for establishing a library is reduced to 50 nanogram and the sensitivity of the original DNA samples is improved by 100 times; and 2) 100% of effective sequencing samples are produced in the process of adapter connection for establishing the library.
Owner:SUZHOU ZHONGXIN BIOTECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com