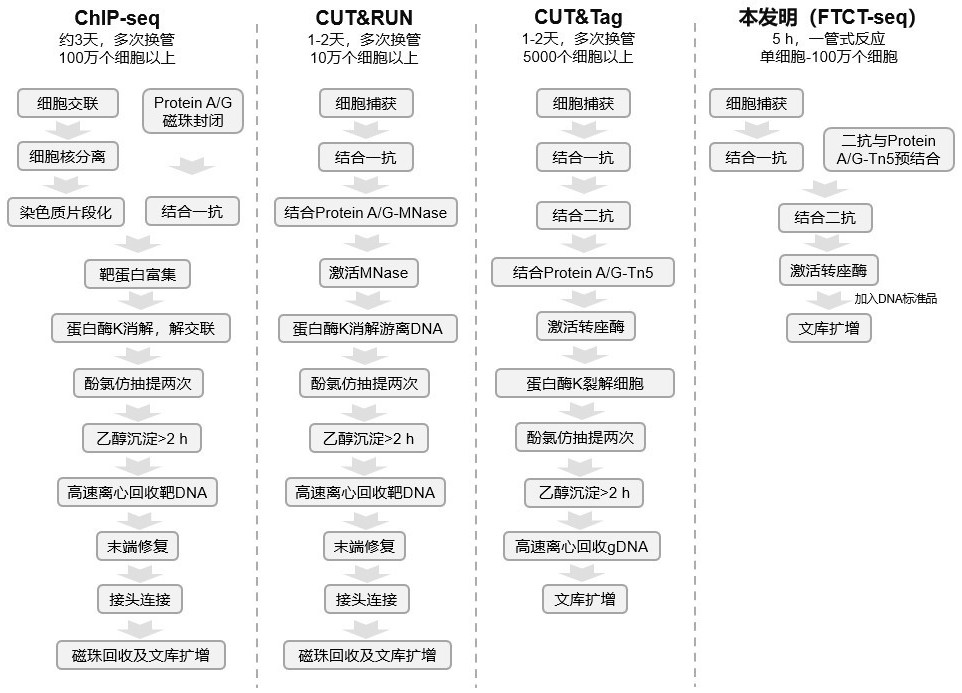
A Rapid Library Construction Method for Identifying Chromatin Binding Profiles of Target Proteins
A technology for chromatin and target identification, which is used in the field of rapid library construction for identifying chromatin binding maps of target proteins. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0090] Example 1: Effect of pre-combination of secondary antibody with recombinant fusion transposase Protein A-Tn5 and Protein AG-Tn5 on library yield.
[0091] See flow chart Figure 7 , the specific process is as follows:
[0092] 1. Harvest the cells. Take about 100,000 293FT cells and centrifuge at 600xg for 5 min at room temperature to collect the cells. Wash 2 times with wash buffer (20 mM HEPES (pH 7.5), 150 mM NaCl, 0.5 mM spermidine, 1x Protease inhibitorscocktail). Resuspend the cells in 90 µL of wash buffer.
[0093] 2. Activate magnetic beads and capture cells. Take 10 μL ConA magnetic beads, add binding buffer (20 mM HEPES (pH7.5), 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2) to wash the magnetic beads twice. Resuspend the magnetic beads in 10 µL of binding buffer. Add the magnetic beads to the cells and incubate with rotation at room temperature for 5-10 min.
[0094] 3. Primary antibody binding. Dilute the three kinds of antibodies at 1:100 with primary antibody ...
Embodiment 2
[0109] Example 2: Effects of different genomic DNA extraction methods on library yield.
[0110] Use the recombinant fusion transposase Protein AG-Tn5 and H3K27me3 antibody to verify the effect of different genomic DNA extraction methods on the library yield. The flow chart is shown in Figure 12 , the specific process is as follows:
[0111] 1. Harvest the cells. Take about 100,000 293FT cells and centrifuge at 600xg for 5 min at room temperature to collect the cells. Wash 2 times with wash buffer (20 mM HEPES (pH 7.5), 150 mM NaCl, 0.5 mM spermidine, 1x Protease inhibitorscocktail). Resuspend the cells in 90 µL of wash buffer.
[0112] 2. Activate magnetic beads and capture cells. Take 10 μL ConA magnetic beads, add binding buffer (20 mM HEPES (pH7.5), 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2) to wash the magnetic beads twice. Resuspend the magnetic beads in 10 µL of binding buffer. Add the magnetic beads to the cells and incubate with rotation at room temperature for 5-10 mi...
Embodiment 3
[0133] Example 3: Library construction effect of Protein A-Tn5 and Protein AG-Tn5 in FTCT-seq.
[0134] According to the improvement of Example 1 and Example 2, we invented a rapid library construction method for identifying the DNA binding profile of the target protein, and named it FTCT-seq. And use recombinant fusion transposase Protein A-Tn5 and Protein AG-Tn5 to verify the effect of building the library of the method process of the present invention, specific process is as follows:
[0135] 1. Harvest the cells. Take about 100,000 293FT cells and centrifuge at 600xg for 5 min at room temperature to collect the cells. Wash 2 times with wash buffer (20 mM HEPES (pH 7.5), 150 mM NaCl, 0.5 mM spermidine, 1x Protease inhibitorscocktail). Resuspend the cells in 90 µL of wash buffer.
[0136] 2. Activate magnetic beads and capture cells. Take 10 μL ConA magnetic beads, add binding buffer (20 mM HEPES (pH7.5), 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2) to wash the magnetic beads twic...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com