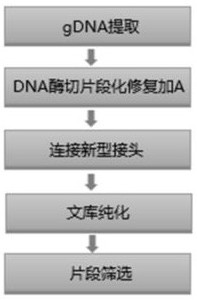
Method and kit for constructing whole genome high-throughput sequencing library
A technology for sequencing libraries and whole genomes, applied in chemical libraries, combinatorial chemistry, recombinant DNA technology, etc., can solve the problems of unusable redundant sequencing data and high sequencing costs, and achieve the effect of simple operation and short time consumption
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0092] Example 1: Using the standard cell line NA12878 genomic DNA, using a common Y-type connector (TruSeq connector), a Y-type reverse connector according to the present invention, and a novel connector combination according to the present invention (Y-type reverse connector + high GC splint) respectively linker) to construct a PCR-free library, and use the PCR library as a control. Sequencing on the machine, data analysis, and comparing the sequencing data of the PCR-free library and the PCR library constructed by three different adapters.
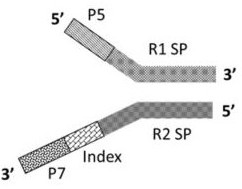
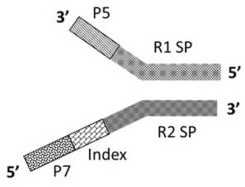
[0093] According to the structure of the Y-type reverse joint of the present invention, such as image 3 Shown; According to the structure of the novel joint combination of the present invention (Y type reverse joint + high GC splint joint) such as Figure 4 shown.
[0094] In this example, NA12878 gDNA was used as a sample to construct PCR-free whole-genome high-throughput sequencing using common Y-type connectors, Y-type reverse con...
Embodiment 2
[0110] Example 2: Human genomic DNA was used to construct a PCR-free library using common Y-joints (TrueSeq joints), Y-type reverse joints, combinations of common Y-joints and high GC splint joints, and novel joints of the present invention, respectively. Sequence together with the phix library, analyze the number of phix sequences detected in the library, calculate the crosstalk ratio of the index, and compare the redundancy under the same amount of data at the same time.
[0111] This example uses phix's principle of testing crosstalk ratios: phix library inserts are derived from viral genomic DNA. Its gene sequence has been precisely known, and its GC ratio is about 40, which is close to the GC ratio of the human genome. Its gene sequence is far from that of human beings and does not contain an index. Therefore, the library to be tested and phix were sequenced together on the computer, the number of phix sequences in the library was analyzed, and the ratio of the sequence ...
Embodiment 3
[0117] Example 3: Using different input amounts of templates to construct a library, perform sequencing on a computer, analyze data, and compare the sequencing data quality and performance analysis results of different input amounts.
[0118] In this example, using the novel linker combination according to the present invention, NA12878 genomic DNA was used as a sample to construct genome-wide high-throughput sequencing libraries with inputs of 200 ng and 300 ng respectively, and the library was subjected to 150PE paired-end sequencing on NovaSeq. Bioinformatics analyzes the sequencing results and analyzes the library quality of libraries constructed with different adapters. The following is the specific scheme.
[0119] Step 1: Prepare the reaction mixture shown in Table 8, prepare four tubes, two tubes with a DNA input volume of 200ng, and the other two tubes with a DNA input volume of 300ng, and then run the interruption, end filling and addition shown in Table 9 together A...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com