Patents
Literature
31results about How to "Lower preference" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Method for optimising gene expressing using synonymous codon optimisation
InactiveUS20060292566A1Improve selection qualityHigh phenotypic preferenceBacteriaMicrobiological testing/measurementBiological bodyNucleotide
The present invention discloses a method for modulating the quality of a selected phenotype that is displayed by an organism or part thereof and that results from the expression of a polypeptide-encoding polynucleotide by replacing at least one codon of that polynucleotide with a synonymous codon that has a higher or lower preference of usage by the organism or part thereof to produce the selected phenotype than the codon it replaces. The present invention is also directed to the use of a codon-modified polynucleotide so constructed for modulating the quality of a selected phenotype displayed by an organism or part thereof.
Owner:THE UNIV OF QUEENSLAND
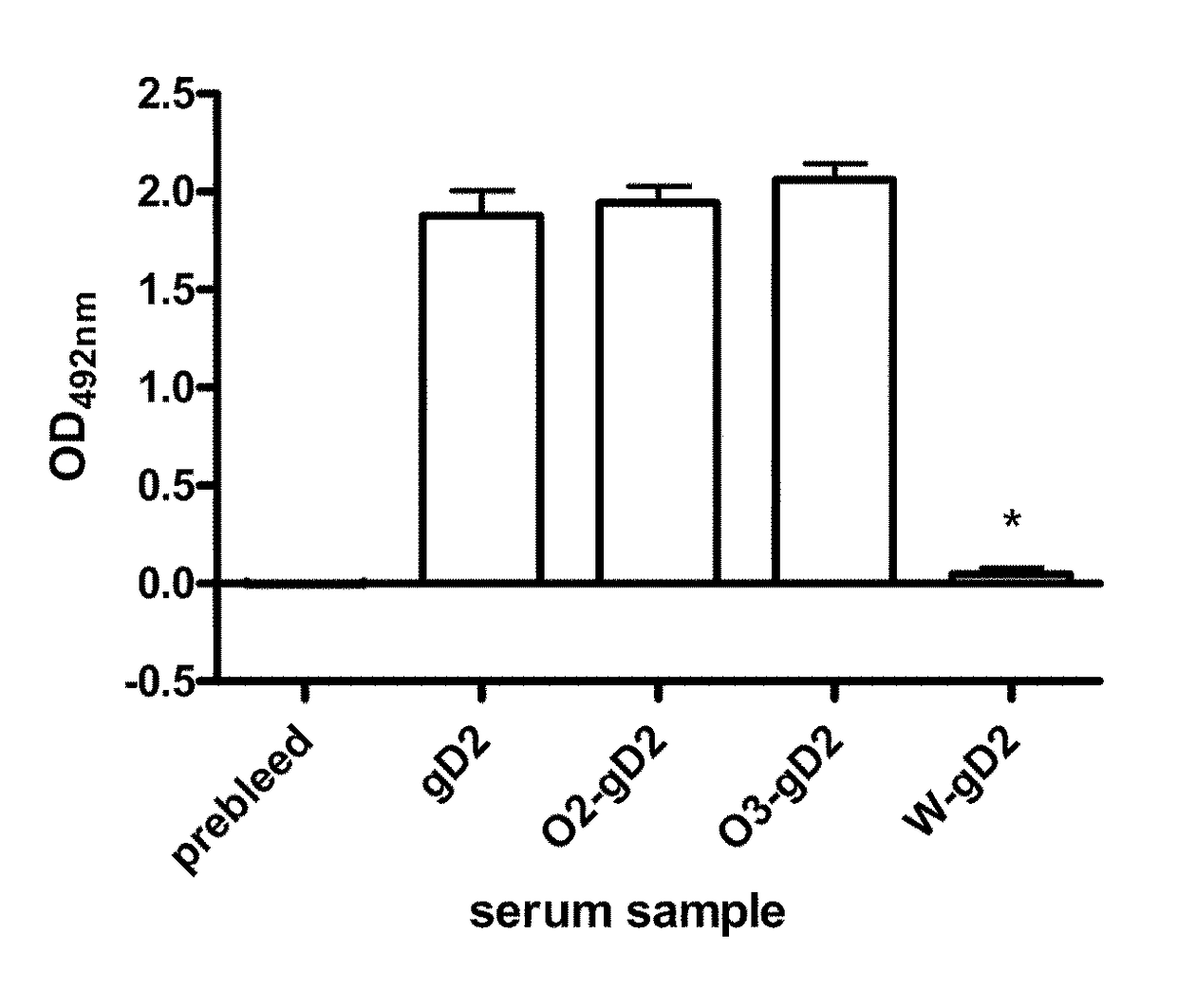
Expression system for modulating an immune response
ActiveUS20110020374A1Enhance immune responseEfficient productionAntibacterial agentsAntimycoticsMammalPolynucleotide
The present invention discloses methods and compositions for modulating the quality of an immune response to a target antigen in a mammal, which response results from the expression of a polynucleotide that encodes at least a portion of the target antigen, wherein the quality is modulated by replacing at least one codon of the polynucleotide with a synonymous codon that has a higher or lower preference of usage by the mammal to confer the immune response than the codon it replaces.
Owner:JINGANG MEDICINE AUSTRALIA PTY LTD
DNA amplification method
ActiveCN105463066AGuaranteed balanceImprove efficiencyMicrobiological testing/measurementA-DNADna amplification
The invention provides a DNA amplification method. The DNA amplification method is characterized by comprising the step that a T7 promoter sequence is added to the target DNA tail end. According to the DNA amplification method, amplification can be conducted on DNA smaller than one nanogram or even a single cell genome DNA on the basis of trace DNA; relatively even amplification is achieved through the target DNA containing the T7 promoter sequence; an error produced in the amplification process can be effectively removed.
Owner:BEIJING INST OF GENOMICS CHINESE ACAD OF SCI CHINA NAT CENT FOR BIOINFORMATION
Linker sequence and method for detecting ultra-low frequency mutation of target sequence
InactiveCN107446996AReduce dosageEfficient removalMicrobiological testing/measurementDNA/RNA fragmentationAnalysis methodGenome
The invention discloses a linker sequence and method for detecting the ultra-low frequency mutation of a target sequence. According to the method, a certain number of labeled molecules can be obtained by conducting terminal repair, A adding, linker connection, fragment enrichment, capture and the like on an interrupted genome or cfDNA, then on-machine sequencing is conducted, positive and negative chains are corrected by means of a later-period biological analysis method, and real mutation information is obtained through analysis.
Owner:IGENETECH BIOTECH (BEIJING) CO LTD
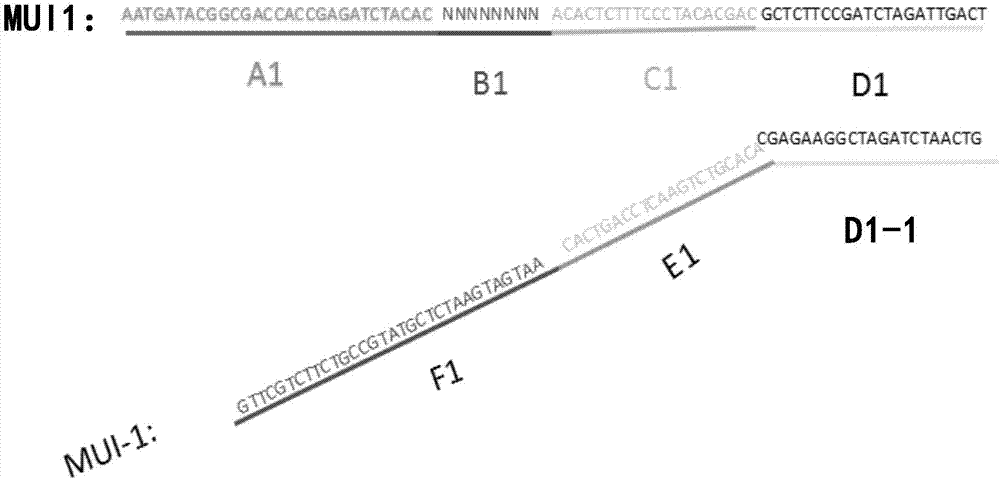
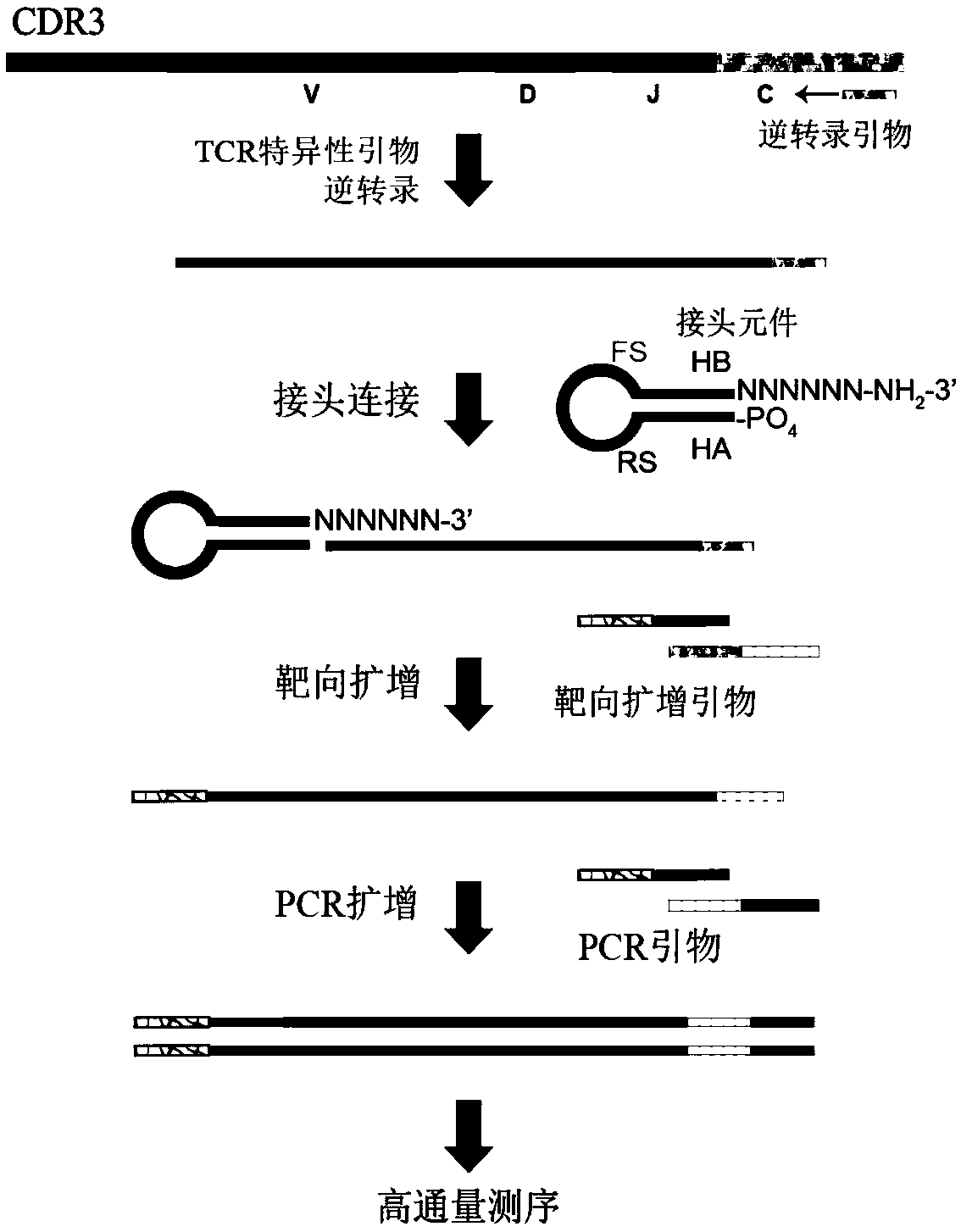
Specific recognition sequence based T cell receptor high-throughput sequencing library construction and sequencing data analysis method
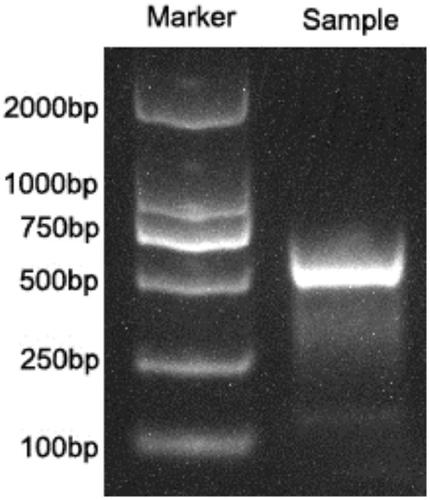
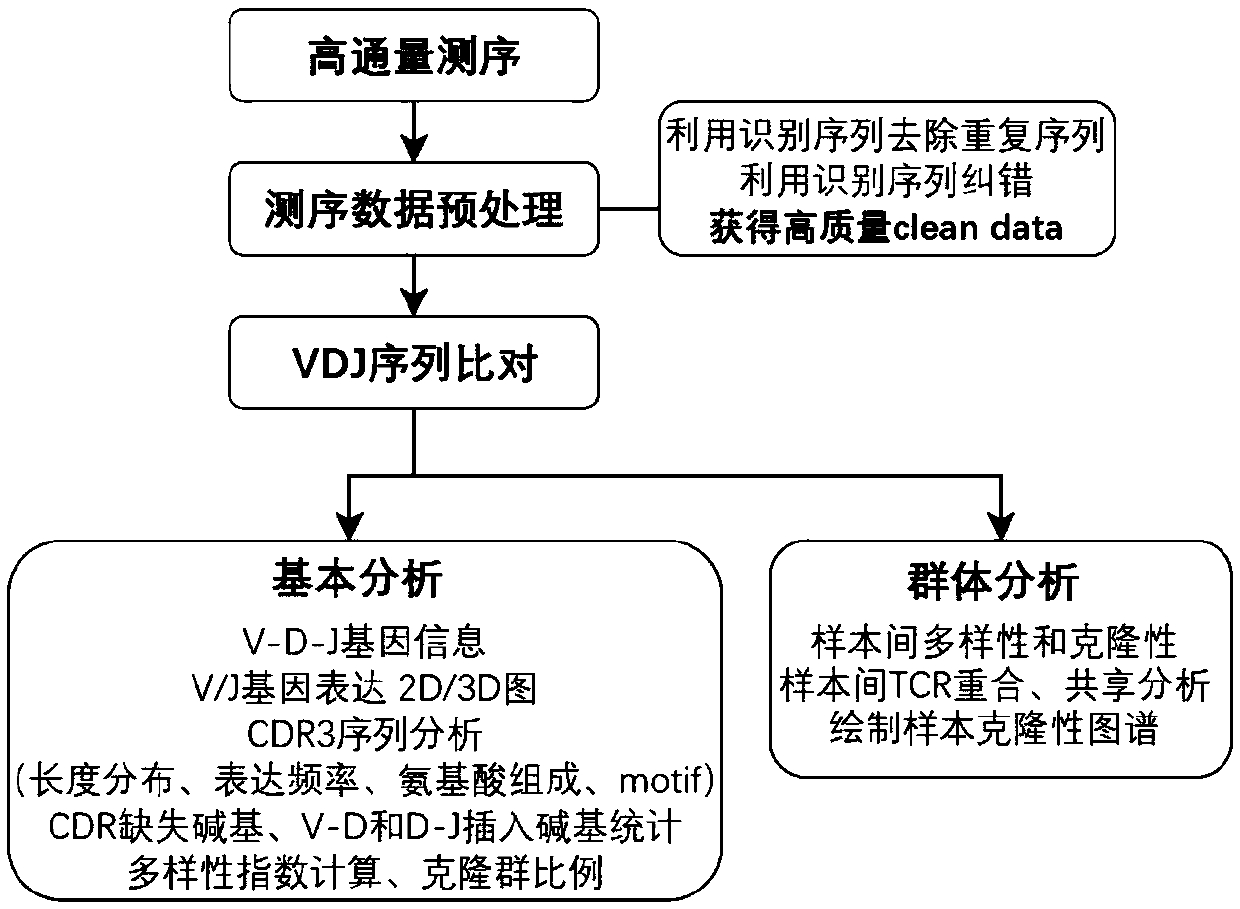
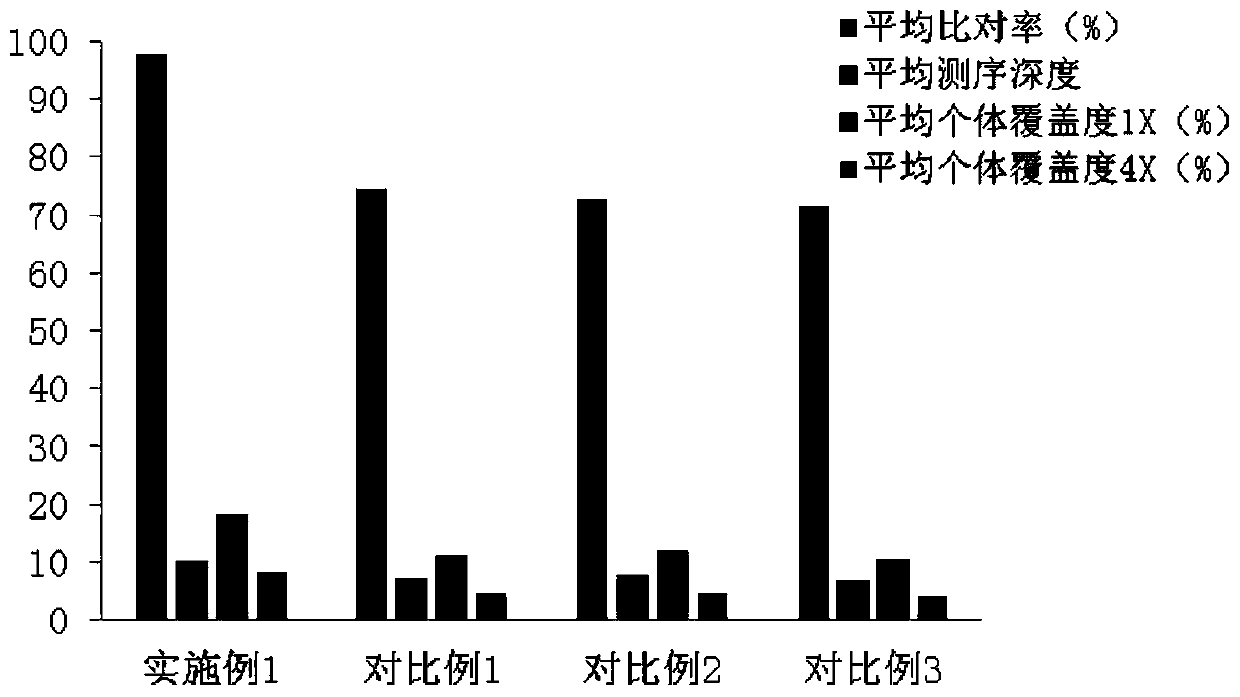
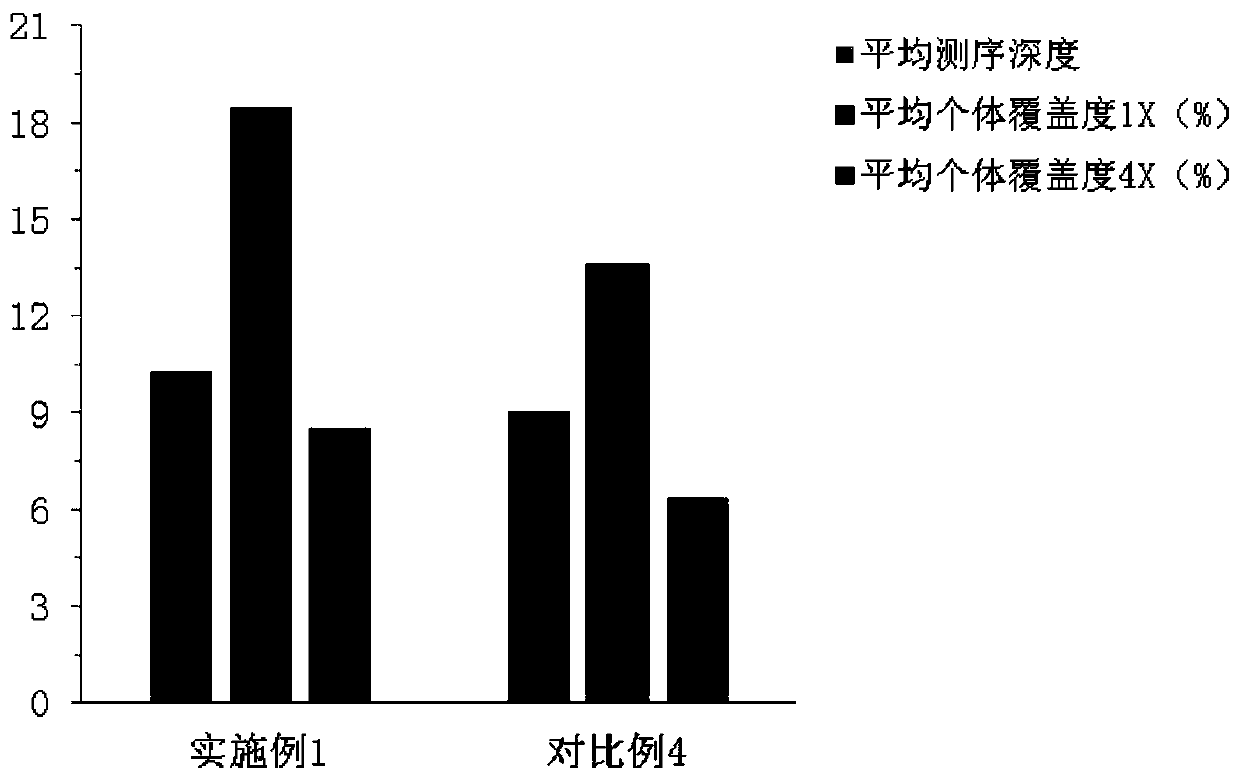
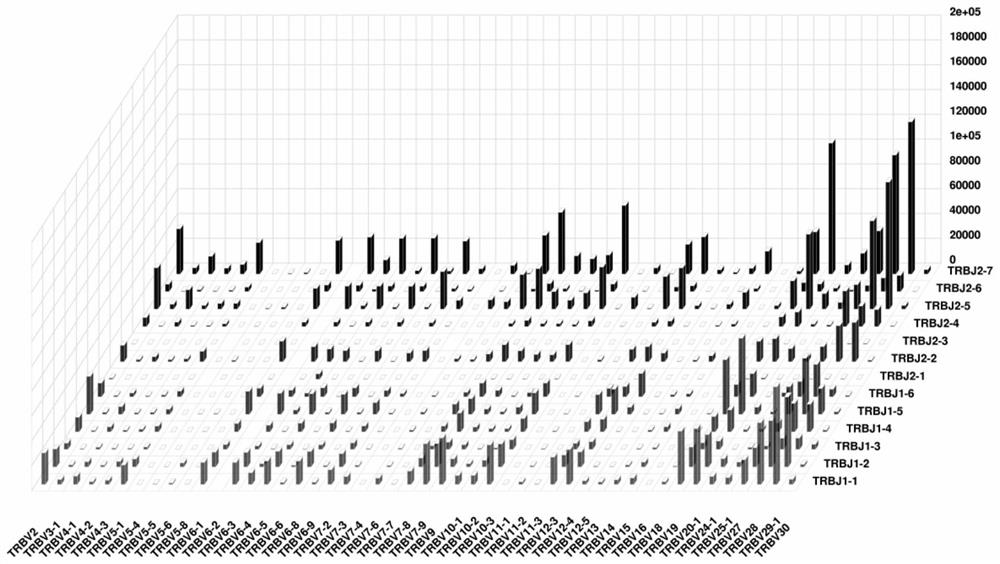
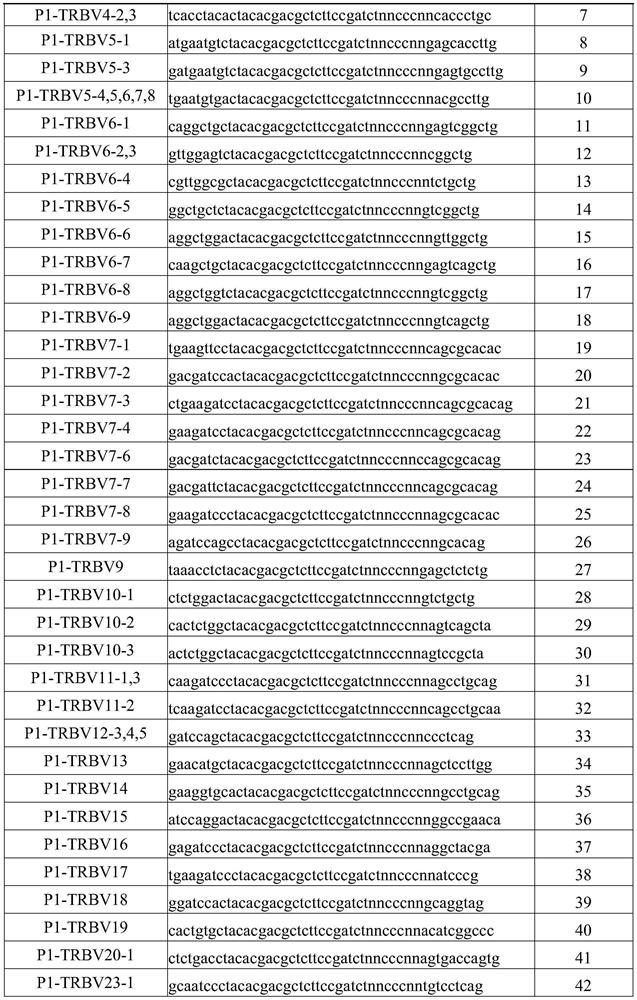
PendingCN111363783AComprehensive and Accurate Diversity InformationLower preferenceMicrobiological testing/measurementSequence analysisA-DNAV region
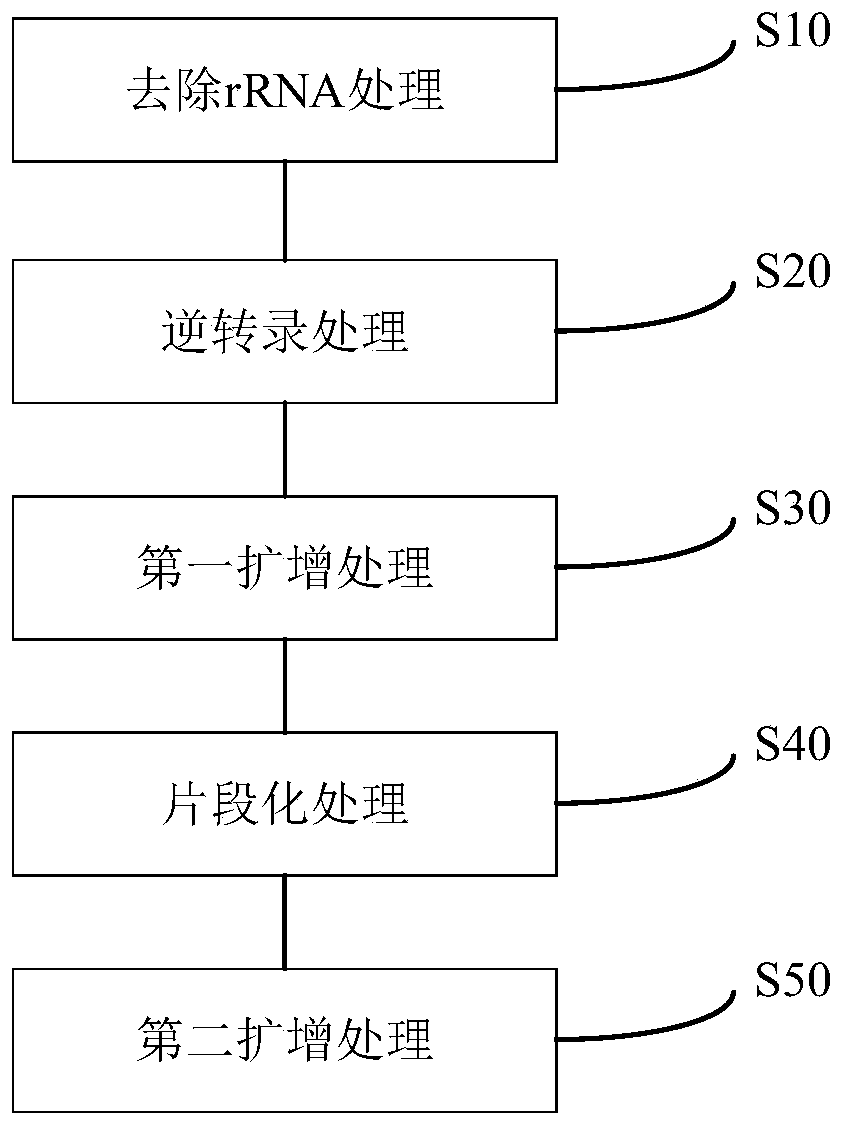
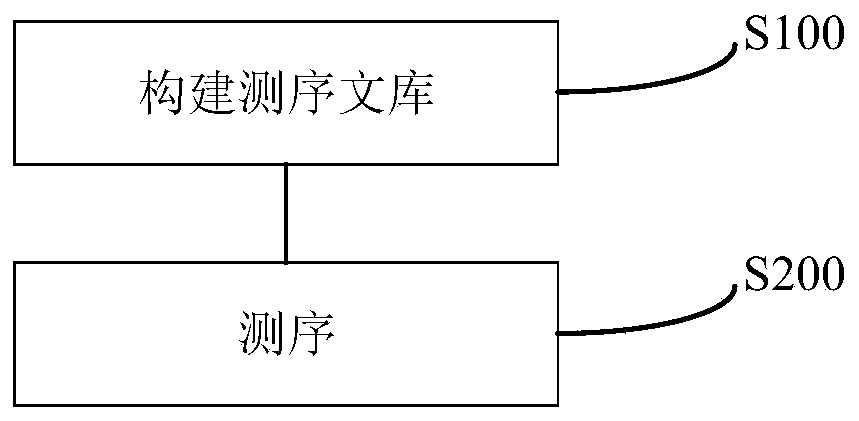
The present invention discloses a specific recognition sequence based T cell receptor high-throughput sequencing library construction and sequencing data analysis method. According to the method, a specific reverse transcription primer is designed for an mRNA sequence of a C region of a TCR constant region, cDNA is obtained by reverse transcription, and the 3' end of the cDNA is connected with a library construction linker with a specific recognition sequence; then the linker with the specific recognition sequence is added by using a splint connection method, and a TCR gene rearrangement sequence is amplified under the action of DNA polymerase by utilizing a gene specific primer with a tag; and finally a DNA library is added with a sequencing linker by PCR amplification to prepare a high-throughput sequencing library, and the high-throughput sequencing library is used for sequencing. The TCR gene diversity is comprehensively analyzed by bioinformatics, and a rearrangement rule of TCR genes including J region, D region and V region genes can be accurately obtained. The method is high in library construction efficiency, few in library construction step, low in required RNA initial quantity and low in library construction cost.
Owner:武汉康测科技有限公司
Expression system for modulating an immune response
ActiveUS20110287039A1Enhance immune responseEfficient productionSsRNA viruses negative-senseSsRNA viruses positive-senseMammalNucleotide
The present invention discloses methods and compositions for modulating the quality of an immune response to a target antigen in a mammal, which response results from the expression of a polynucleotide that encodes at least a portion of the target antigen, wherein the quality is modulated by replacing at least one codon of the polynucleotide with a synonymous codon that has a higher or lower preference of usage by the mammal to confer the immune response than the codon it replaces.
Owner:JINGANG MEDICINE AUSTRALIA PTY LTD
Sebastiscus marmoratus gene screening and mining method based on simplified genome sequencing technology
ActiveCN110283892AQuality improvementIncreased enzyme cleavage rateMicrobiological testing/measurementClimate change adaptationGenomic sequencingNucleotide
The invention provides a sebastiscus marmoratus gene screening and mining method based on a simplified genome sequencing technology, and belongs to the technical field of molecular marker development. The method comprises steps as follows: genome DNA extraction; genome DNA double digestion; linkage connection; PCR (polymerase chain reaction) amplification; sequence fragment recovery and high-throughput sequencing; data analysis and screening and gene locus mining. According to the method, extracted DNA has higher quality, enzyme cleavage rate is effectively increased, primer design difficulty is reduced, recovered fragments have more uniform size, the sequencing amount is larger, and a sequencing result is more accurate; areas such as a high-repetition area and the like with relatively higher methylation degree can be avoided, so that enzyme cleavage fragments with more specific sequences are collected, and marking effectiveness of SNPs (single nucleotide polymorphisms) for following identification is higher; obtained nucleic acid molecules have quite low preference and high complexity, and therefore, the maximum gene coverage is obtained with the least sequencing data during sequencing.
Owner:ZHEJIANG OCEAN UNIV
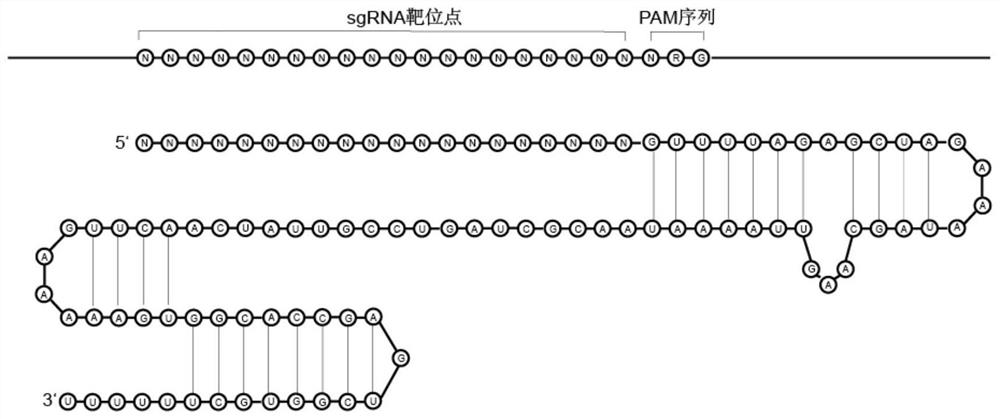
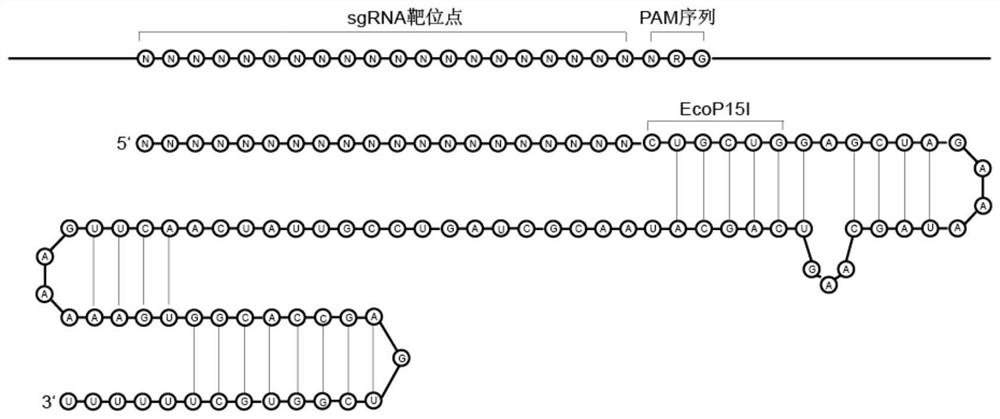
Preparation method of target sequence random sgRNA full-coverage group
The invention provides a preparation method of a target sequence random sgRNA full-coverage group. The preparation method comprises the following steps: cutting a PAM region of a target sequence by using restriction enzyme; connecting an upper sgRNA skeleton; obtaining a sequence of a targeting original spacer region through a side-cut activity restriction enzyme site on the sgRNA skeleton; connecting a T7 promoter; performing amplification to obtain an sgRNA library with a T7 promoter; and carrying out in-vitro transcription to obtain a target sequence sgRNA library. The invention also discloses a preparation method of the sgRNA library of the ribosomal RNA, a method for removing the ribosomal RNA in the RNA library, and a method for removing a human whole genome from a host genome. The method disclosed by the invention has the advantages of low cost, simplicity in manufacturing, uniform coverage, small preference, no limitation by the length of a target sequence, no need of designing sgRNA on a large scale and the like.
Owner:YEASEN BIOTECHNOLOGY (SHANGHAI) CO LTD
Construction method of single cell transcriptome sequencing library and applications thereof
ActiveCN113444770AMeet the requirements for sequencing developmentLower preferenceMicrobiological testing/measurementLibrary creationTransposaseEnzyme digestion
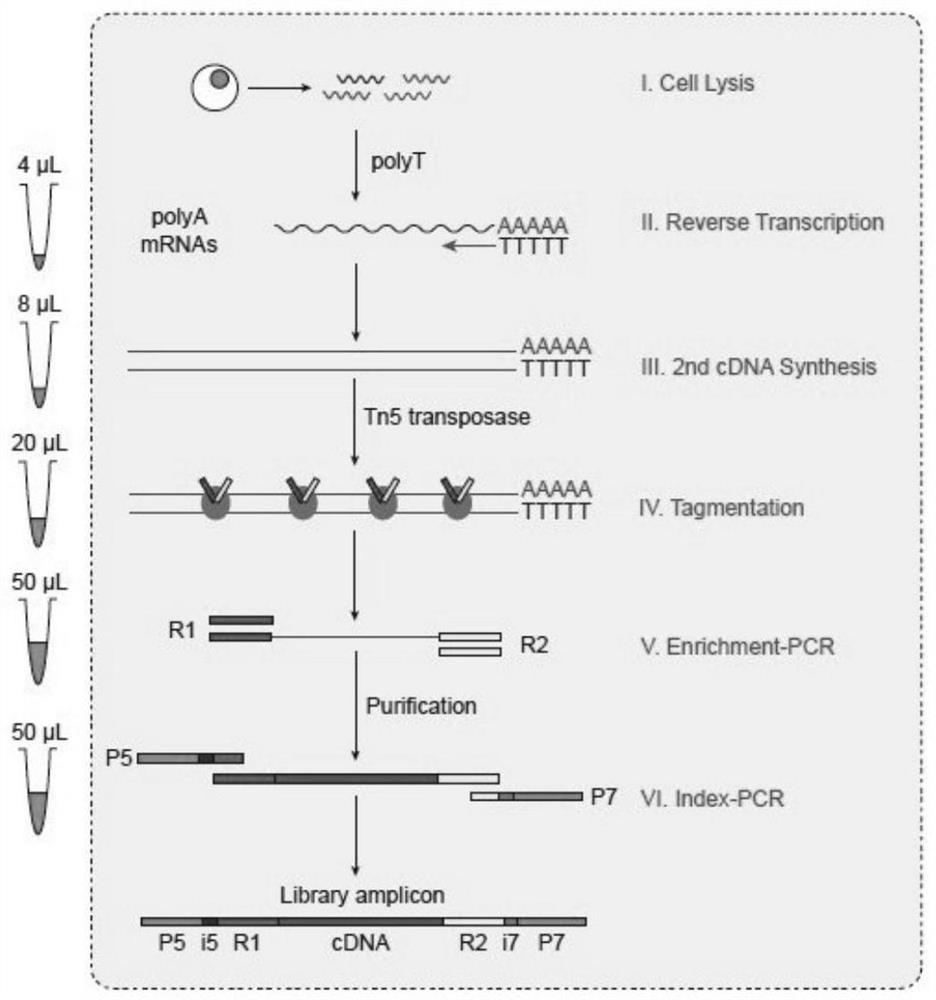
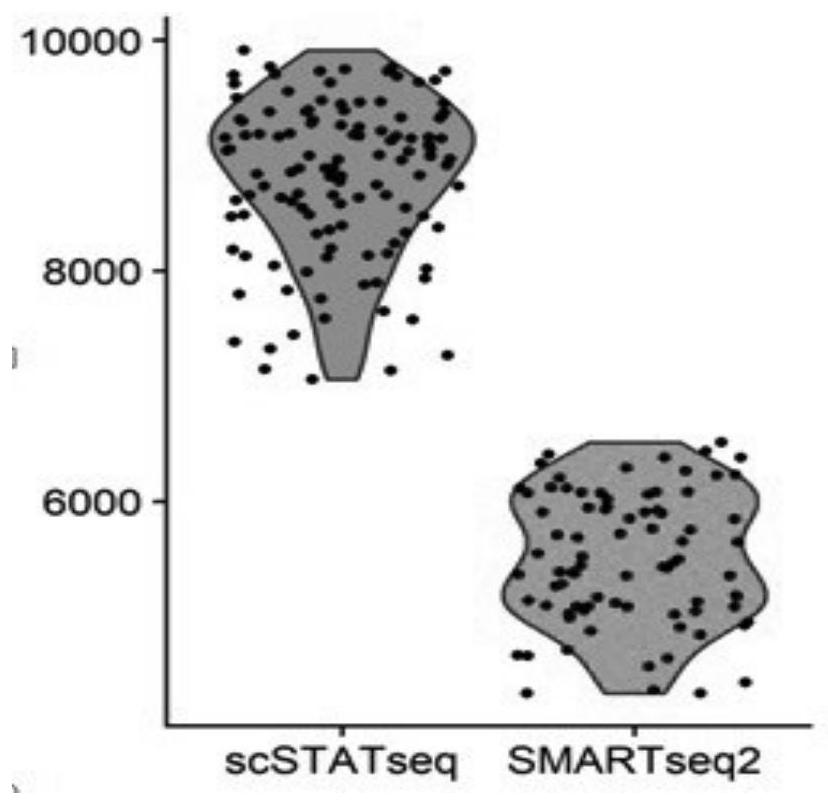
The invention belongs to the technical field of biology, and particularly discloses a construction method of a single cell transcriptome sequencing library and applications thereof. The method comprises the following steps: splitting sorted single cells in a pore plate, and carrying out reverse transcription by using a reverse transcription primer to synthesize a first chain cDNA; synthesizing a second chain cDNA through a replacement synthesis reaction; carrying out fragmentation on the double-chain cDNA by using Tn5 transposase; enriching a fragmented template by using PCR; and labeling the enriched fragment by using Index PCR, and performing purification to obtain the single cell transcriptome sequencing library. The Tn5 transposase is used for directly performing enzyme digestion on the double-stranded cDNA, and PCR pre-amplification on the cDNA is not needed, so that the library preference can be reduced, and more accurate transcriptome information can be obtained; and meanwhile, by using the Tn5 transposase, the library establishment efficiency can be improved, and the library establishment time can be shortened. The method is simple in library establishment steps, more genes can be detected, and the proportion of low and medium abundance gene dropout is effectively reduced. Therefore, library establishment time and costs are reduced, and more detailed research on the single cell level is facilitated.
Owner:ARMY MEDICAL UNIV
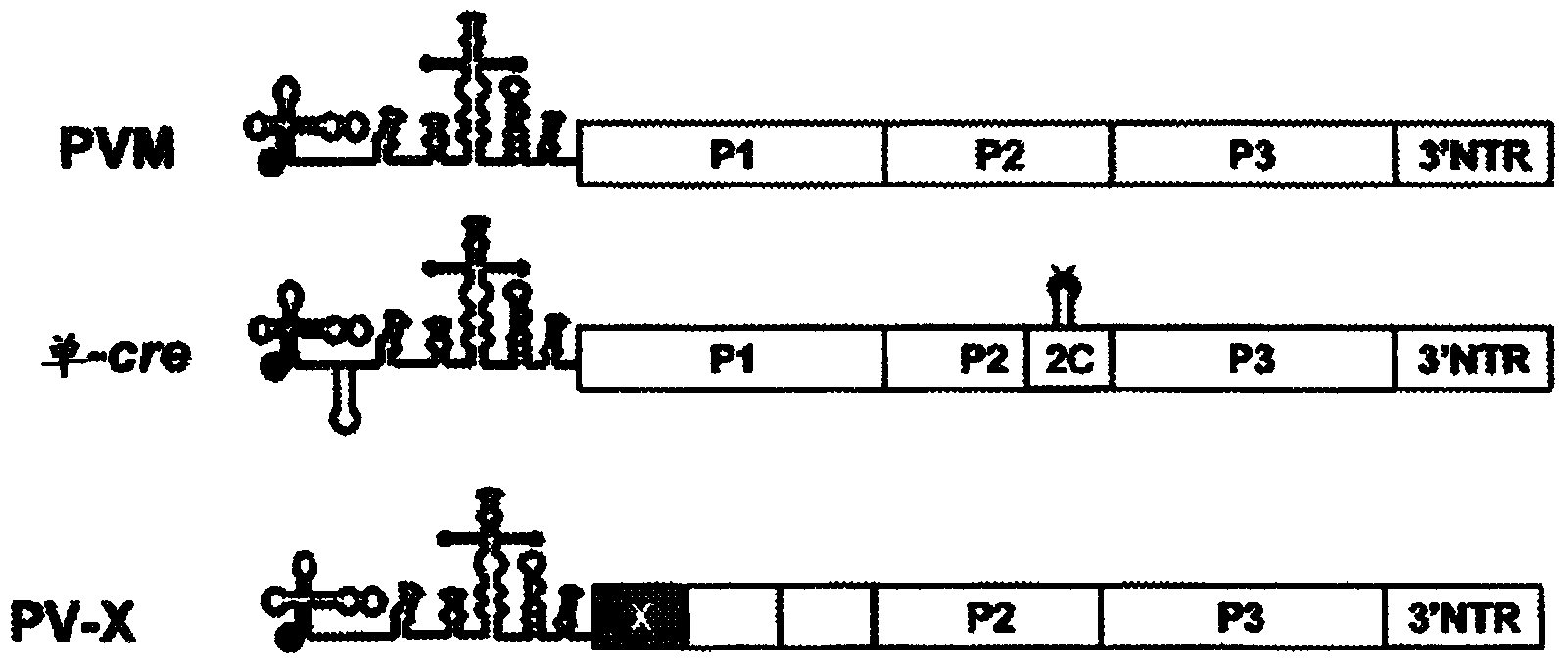
Novel attenuated poliovirus: PV-1 mono-cre-X
InactiveCN104204196ALower preferenceSsRNA viruses positive-senseInactivation/attenuationNucleic acid sequencingCoding region
A novel and stable attenuated poliovirus is produced by engineering an indigenous replication element (cre), into the 5′ non-translated genomic region (with inactivation of the native ere element located in the coding region of 2C (mono-crePV), and replacing the nucleic acid sequence of all or part of the capsid coding region (PI) with a substitute PI coding region having reduced codon pair bias. The stably attenuated poliovirus is effective for vaccines and immunization.
Owner:THE RES FOUND OF STATE UNIV OF NEW YORK
Method for constructing low-initial-quantity PCR-free high-throughput sequencing library
PendingCN109609597AOmit the purification stepOptimize the reaction systemMicrobiological testing/measurementLibrary creationNon invasiveBlood plasma
The present invention relates to the field of bioinformatics, and specifically provides a method for constructing a low-initial-quantity PCR-free high-throughput sequencing library. The method includes the following steps: (1) extracting cfDNA from plasma, carrying out terminal repair and 5' terminal phosphorylation, and purifying and recovering terminal repair products with magnetic beads; (2) adding "A" bases to 3' terminal of the purified terminal repair products to be integrated into one system with a joint connection system, and carrying out a reaction; and (3) purifying joint products with magnetic beads to obtain the sequencing library. The constructing method has the advantages of low cost, short time-consuming, no need of PCR amplification for library construction, maintained stable data output and low requirements for the initial DNA quantity. Only 1-2ng of cfDNA can meet the needs of library construction, and the method can be used for effective sequencing, and has very broad application prospects in high-throughput sequencing, non-invasive diagnosis and other research.
Owner:BEIJING USCI MEDICAL LAB CO LTD
Construct system and uses therefor
InactiveUS20120040367A1Improve preferenceHigh sensitivitySsRNA viruses negative-senseAntibacterial agentsBiological bodyTranslational efficiency
The present invention discloses construct systems and methods for comparing different iso-accepting codons according to their preference for translating RNA transcripts into proteins in cell or tissues of interest or for producing a selected phenotype in an organism of interest or part thereof. The codon preference comparisons thus obtained are particularly useful for modifying the translational efficiency of protein-encoding polynucleotides in cells or tissues of interest or for modulating the quality of a selected phenotype conferred by a phenotype-associated polypeptide upon an organism of interest or part thereof.
Owner:CORIDON
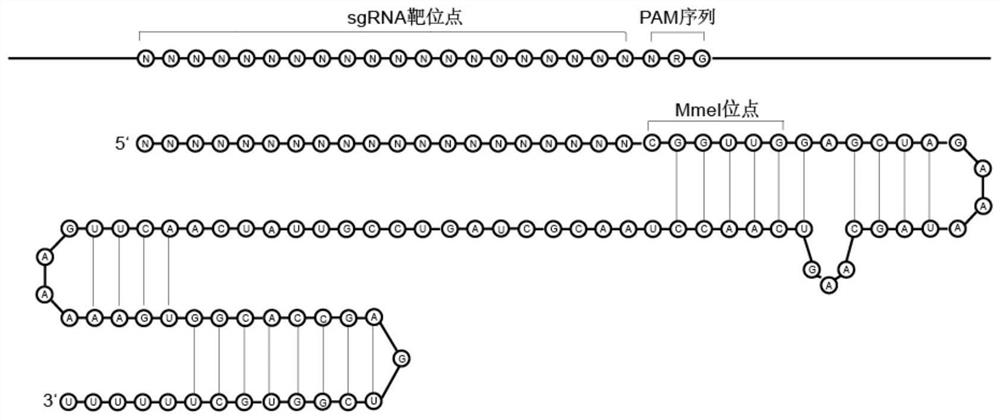
Preparation method of random sgRNA library of enzymic method target sequence
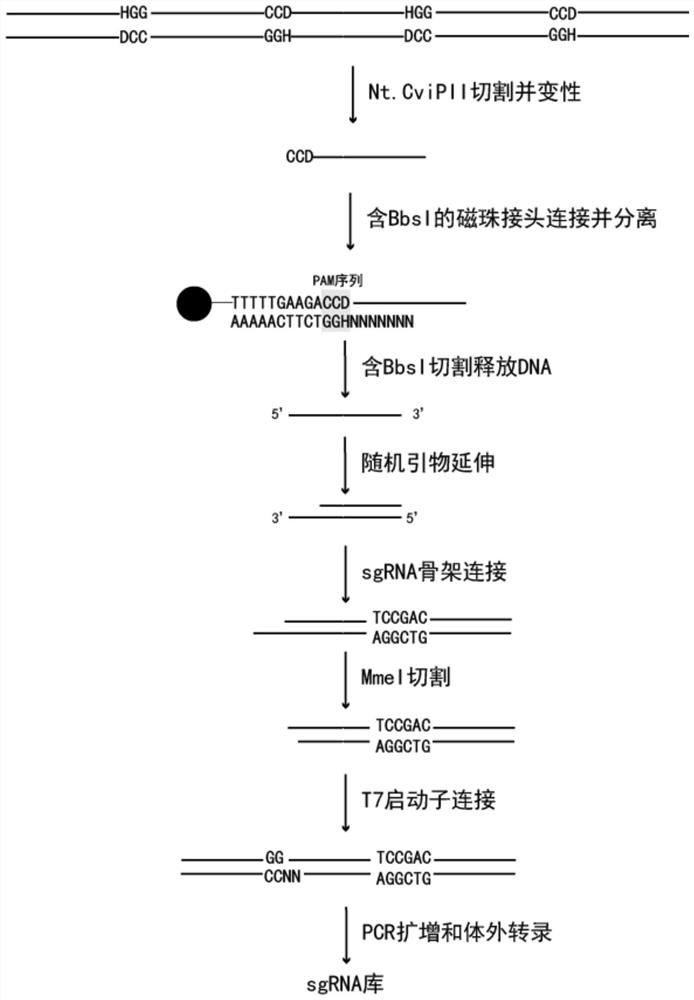
PendingCN114293264AMade evenlyUniform coverageNucleotide librariesLibrary creationRestriction Enzyme Cut SitePromoter
The invention provides a preparation method of an enzymic method target sequence random sgRNA library. The preparation method is characterized by comprising the following steps: nicking a PAM region of a sample DNA target sequence by adopting a random nicking enzyme Nt.CviPII; linker magnetic beads containing BbsI restriction enzyme cutting sites are connected with the fragmented DNA; carrying out BbsI cleavage to release DNA; performing random primer extension; connecting an sgRNA skeleton containing an MmeI restriction enzyme cutting site; performing MmeI cutting to release the original interval region; connecting a T7 promoter sequence; performing amplification to obtain an sgRNA library template containing the T7 promoter; and carrying out in-vitro transcription to obtain the sgRNA library. According to the preparation method of the enzyme-method target sequence random sgRNA library, all sgRNA groups which randomly cover a target area can be prepared at one time, and the preparation method has the advantages that the cost is low, the preparation is simple, the coverage is uniform, the preference is small, the method is not limited by the length of the target sequence, the sgRNA does not need to be designed on a large scale and the like, and has an important application value in capture and removal of target genes.
Owner:YEASEN BIOTECHNOLOGY (SHANGHAI) CO LTD
Expression system for modulating an immune response
ActiveUS9795658B2Enhance immune responseEfficient productionSsRNA viruses negative-senseSsRNA viruses positive-senseMammalPolynucleotide
The present invention discloses methods and compositions for modulating the quality of an immune response to a target antigen in a mammal, which response results from the expression of a polynucleotide that encodes at least a portion of the target antigen, wherein the quality is modulated by replacing at least one codon of the polynucleotide with a synonymous codon that has a higher or lower preference of usage by the mammal to confer the immune response than the codon it replaces.
Owner:JINGANG MEDICINE AUSTRALIA PTY LTD
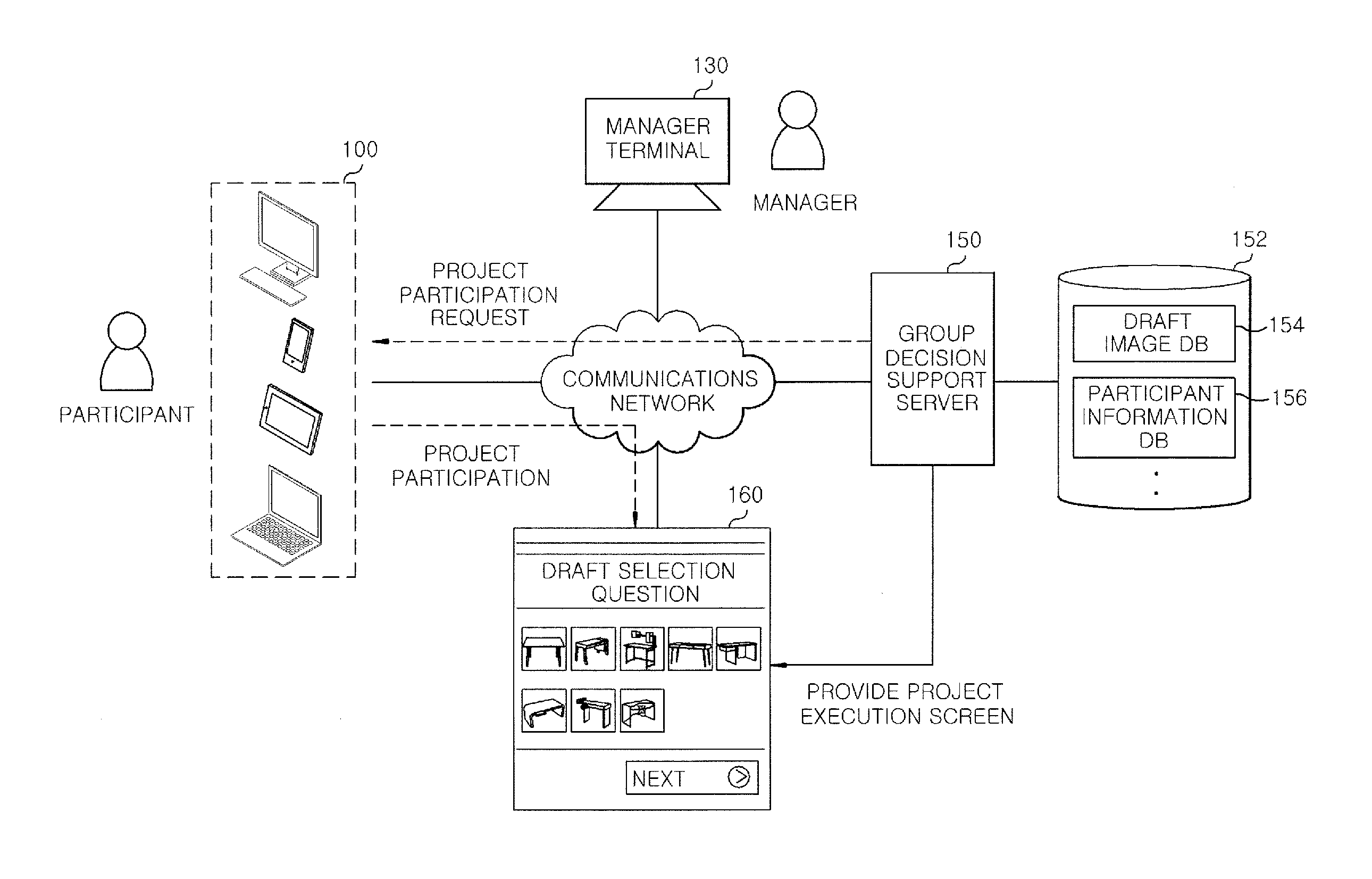
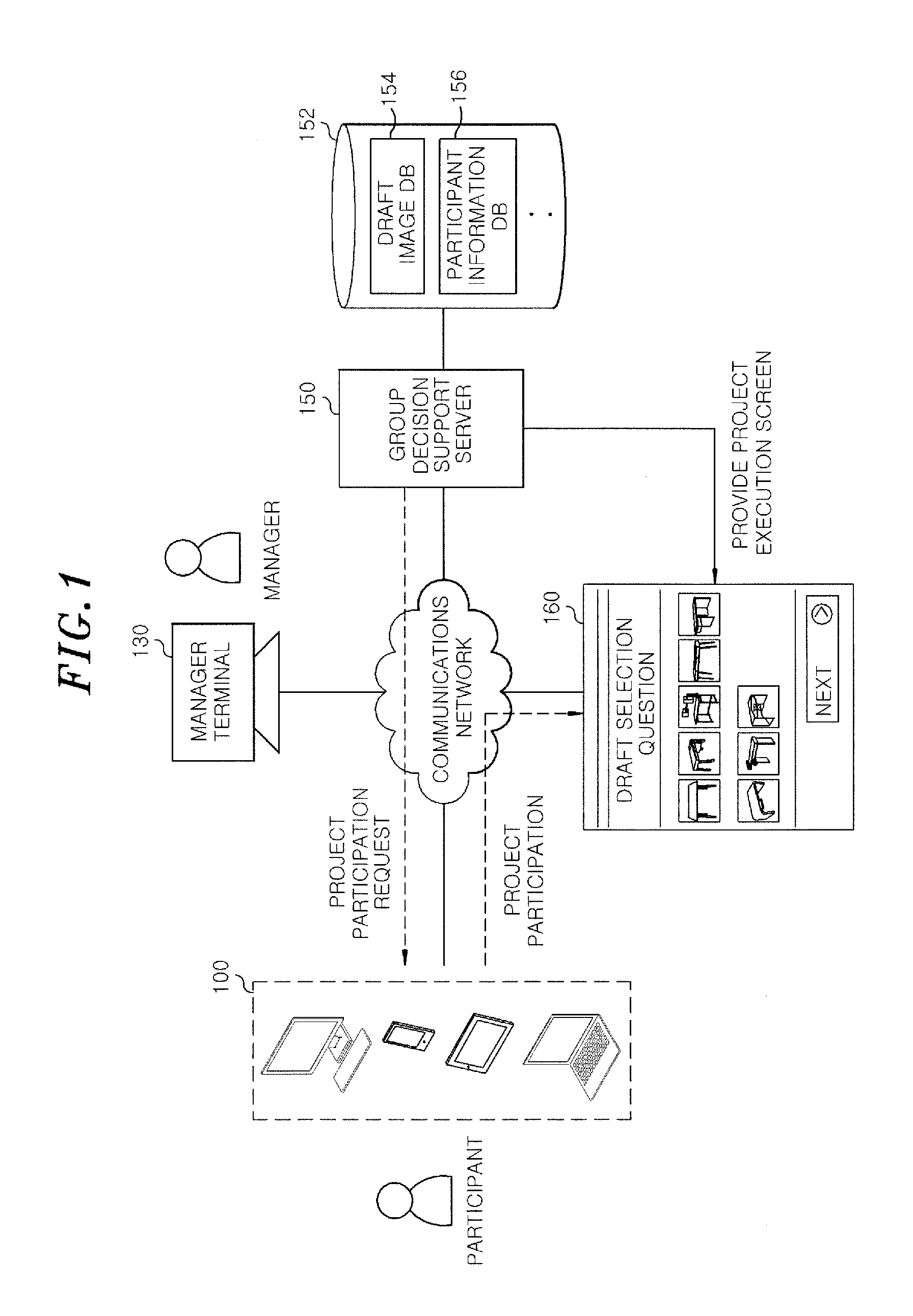
Server for providing information for group decision and method thereof
InactiveUS20160335597A1Reliable assessmentLower preferenceDigital data information retrievalOffice automationDecision takingComputer science
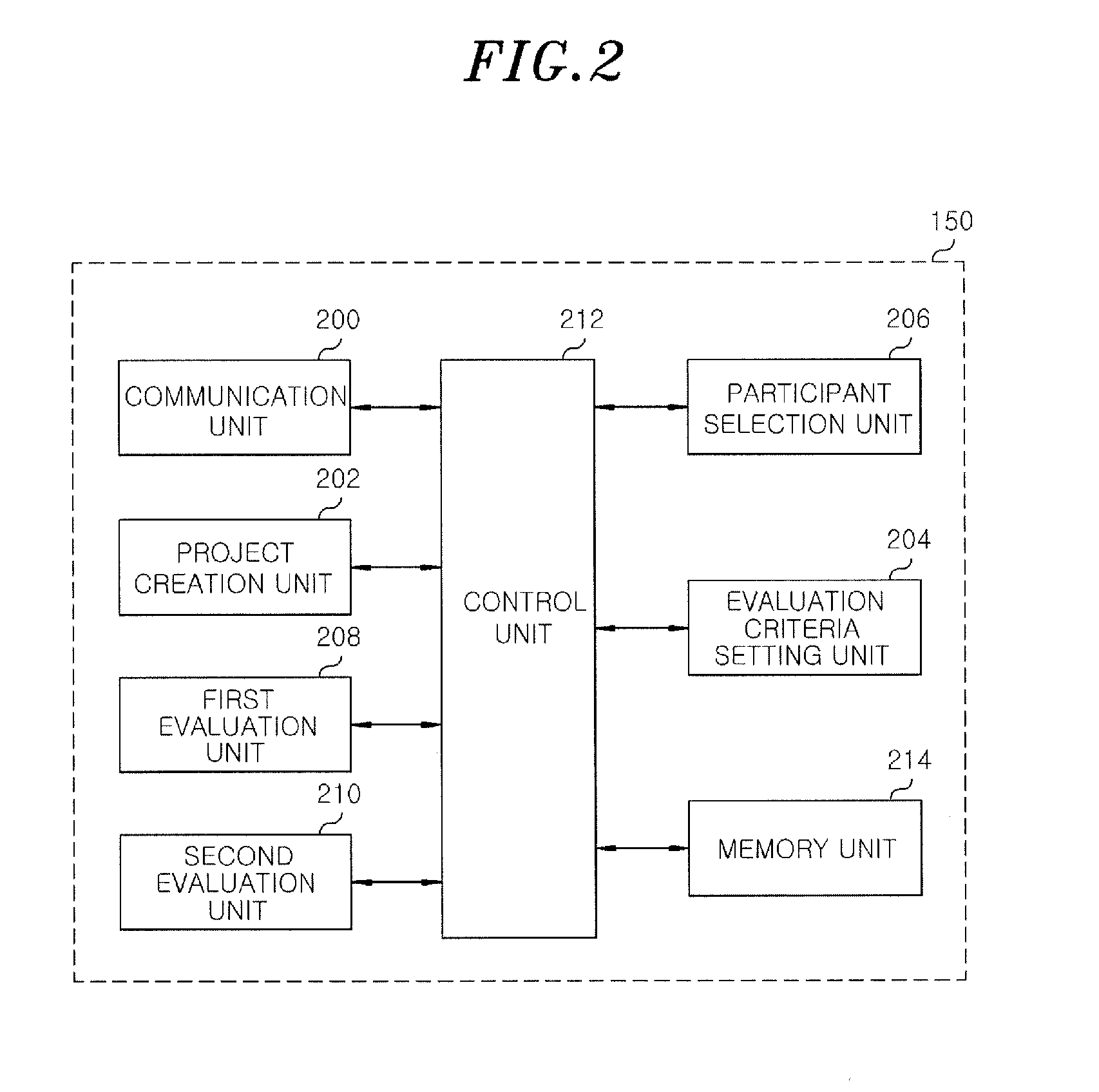
A group decision support server includes a project creation unit configured to register a first group of drafts including at least one or more draft and create a project for evaluating the drafts; a first evaluation configured to yield a first evaluation result based on evaluation information of a first preference base inputted from the participant when the participant participates in the project; a second evaluation unit configured to select a final draft that has the most favorable evaluation based on the first evaluation result and an evaluation information of a second weighted value base inputted from the participant.
Owner:PERCEPTION
Method and related equipment for sending signaling
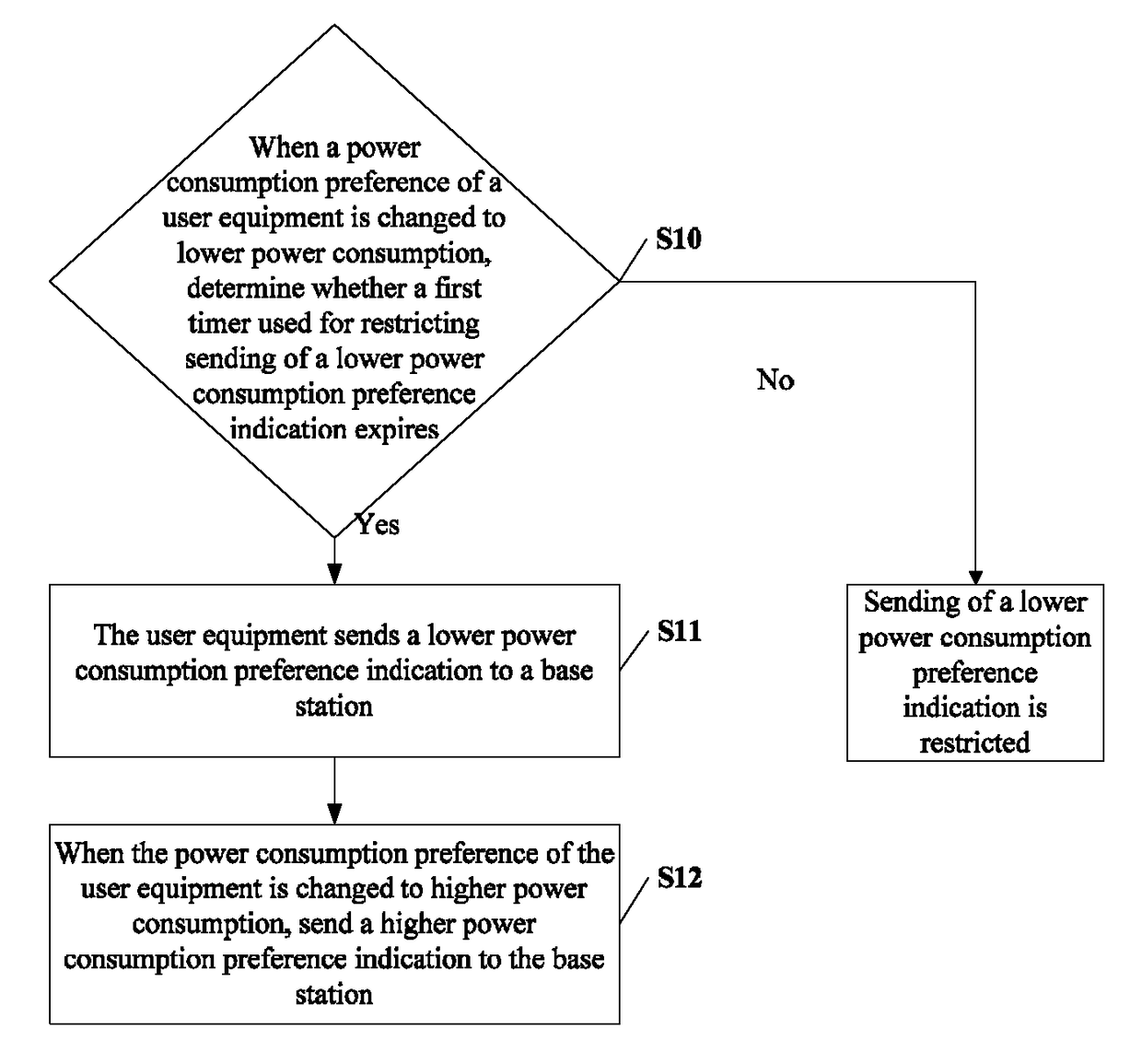
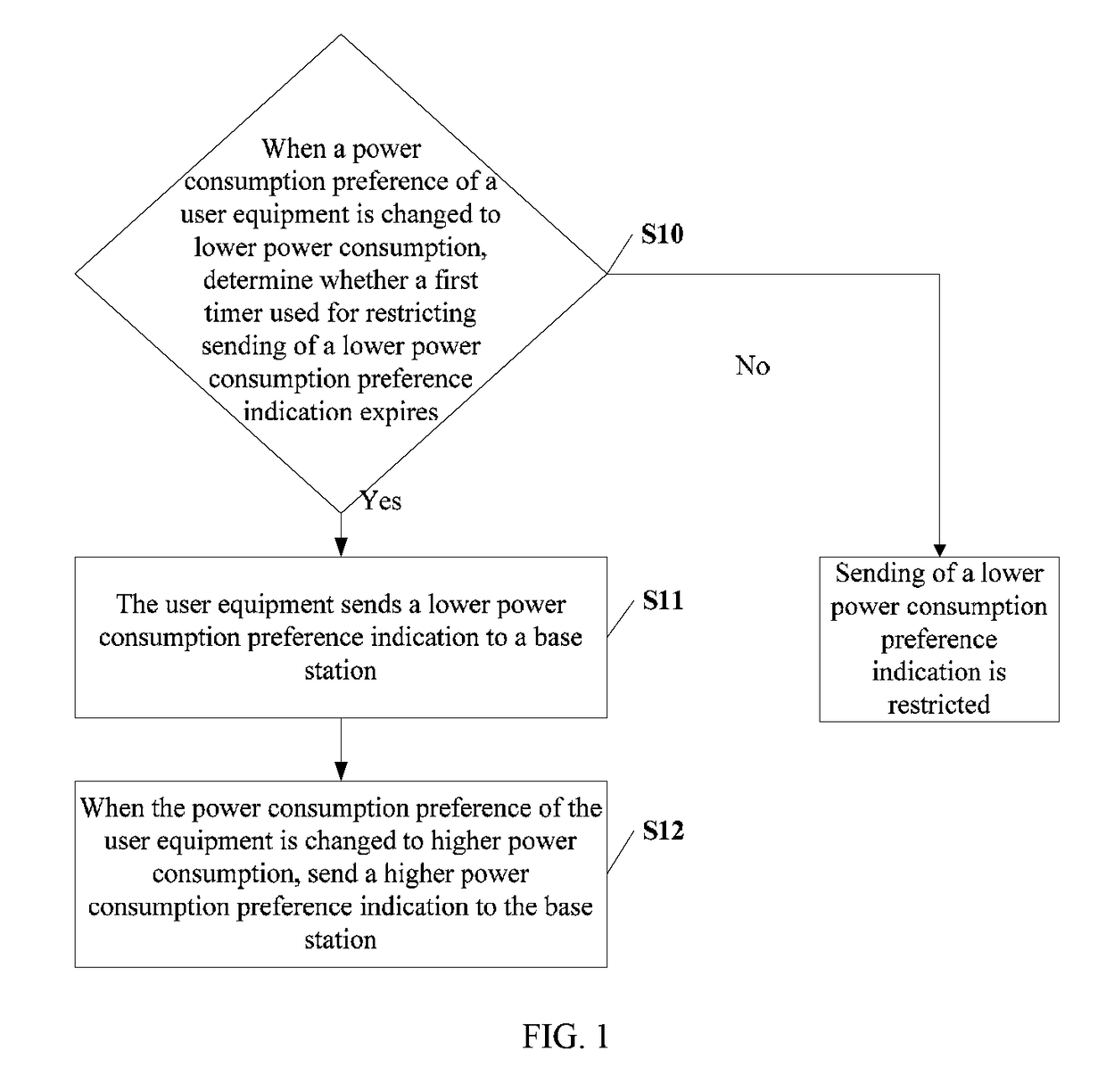
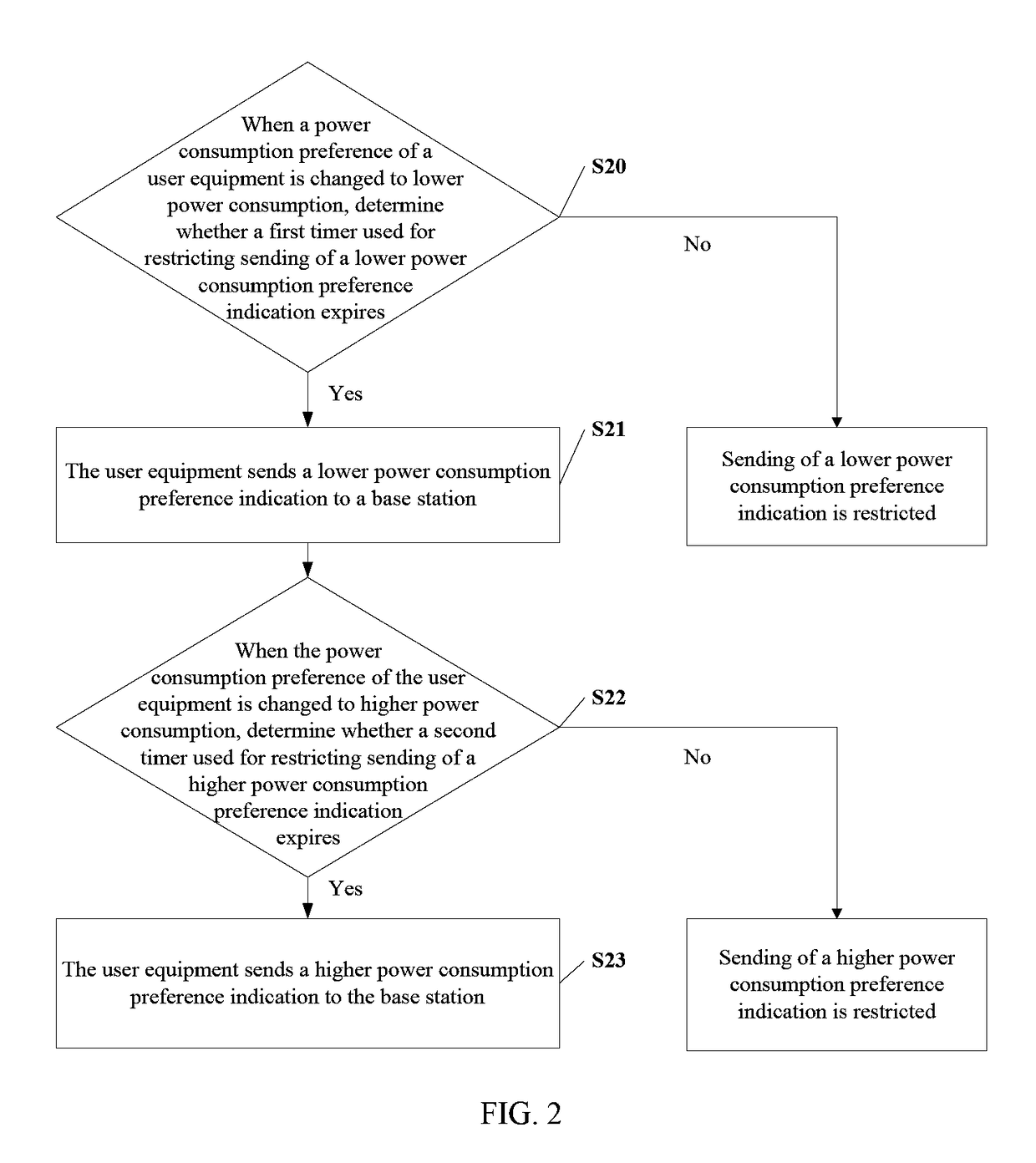
ActiveUS9681376B2Lower preferenceImprove preferenceEnergy efficient ICTPower managementEngineeringTimer
The present invention relates to the field of communications, and in particular, to a method and related equipment for sending signaling. The method includes: when a power consumption preference of a user equipment is changed to lower power consumption, determining whether a first timer used for restricting sending of a lower power consumption preference indication expires, and if a determination result is yes, sending a lower power consumption preference indication to a base station; and when the power consumption preference of the user equipment is changed to higher power consumption, sending a higher power consumption preference indication to the base station, or determining whether a second timer used for restricting sending of a higher power consumption preference indication expires, and if a determination result is yes, sending a higher power consumption preference indication to the base station. The present invention may improve user experience.
Owner:HUAWEI TECH CO LTD
Method for accurately detecting T cell immune repertoire based on high-throughput sequencing and primer system thereof
ActiveCN113122618AEasy to handleRealize accurate quantificationMicrobiological testing/measurementSequence analysisCell immunityT cell
The invention provides a method for accurately detecting a T cell immune repertoire based on high-throughput sequencing and a primer system thereof, the primer system comprises a PCR1 primer and a PCR2 primer, the PCR1 primer sequence and the PCR2 primer sequence comprise an upstream V primer and a downstream J primer, and a sequencing linker sequence and a molecular tag sequence are respectively inserted into target areas of the upstream primer and the downstream primer of the PCR1; the PCR2 primer takes a PCR1 product as a template, and a sequencing primer sequence and an index tag are introduced. The primer system and the method for accurately detecting a T cell immune repertoire based on high-throughput sequencing have the characteristics of simplicity and convenience in sample treatment, rapidness in experimental operation and accurate quantification of TCR cloning clusters.
Owner:武汉弘康医学检验实验室股份有限公司
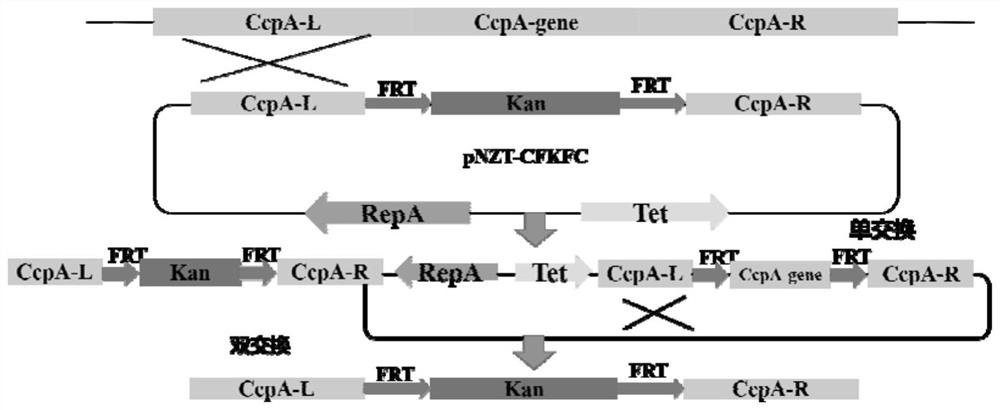
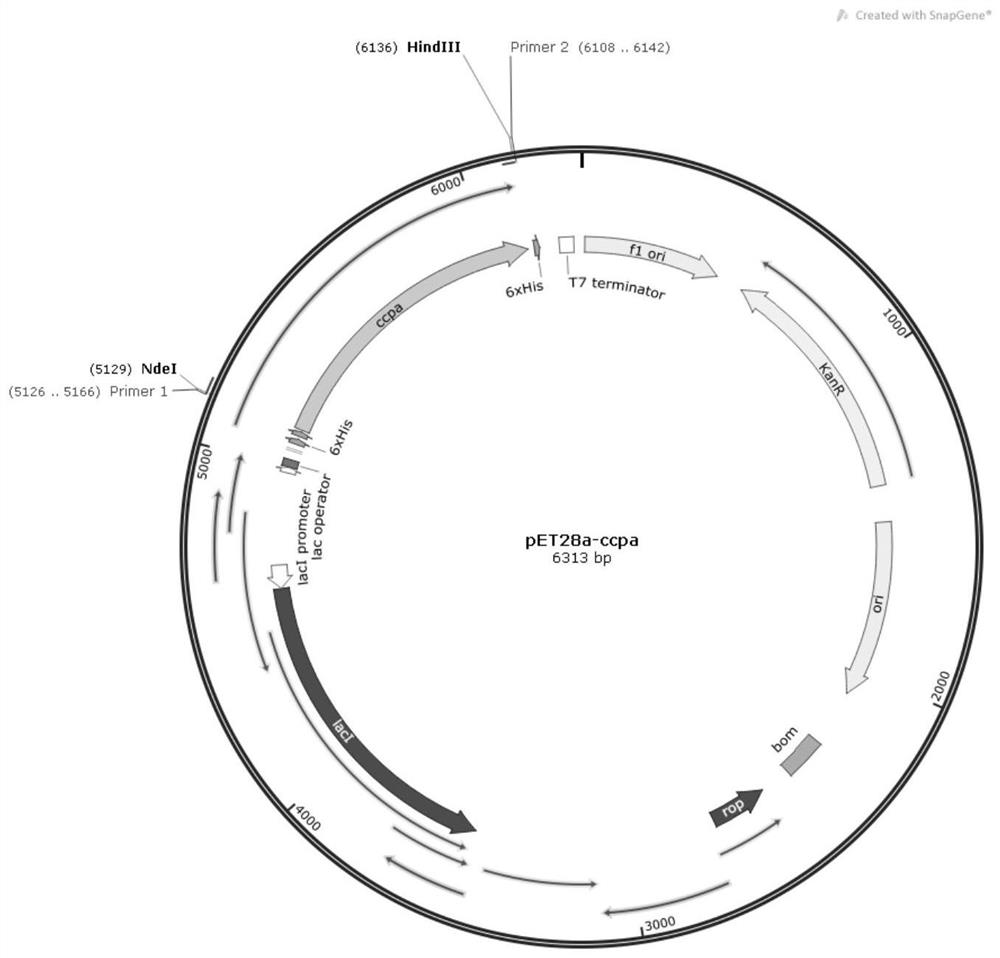
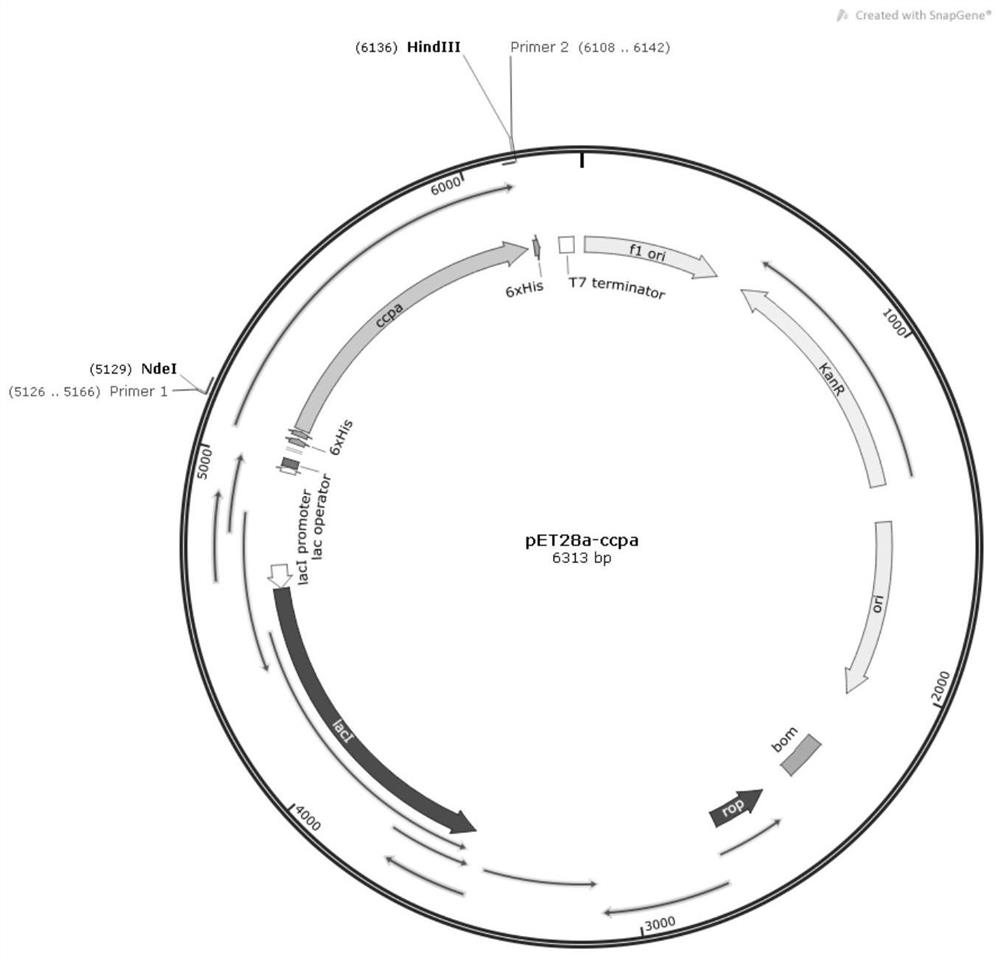
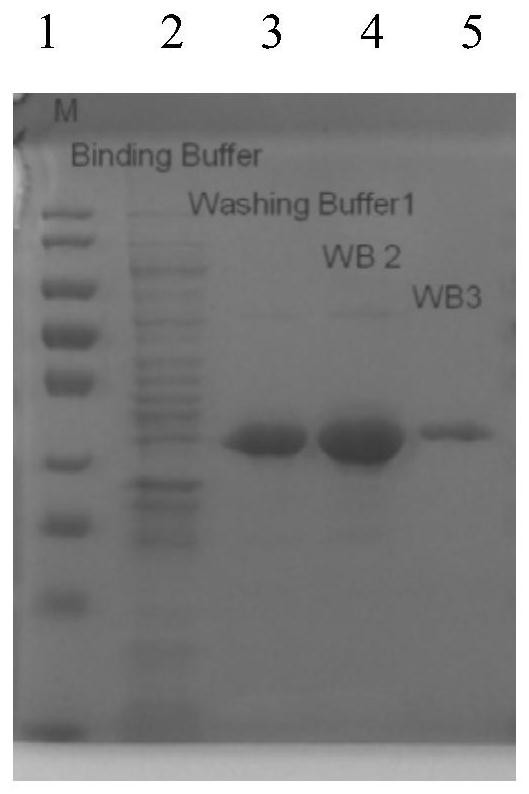
A carbon catabolism regulatory protein CCPA mutant K31A
ActiveCN112062821BAltered catabolic repressionLower preferenceBacteriaMicroorganism based processesBacillus licheniformisNucleotide
The invention discloses a carbon catabolism regulating protein CcpA mutant K31A, which belongs to the technical field of protein engineering. The present invention mutates the CcpA gene cloned from the Bacillus licheniformis genome to obtain a carbon catabolism regulatory protein CcpA mutant K31A, whose amino acid sequence is shown in SEQ ID NO.2, and the gene nucleotide sequence encoding it is shown in Shown in SEQ ID NO.1. The present invention constructs a recombinant expression vector containing the above-mentioned CcpA mutant K31A gene, and transforms the constructed expression vector into a CcpA gene-deficient strain of Bacillus licheniformis to obtain the recombinant Bacillus licheniformis. The obtained recombinant bacillus licheniformis can obviously change the phenomenon of carbon catabolism repression caused by the presence of glucose, and can reduce the preference of the bacillus licheniformis to xylose.
Owner:JIANGNAN UNIV
Mutant Tn5 transposase and kit
ActiveCN114807084ALower preferenceImprove uniformityMicrobiological testing/measurementTransferasesA-DNAWild type enzyme
The invention provides a mutant Tn5 transposase and a kit, and belongs to the technical field of biology. On the basis of a wild type Tn5 transposase, the wild type Tn5 transposase has five amino acid site mutations: R26W, E54K, R62P, D156K and L372P, and at least two of three mutation sites: S19R, N204K and S267T, and the amino acid sequence of the wild type Tn5 transposase is as shown in SEQ ID NO: 1. The invention also relates to an application of the mutant Tn5 transposase in library construction. Compared with a wild type Tn5 enzyme, the mutant Tn5 transposase provided by the invention has the advantages that the activity is obviously improved, the preference on a DNA sequence is obviously reduced, dsDNA and DNA / RNA hybrid chains can be recognized without preference, any (0.1-50ng) input amount can be used for building a library within about 2.5 hours, and the requirements of various high-throughput sequencing library building can be met.
Owner:YEASEN BIOTECHNOLOGY (SHANGHAI) CO LTD
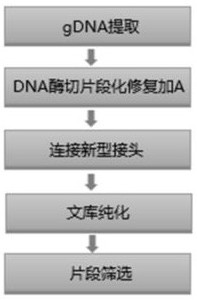
Method and kit for constructing whole genome high-throughput sequencing library
ActiveCN112301432ASimple and fast operationShort timeMicrobiological testing/measurementLibrary creationEnzyme digestionGenome
The invention discloses a method for constructing a whole genome high-throughput sequencing library. The method comprises the following steps: 1) extracting a sample gDNA; 2) carrying out enzyme digestion fragmentation on the sample gDNA, filling in the tail end, and adding A to obtain gDNA added with A; 3) connecting the gDNA added with A with a linker combination to obtain a connection product,the linker combination comprising two parts: a Y-shaped reverse linker and a high GC splint linker; 4) purifying the connection product to obtain a purified product, and 5) performing fragment screening on the purified product to obtain the sequencing library. The invention also discloses a kit for constructing the whole genome high-throughput sequencing library.
Owner:BERRYGENOMICS CO LTD
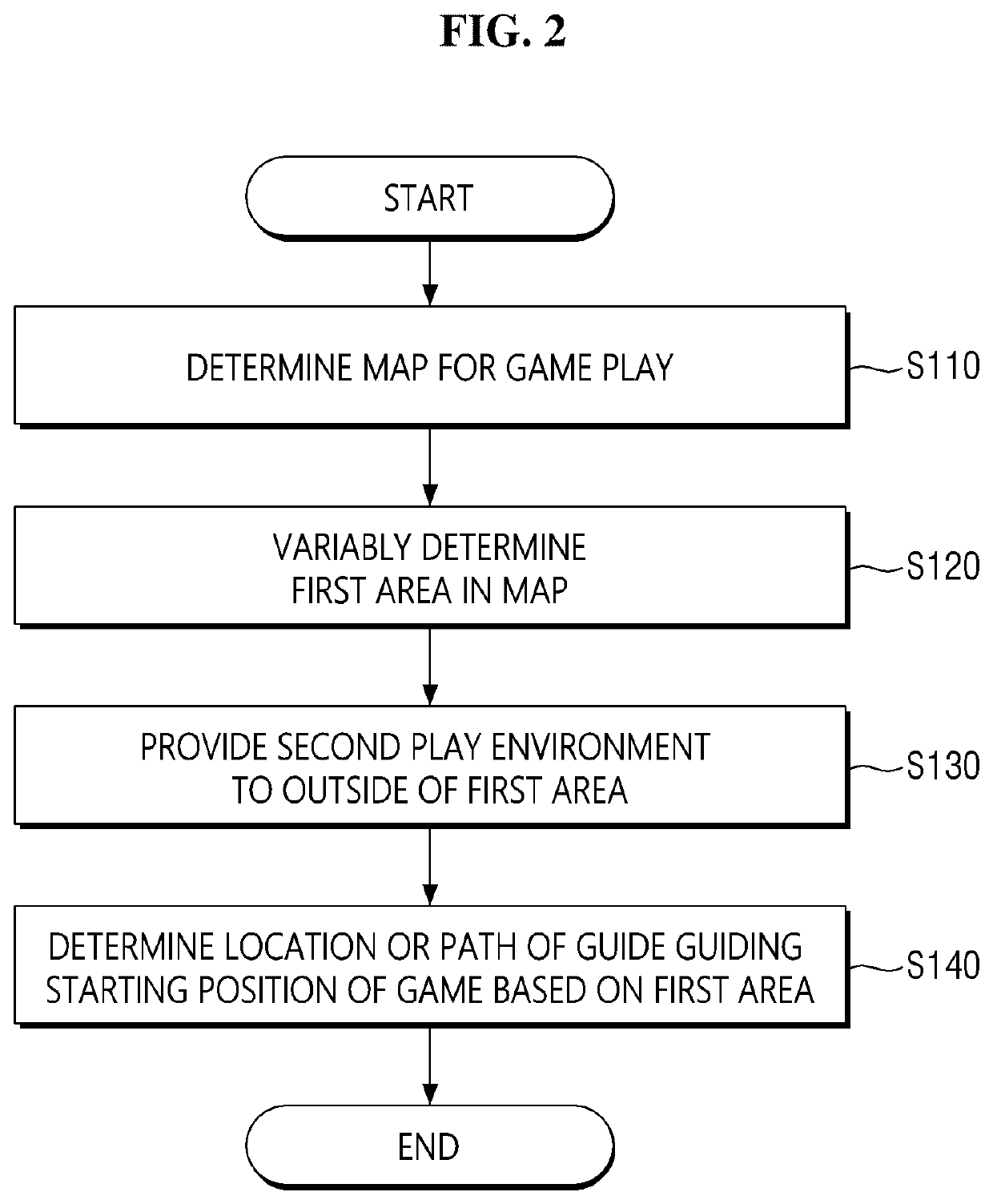
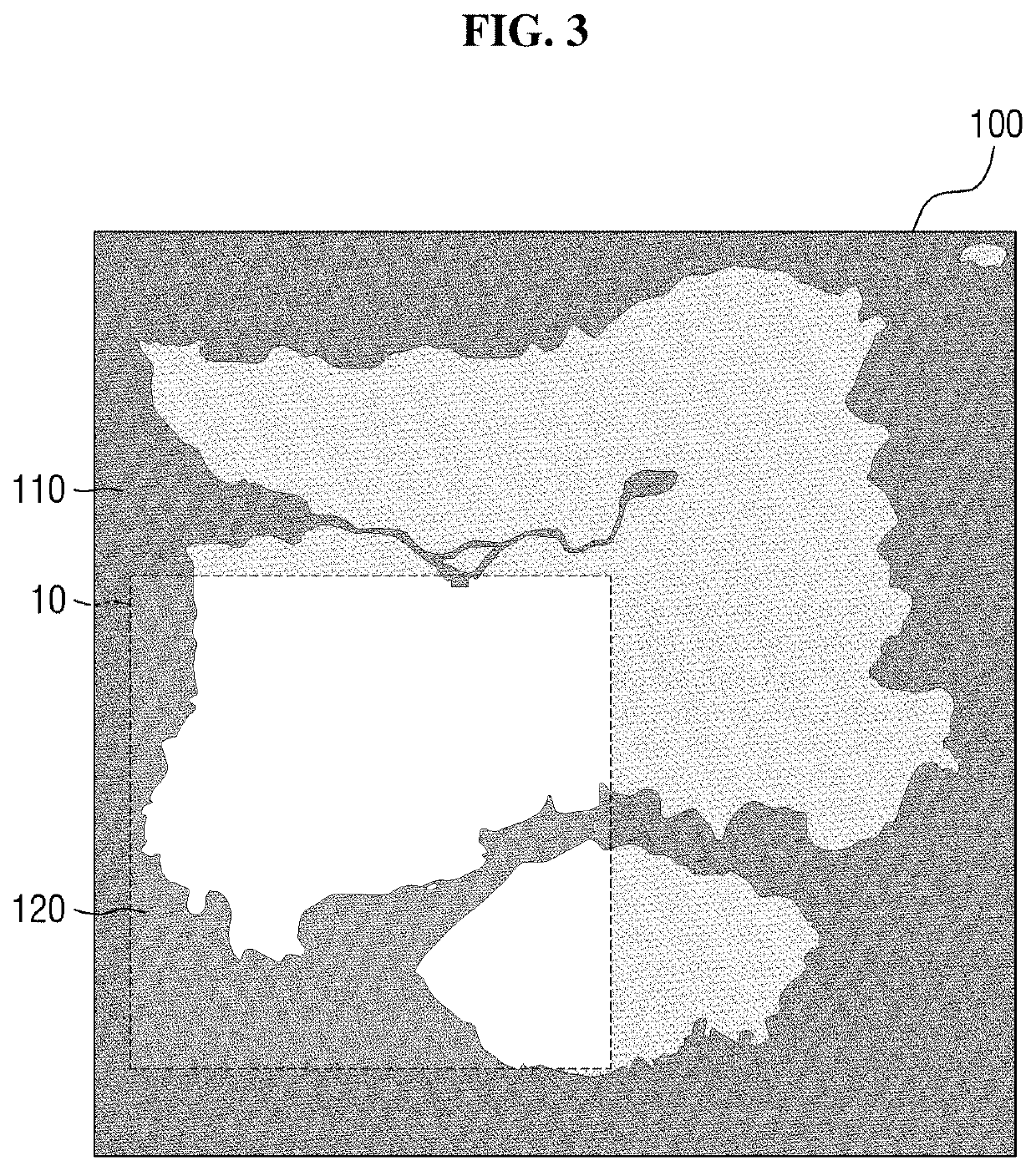
Method and apparatus for providing a game with enhanced unexpectedness
A method and apparatus for providing a game are provided. According to embodiments of the present, the method for providing a game comprises determining a map for game play, variably determining a first area in the map, and providing a second play environment different from a first play environment of the first area to outside the first area, wherein an item or vehicle is not spawned or a spawn rate of the item or vehicle is lower than the first play environment in the second play environment. According to the method, an In-Game play environment is variably provided each time a game is played, thereby the unexpectedness of the game play can be enhanced and the utilization and lifespan of the game map can be remarkably improved.
Owner:KRAFTON INC
Carbon catabolism regulation protein CcpA mutant I42A
ActiveCN112062822AAltered catabolic repressionLower preferenceBacteriaMicroorganism based processesBacillus licheniformisNucleotide
The invention discloses a carbon catabolism regulation protein CcpA mutant I42A, and belongs to the technical field of protein engineering. A CcpA gene cloned from a bacillus licheniformis genome is mutated to obtain a carbon catabolism regulation protein CcpA mutant I42A, wherein the amino acid sequence of the mutant I42A is as shown in SEQ ID NO.2, and the nucleotide sequence of the gene for encoding the mutant I42A is as shown in SEQ ID NO.1. According to the invention, a recombinant expression vector containing the CcpA mutant I42A gene is constructed, and the constructed expression vectoris transformed into a bacillus licheniformis CcpA gene defective strain to obtain recombinant bacillus licheniformis. The obtained recombinant bacillus licheniformis can obviously change the carbon catabolism repression phenomenon caused by the existence of glucose, and can reduce the preference of bacillus licheniformis to xylose.
Owner:JIANGNAN UNIV
A carbon catabolism regulatory protein ccpa mutant i42a
ActiveCN112062822BAltered catabolic repressionLower preferenceBacteriaMicroorganism based processesBacillus licheniformisNucleotide
Owner:JIANGNAN UNIV
Method for constructing a DNA library and application thereof
ActiveCN111073952AImprove efficiencyLower preferenceMicrobiological testing/measurementLibrary creationEngineeringA-DNA
The invention discloses a method for constructing a DNA library and application thereof. The method comprises the following steps: carrying out terminal repairing on a DNA sample, and carrying out adenine nucleotide tailing treatment on the DNA sample by using high-temperature-resistant polymerase so as to obtain a tailed DNA under the condition of performing the terminal repairing and the adeninenucleotide tailing treatment continuously; and performing linker ligation treatment on the tailed DNA so as to obtain a linking product. In the DNA library, the conditions of terminal repairing and adenine nucleotide tailing treatment are as follows: the temperature is 27-37 DEG C and the time is 5-30 minutes; and the temperature is 70-75 DEG C, and the time is 10-20 minutes. According to the method, tail end repairing and adenine nucleotide tail adding treatment are continuously carried out, purification treatment is not needed in the process, stability is good; meanwhile, reaction time is remarkably shortened, and the reaction process is remarkably simplified.
Owner:ZHEJIANG ANNOROAD BIO TECH CO LTD +1
Carbon catabolism regulatory protein CcpA mutant K31A
ActiveCN112062821AAltered catabolic repressionLower preferenceBacteriaMicroorganism based processesBacillus licheniformisNucleotide
The invention discloses a carbon catabolism regulatory protein CcpA mutant K31A, and belongs to the technical field of protein engineering. A CcpA gene cloned from a bacillus licheniformis genome is mutated to obtain the carbon catabolism regulatory protein CcpA mutant K31A, the amino acid sequence of the carbon catabolism regulatory protein CcpA mutant K31A is shown as SEQ ID NO.2, and the nucleotide sequence of the gene encoding the carbon catabolism regulatory protein CcpA mutant K31A is shown as SEQ ID NO. 1. A recombinant expression vector containing the gene encoding the CcpA mutant K31Ais constructed, and the constructed expression vector is transformed into a bacillus licheniformis CcpA gene defect strain to obtain recombinant bacillus licheniformis. The obtained recombinant bacillus licheniformis can obviously change the carbon catabolism repression phenomenon caused by the existence of glucose, and the preference of the bacillus licheniformis to xylose can be reduced.
Owner:JIANGNAN UNIV
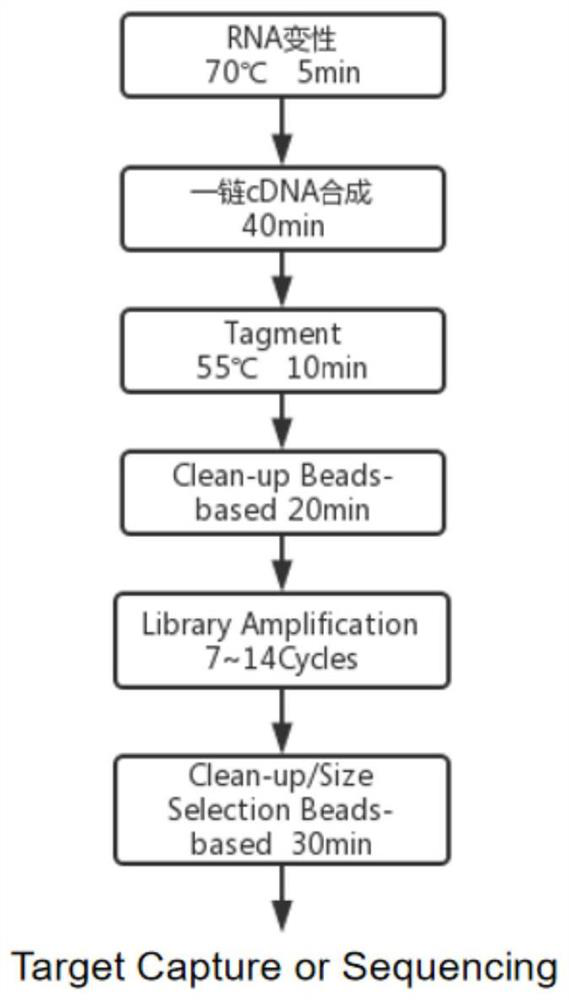
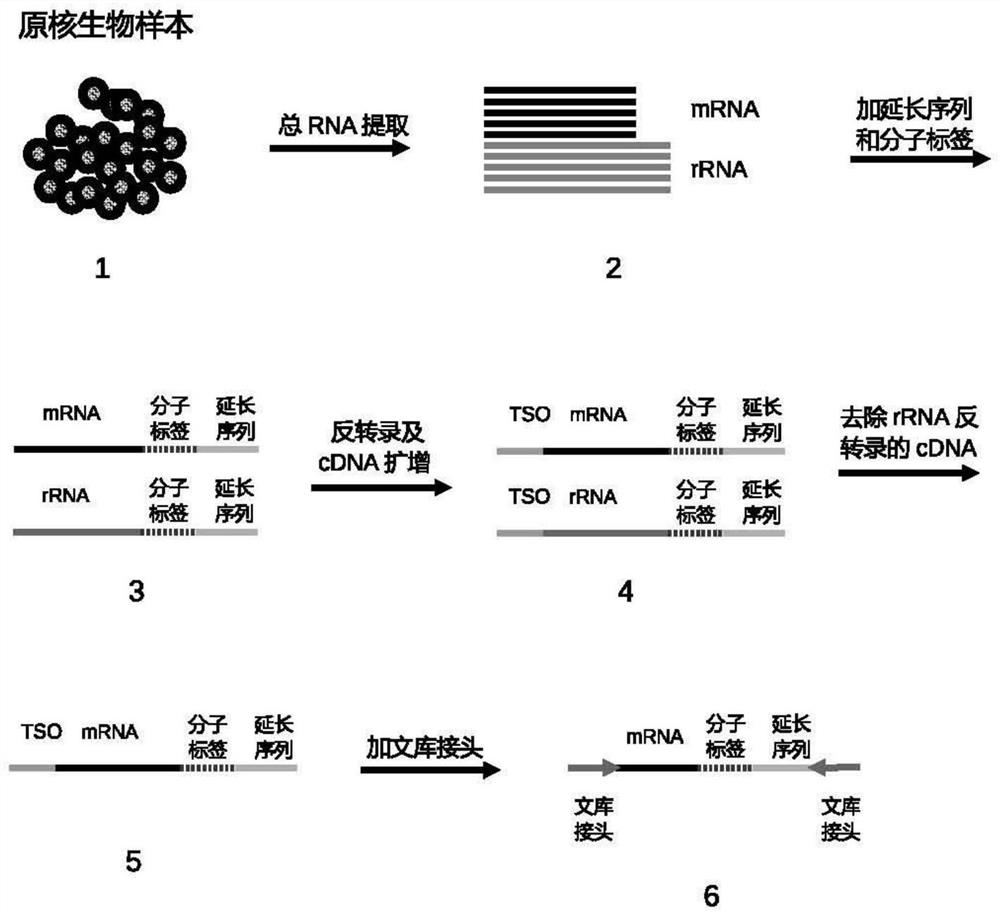
Method for constructing prokaryote RNA sequencing library
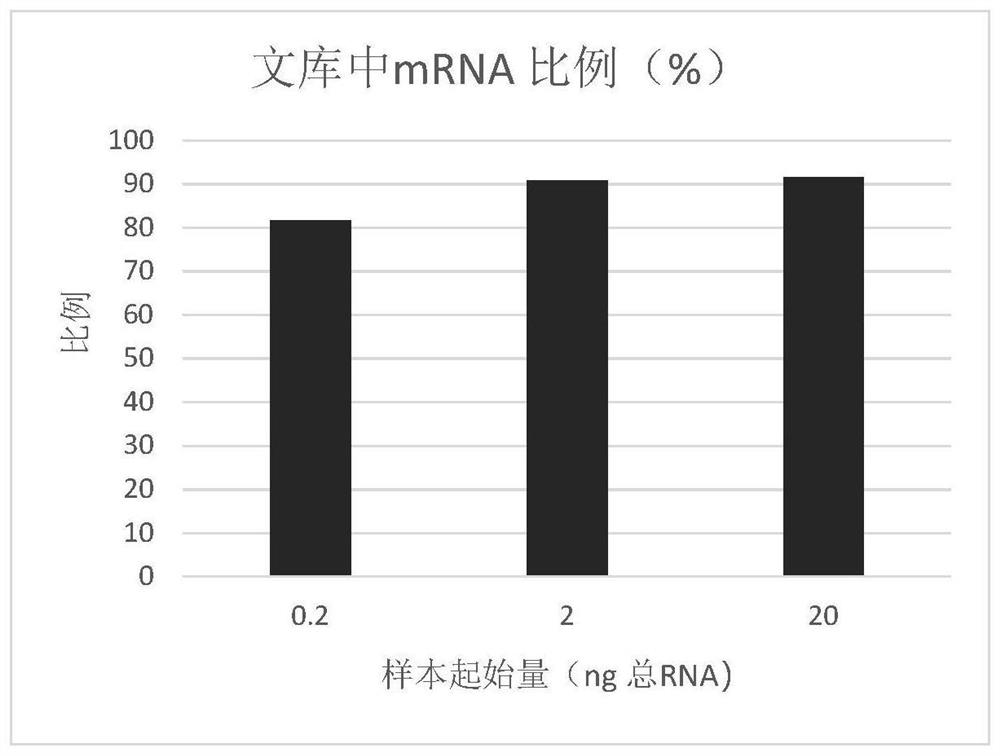
PendingCN114657646ALower preferenceRealize preferenceNucleotide librariesMicrobiological testing/measurementProkaryoteLibrary
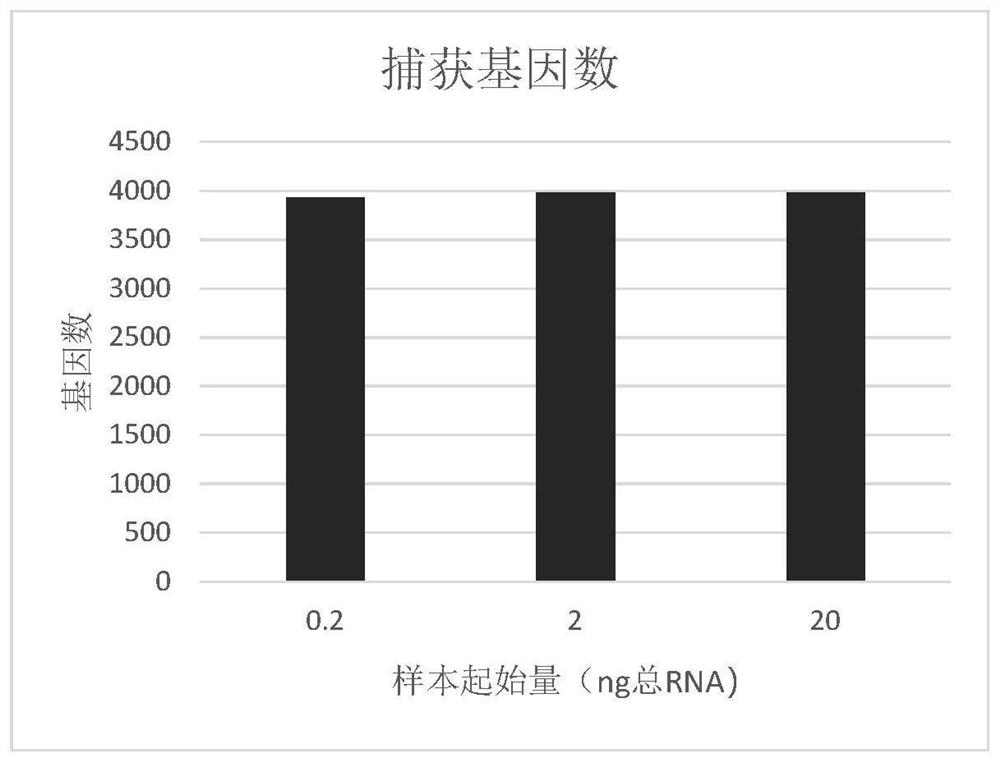
The invention provides a method for constructing a prokaryote RNA sequencing library. According to the method, an extension sequence can be added to prokaryote mRNA, and prokaryote RNA can be efficiently called and reversely transcribed by using the extension sequence; meanwhile, according to the method, a molecular tag can be marked for each prokaryote original mRNA molecule, so that the influence of PCR preference on the detection accuracy is avoided, and a prokaryote mRNA sequencing library with low preference and high accuracy is obtained. Compared with a traditional prokaryote RNA sequencing library construction method, the method has the advantages that the constructed library is low in preference, meanwhile, the method is easy and convenient to operate, the success rate is high, and the method is also suitable for detection of samples with extremely low initial quantity (smaller than 2ng total RNA).
Owner:BEIJING STOMATOLOGY HOSPITAL CAPITAL MEDICAL UNIV
Method for constructing prokaryote single cell RNA sequencing library
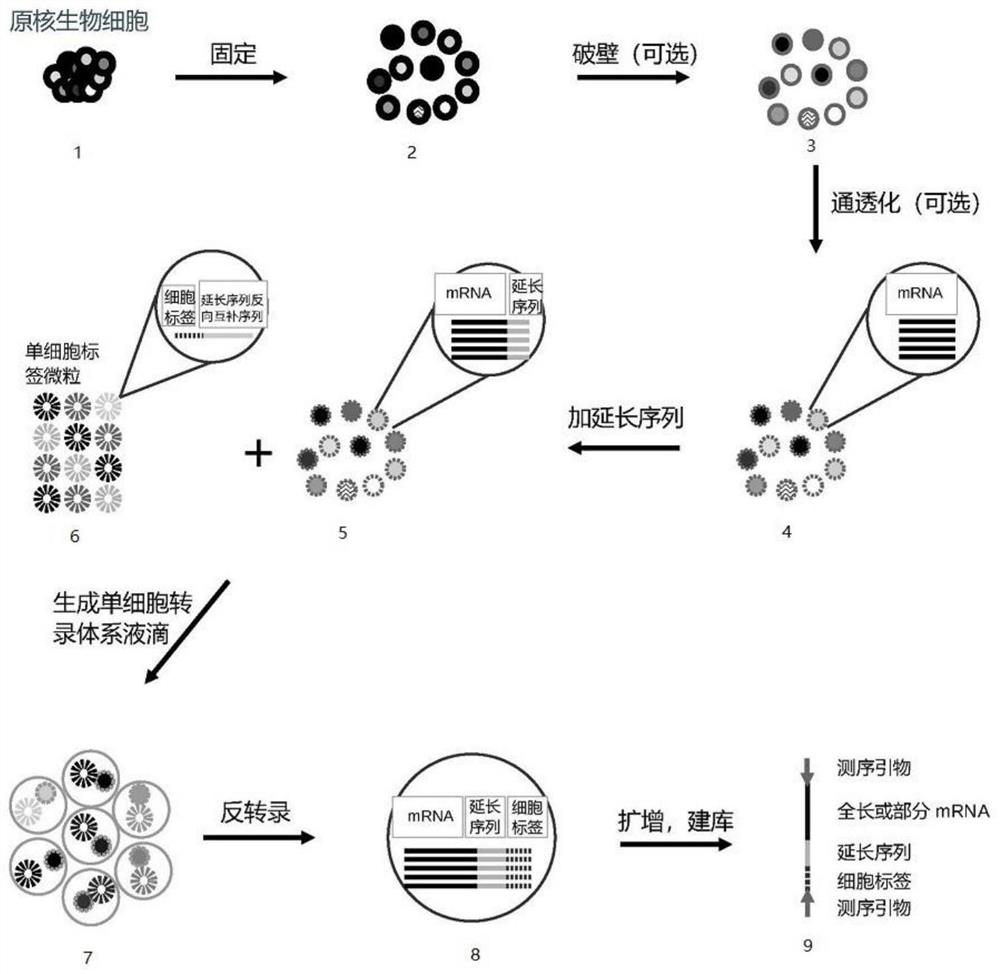
PendingCN114807305ARealize identificationLower preferenceMicrobiological testing/measurementMicroorganism librariesMRNA SequencingProkaryote organisms
The invention provides a method for constructing a prokaryote single-cell RNA sequencing library. According to the method, an extension sequence can be added to mRNA under the condition that prokaryote cells are not cracked, prokaryote mRNA can be efficiently called by utilizing the extension sequence, different cell tags are marked for mRNA from different cell sources, and a low-preference prokaryote single cell mRNA sequencing library is obtained.
Owner:BEIJING STOMATOLOGY HOSPITAL CAPITAL MEDICAL UNIV
A method and kit for constructing a library for whole-genome high-throughput sequencing
ActiveCN112301432BReduce crosstalkReduce redundancyMicrobiological testing/measurementLibrary creationHigh fluxGenetics
The invention discloses a method for constructing a whole-genome high-throughput sequencing library, comprising the following steps: 1) extracting sample gDNA; 2) enzymatically cleaving the sample gDNA into fragments, filling in the ends, and adding A to obtain the added A 3) Ligate the A-added gDNA with an adapter combination to obtain a ligation product, the adapter combination includes two parts: a Y-shaped reverse adapter and a high GC splint adapter; 4) Purify the ligation product, obtaining a purified product; and 5) performing fragment screening on the purified product to obtain a sequencing library. The invention also discloses a kit for constructing a whole genome high-throughput sequencing library.
Owner:BERRYGENOMICS CO LTD
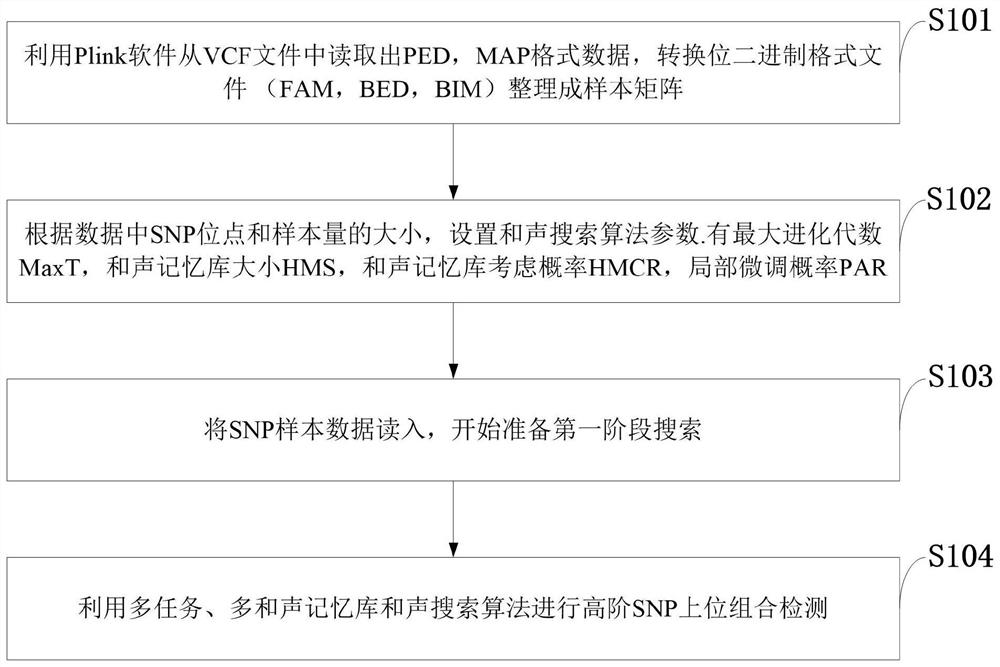
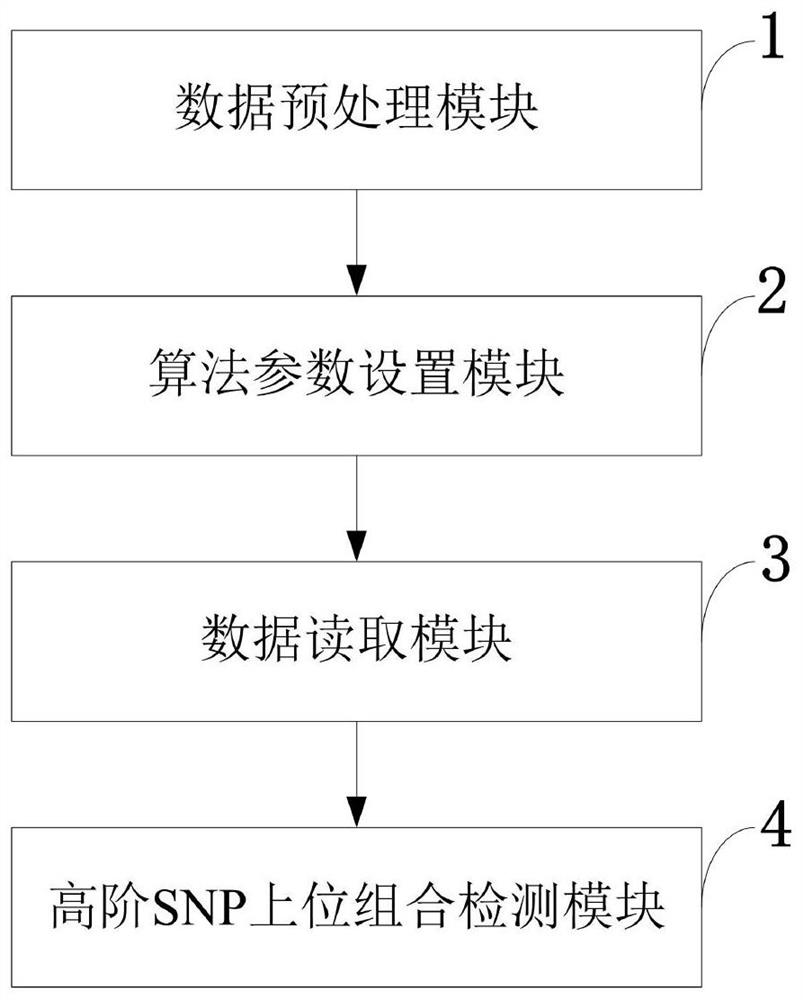
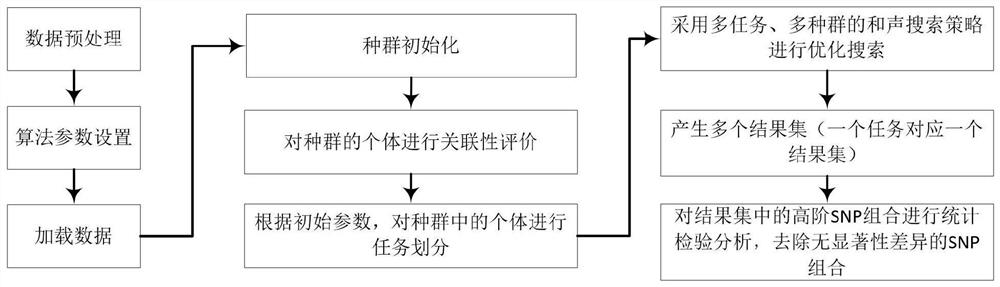
Multi-task high-order SNP upper detection method and system, storage medium and equipment
The invention belongs to the technical field of single nucleotide polymorphism upper detection, and discloses a multi-task high-order SNP upper detection method and a system, a storage medium and equipment. The multi-task high-order SNP upper detection method comprises the following steps: reading PED and MAP format data from a VCF file by using Plink software, converting a bit binary format file,and arranging the bit binary format file into a sample matrix; setting search algorithm parameters according to the sizes of the SNP sites and the sample size in the data; reading in SNP sample data,and preparing first-stage search; carrying out high-order SNP upper combination detection by utilizing a multi-task and multi-harmony memory bank harmony search (HS) algorithm. According to the multi-task harmony search detection method provided by the invention, a plurality of harmony memory banks are adopted, SNP combinations of different orders are respectively stored, and a multi-task technology is applied, so that high-order SNP upper detection of a plurality of different orders can be carried out at the same time, mutual learning among individuals in a population is promoted, the diversity of the population is enhanced, and the global search capability is further improved.
Owner:XIAN UNIV OF POSTS & TELECOMM
A kind of micro-degradation genomic DNA methylation library sequencing method and its kit
ActiveCN107858409BFragment length reductionAccurate detectionMicrobiological testing/measurementLibrary creationDNA methylationGenomic Segment
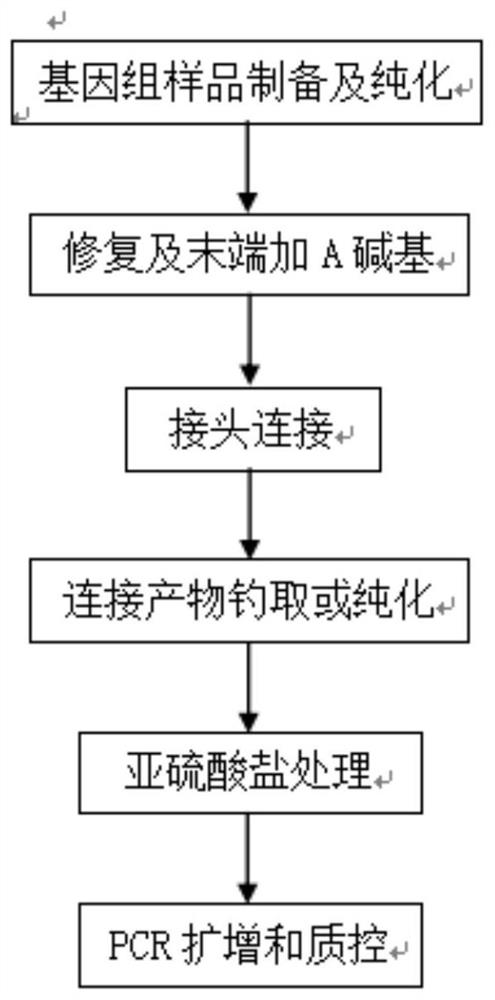
The invention discloses a method for building and sequencing a micro-degraded genomic DNA methylation library. The invention tests different library building technologies and optimizes the reaction process to establish a methylation library that can detect as little as 1ng of severely degraded genomic DNA. technology. By testing different enzyme combinations, the end repair of degraded DNA, the addition of A at the 3-end, and the repair of DNA double-strand gaps due to degradation were completed in the same reaction system; by optimizing the reaction system, the sequencing adapters were connected in the original reaction solution; After further testing the biotin labeling of the adapter, through the specific binding of biotin and streptomycin magnetic beads, the capture of the genome fragments of the ligated adapter before the subsequent sulfite treatment was realized, and the loss of the target fragment was reduced; in addition, the establishment of sulfite Salt reaction system and reaction time reduce DNA damage during processing, ensure methylation conversion rate, and realize methylation library construction and sequencing of micro-degraded samples.
Owner:SHENZHEN E GENE TECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com