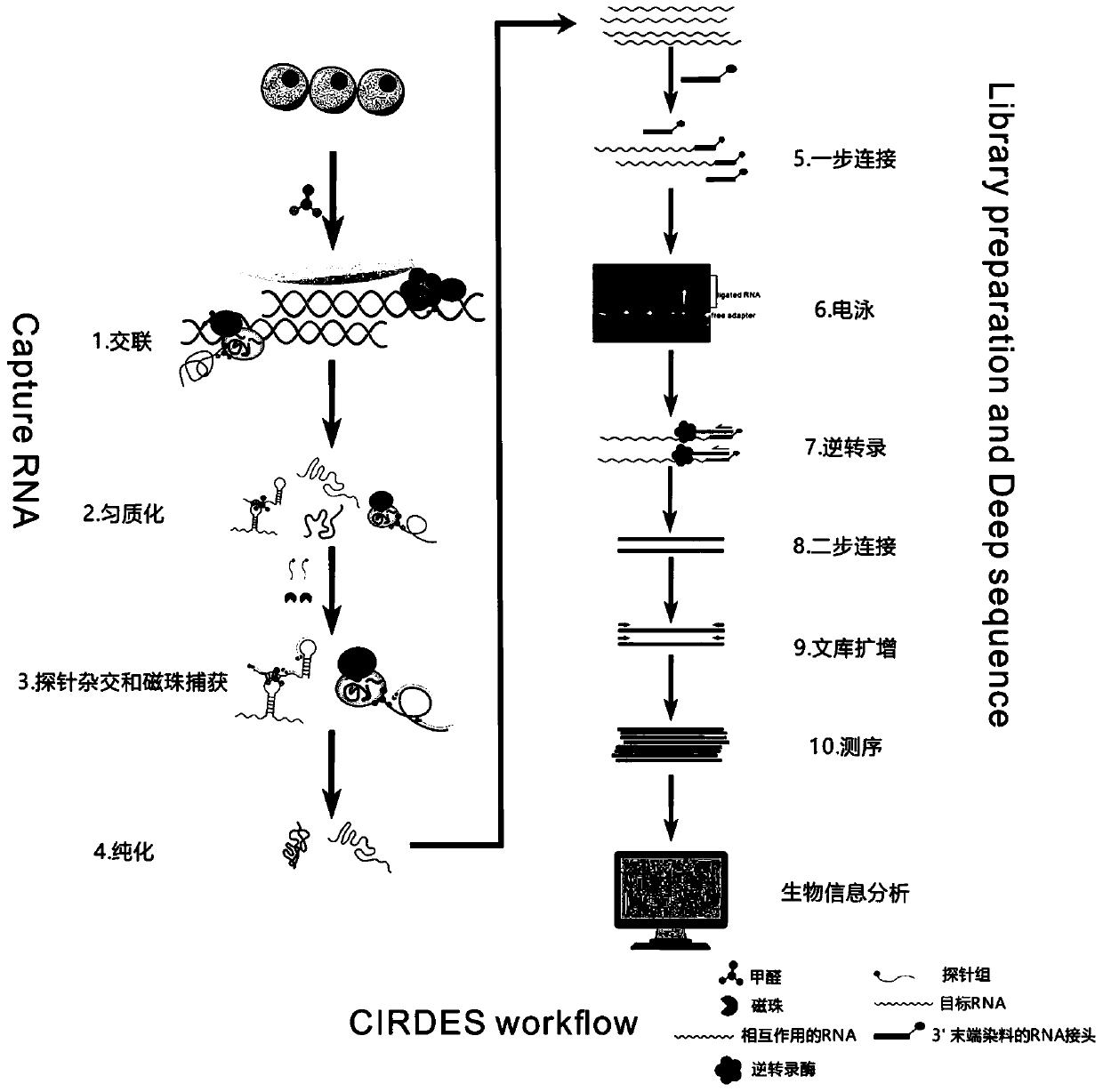
High-throughput sequencing method for efficient research of RNA interaction interactome and application of high-throughput sequencing method
A high-throughput and efficient technology, applied in the field of RNA interactomics, which can solve the problems of underestimation, difficult detection of RNA interactions, and mixing.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0268] 1. Design reverse complementary probes for U6 snRNA using CIRDES design rules
[0269] First query the human U6snRNA sequence (>NC_000015.10:c67840045-67839940, Homo sapiens chromosome 15, GRCh38.p12 Primary Assembly) through the NCBI database (https: / / www.ncbi.nlm.nih.gov / gene / ), the result See Table 1; then design according to the above-mentioned CIRDES probe design principles, because the length of the RNA fragment is less than 200nt, design two probes, the results are shown in Table 1.
[0270] Table 1: U6 snRNA sequence and U6 probe sequence designed by CIRDES technology
[0271]
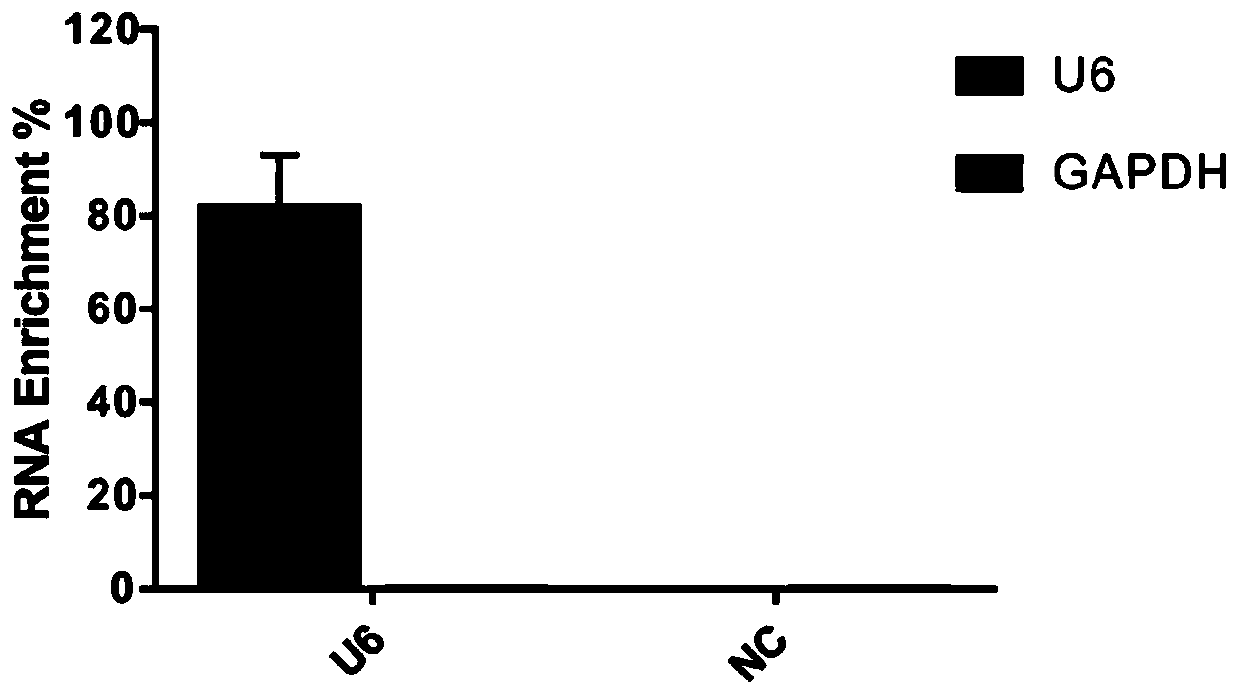
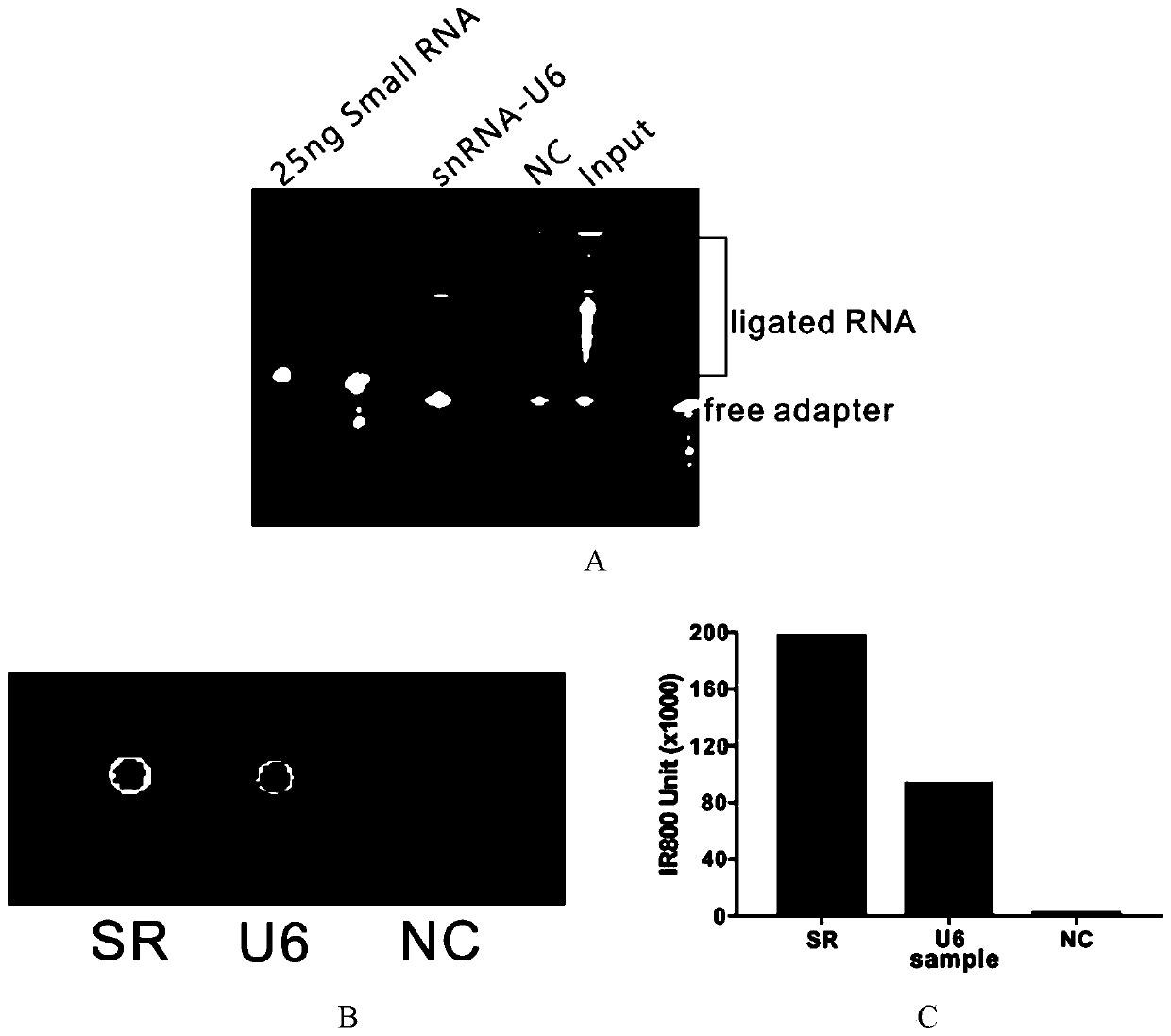
[0272] 2. CIRDES technology enriches U6 snRNA and its interacting RNA
[0273] Culture HepG2 cells in a 15cm dish according to the above method, and then use 2% (v / v) formaldehyde solution, according to 1×10 7 Cells: 10mL formaldehyde solution was cross-linked and fixed, and after 80 cycles of ultrasonic homogenization, the nucleic acid in the cell lysate was fragmented into a lengt...
Embodiment 2
[0292] With reference to the experimental procedure of Example 1, the difference is that the concentration and consumption of crosslinking agent formaldehyde are different, and it is divided into the following five groups of experiments:
[0293] (1) No cross-linking agent formaldehyde (named: -Formaldehyde);
[0294] (2) Use 1% (v / v) formaldehyde solution, according to 1×10 7 Each cell: 2.5mL formaldehyde solution cross-linked and fixed (named: 1%-Formaldehyde-2.5mL);
[0295] (3) Use 2% (v / v) formaldehyde solution, according to 1×10 7 Each cell: 2.5mL formaldehyde solution cross-linked and fixed (named: 2%-Formaldehyde-2.5mL);
[0296] (4) Use 2% (v / v) formaldehyde solution, according to 1×10 7 Cells: 10mL formaldehyde solution cross-linked and fixed (named: 2%-Formaldehyde-10mL);
[0297] (5) Use 3% (v / v) formaldehyde solution, according to 1×10 7 One cell: 2.5mL formaldehyde solution cross-linked and fixed (named: 3%-Formaldehyde-2.5mL).
[0298] The abundance of U4R...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com