Patents
Literature
127 results about "Host genome" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
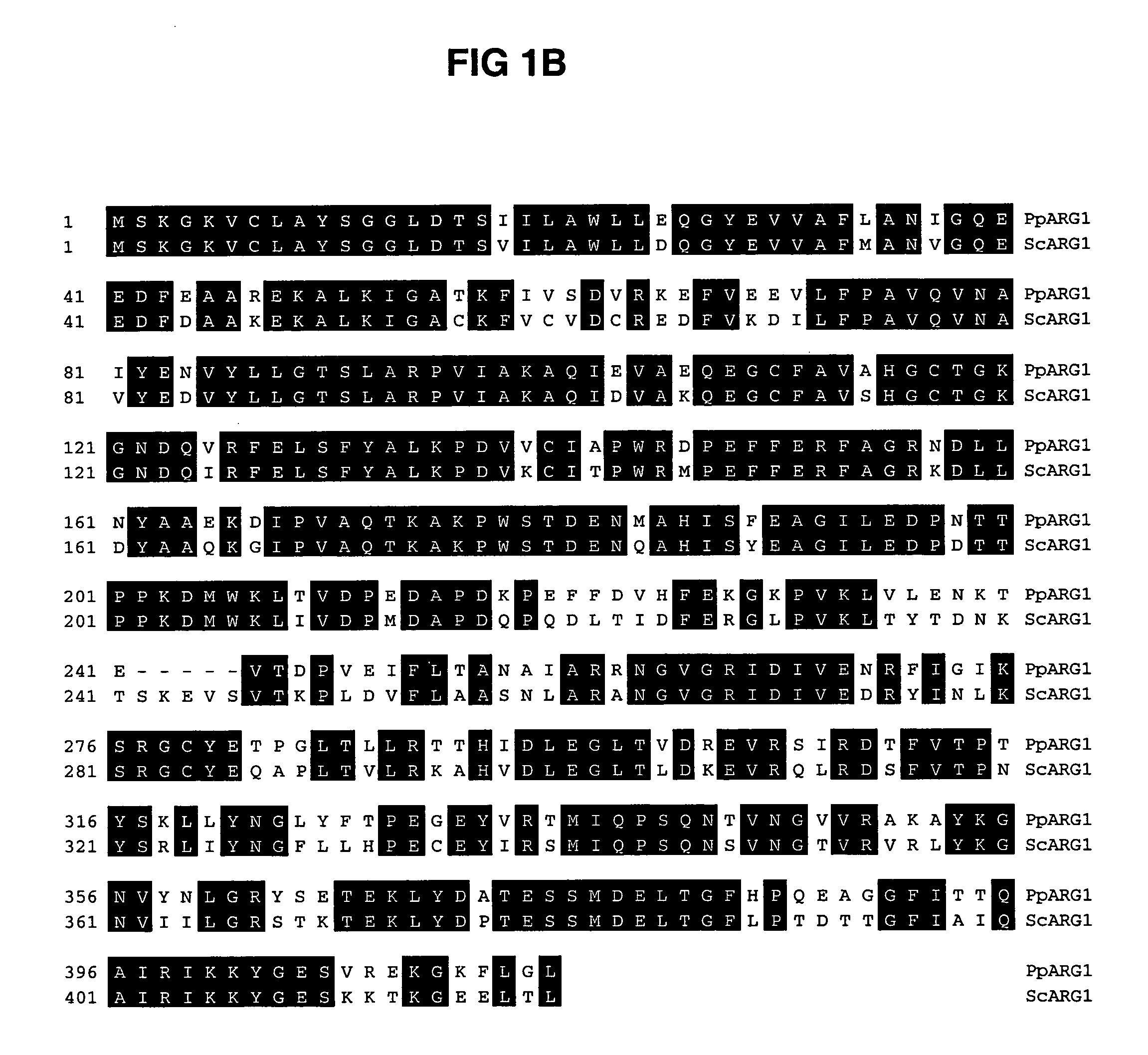
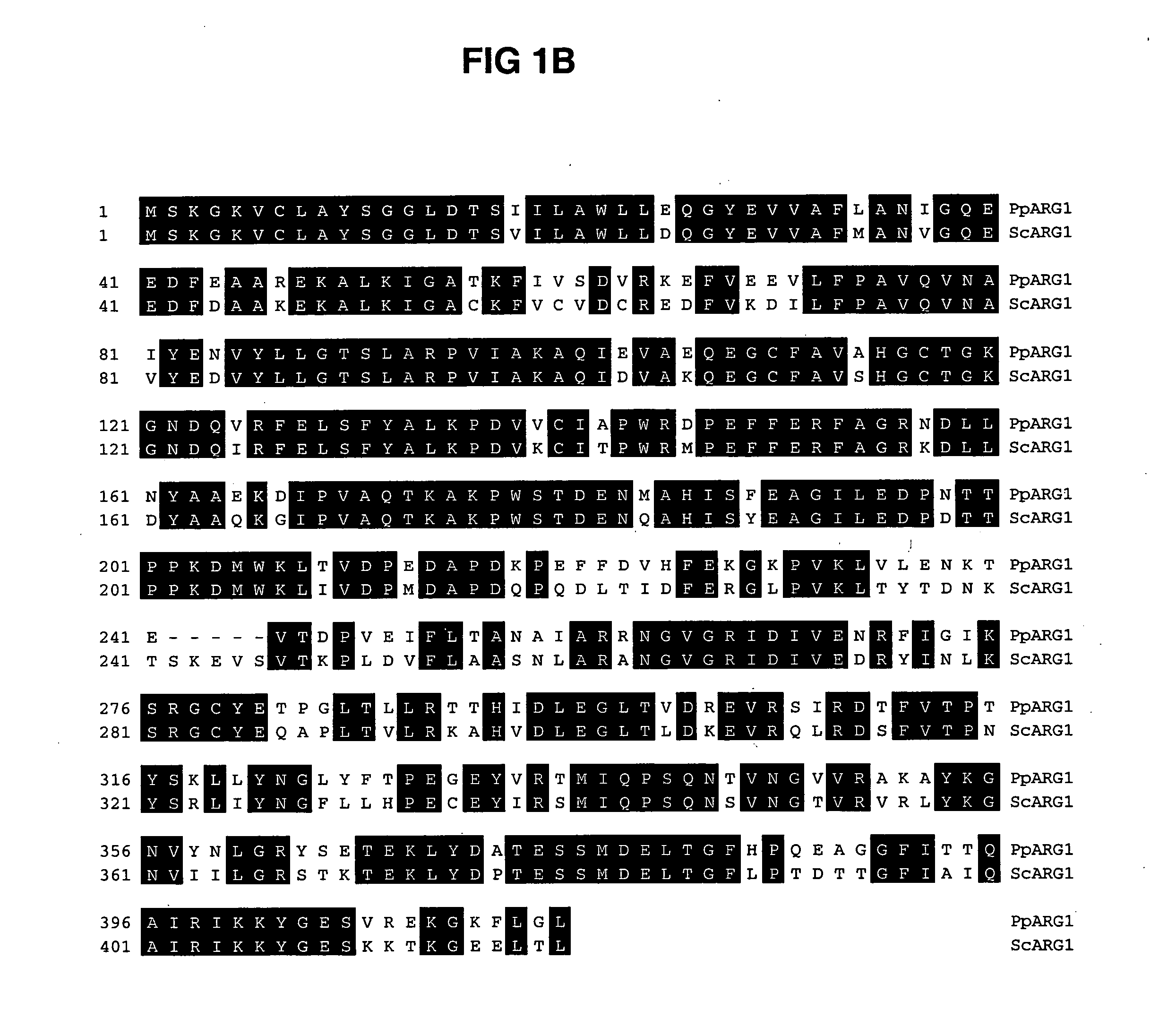
ARG1, ARG2, ARG3, HIS1, HIS2, HIS5, HIS6 genes and methods for stable genetic integration
Novel genes encoding P. pastoris ARG1, ARG2, ARG3, HIS1, HIS2, HIS5 and HIS6 are disclosed. A method for inactivating alternately at least two biosynthetic pathways in a methylotrophic yeast is provided. A method for producing and selecting yeast strains characterized as being capable of genetic integration of heterologous sequences into the host genome using the genes involved in the biosynthetic pathways is also disclosed.
Owner:GLYCOFI
ARG1, ARG2, ARG3, HIS1, HIS2, HIS5, HIS6 genes and methods for stable genetic integration
Novel genes encoding P. pastoris ARG1, ARG2, ARG3, HIS1, HIS2, HIS5 and HIS6 are disclosed. A method for inactivating alternately at least two biosynthetic pathways in a methylotrophic yeast is provided. A method for producing and selecting yeast strains characterized as being capable of genetic stable integration of heterologous sequences into the host genome using the genes involved in the biosynthetic pathways is also disclosed.
Owner:GLYCOFI
gRNA target sequences for endogenous overexpression of 1ncRNA-XIST and application thereof
ActiveCN107513531AImprove efficiencyFermentationVector-based foreign material introductionFetal growthCell migration
The invention discloses gRNA target sequences for endogenous overexpression of 1ncRNA-XIST, a CRISPR / dCas9 lentivirus system and application thereof. The gRNA target sequences are respectively as shown in SED ID No. 1, SED ID No. 2 and SED ID No. 3. The CRISPR / dCas9 lentivirus system comprises the gRNA target sequences for endogenous overexpression of 1ncRNA-XIST. A method for screening stable strains according to the characteristics that lentivirus must be integrated into a host genome is cooperated with CRISPR / dCas9 to realize the endogenous overexpression of the large fragment gene 1ncRNA-XIST, so that the defect of incapability of stably expressing the large fragment gene 1ncRNA-XIST in a traditional method is overcome, the efficient overexpression stable cell strain of the large fragment gene 1ncRNA-XIST can be obtained in a short time, or cells obtained by screening can stably express target genes, thereby obtaining stably silenced 1ncRNA-XIST downstream specific gene cell strain. The gRNA target sequences and the CRISPR / dCas9 lentivirus system have important guiding significance on the research of the function of 1ncRNA-XIST in the trophocyte migration and the effect of 1ncRNA-XIST in the proliferation disorder and fetal growth restriction process.
Owner:WUXI MATERNAL & CHILD HEALTH HOSPITAL
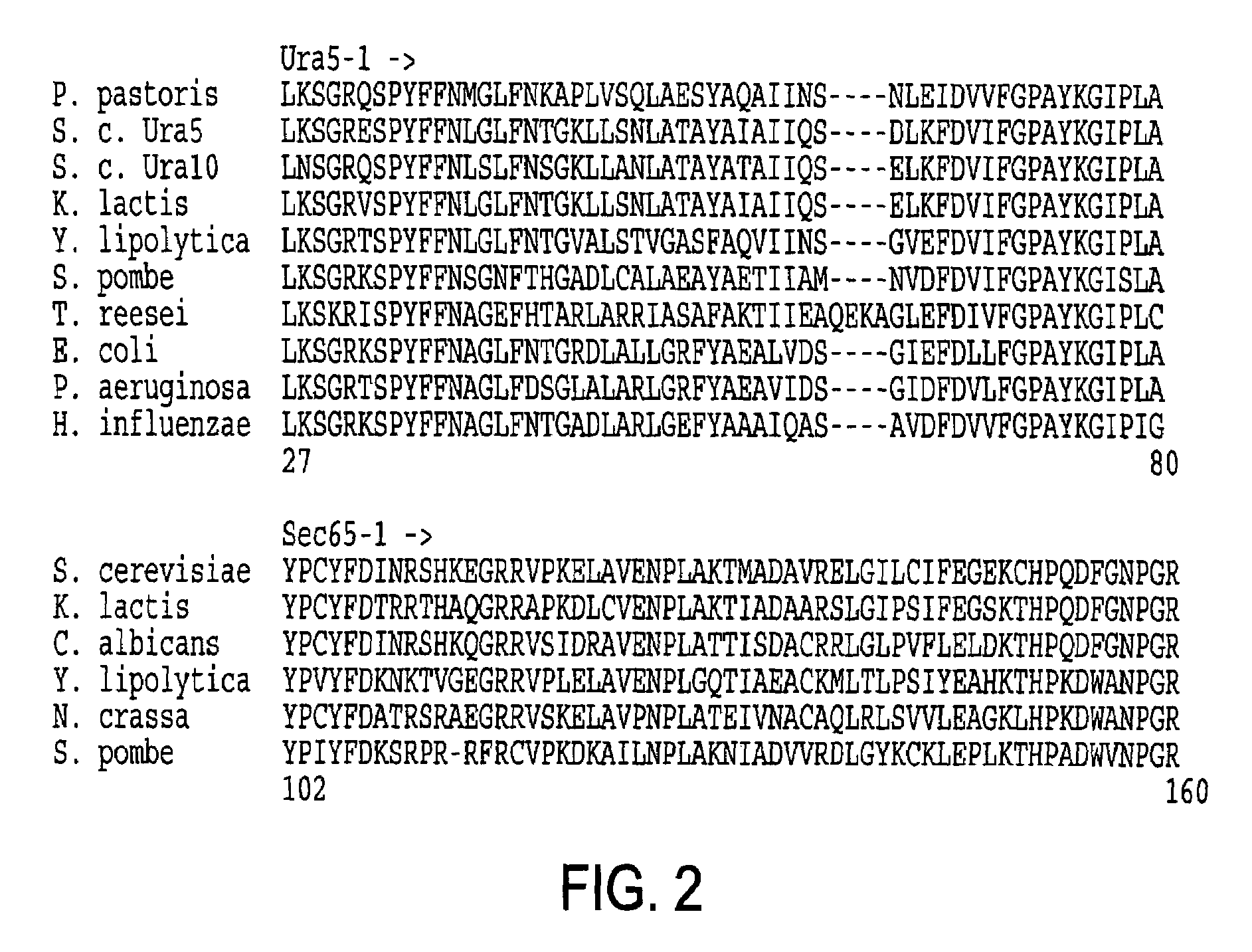
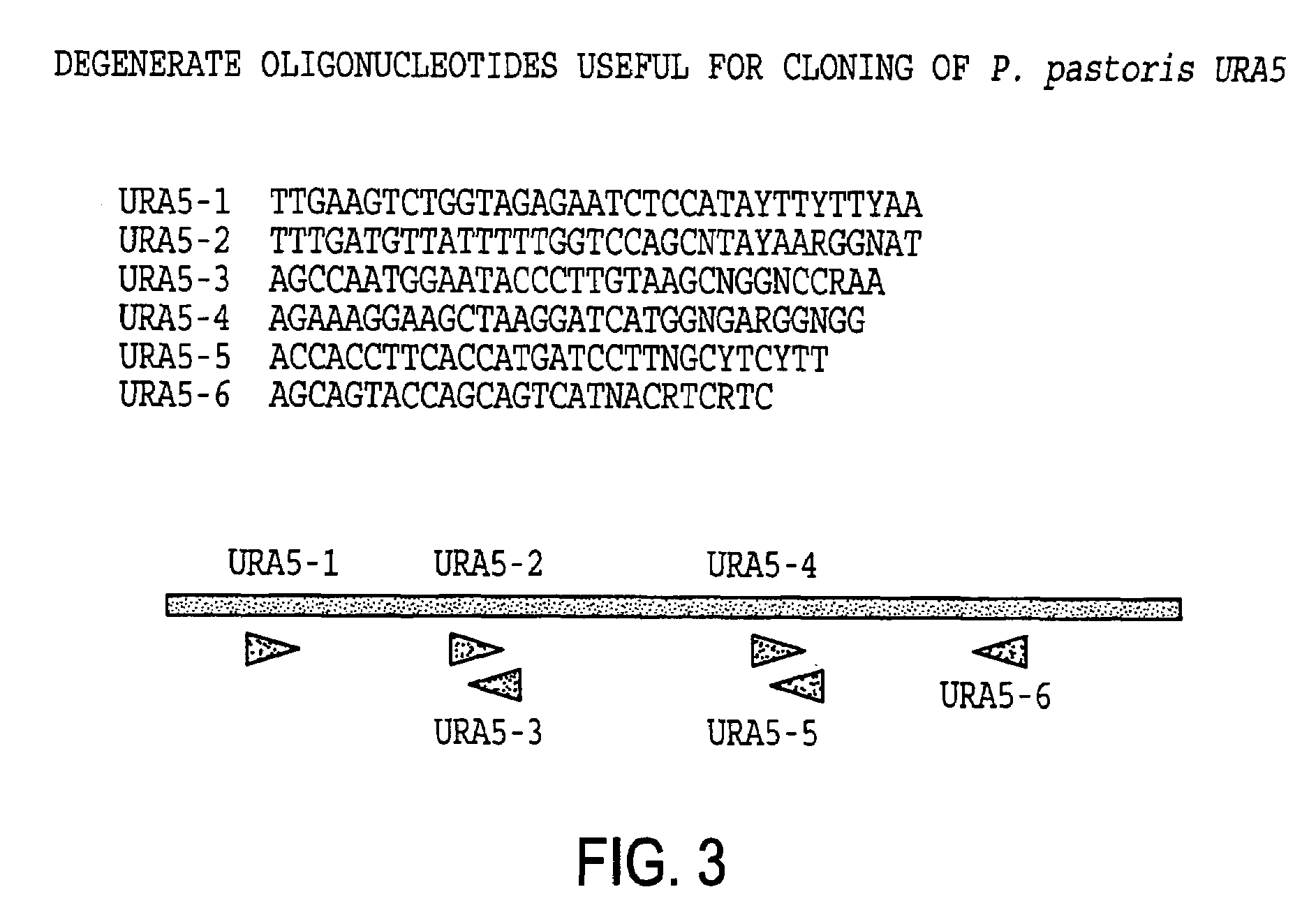
URA5 gene and methods for stable genetic integration in yeast
A novel gene encoding P. pastoris orotate-phosphoribosyl transferase (URA5) is disclosed. Methods for producing and selecting yeast strains capable of stable genetic integration of heterologous sequences into the host genome are also provided.
Owner:GLYCOFI
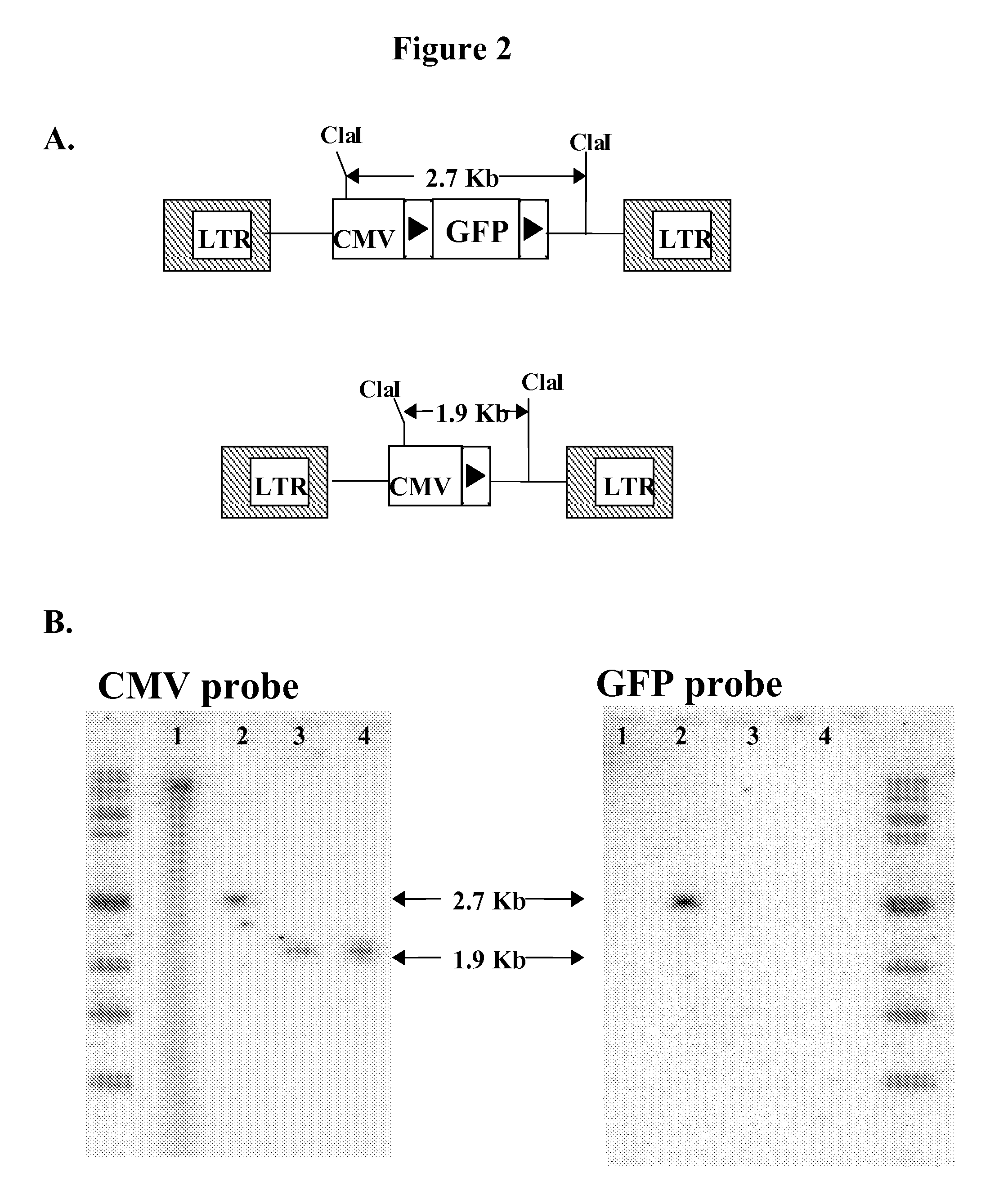
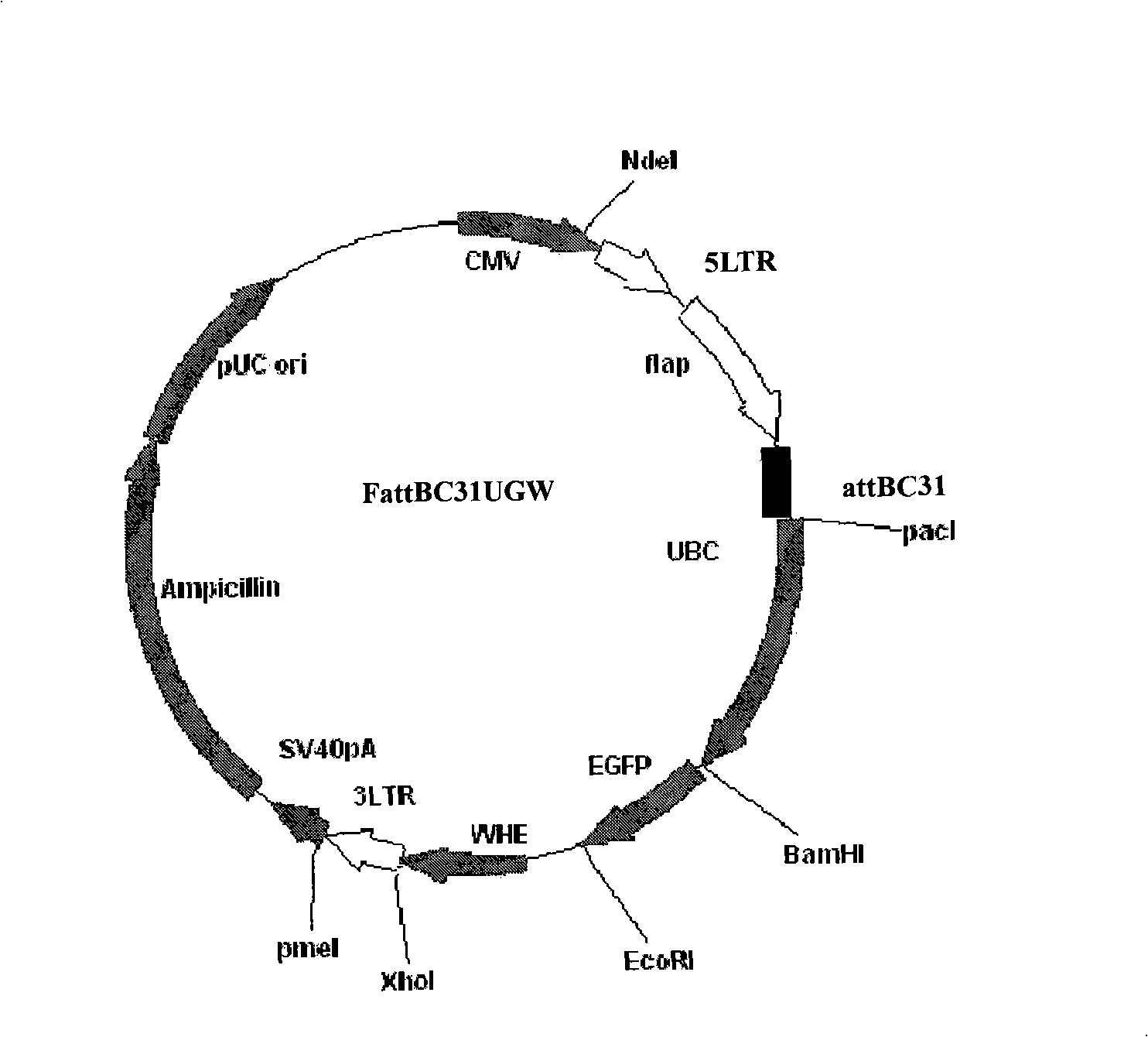
Novel lentiviral vectors for site-specific gene insertion
ActiveUS20080200663A1Easy to reorganizeSugar derivativesGenetic material ingredientsGene deliveryTherapy Trial
Murine leukemia virus (MLV) and lentivirus vectors have been used previously to deliver genes to hematopoietic stem cells (HSCs) in human gene therapy trials. However, these vectors integrate randomly into the host genome, leading to disruption or inactivation of vital host genes. The present invention discloses a novel lentiviral vector system that overcomes this problem by integrating into a host genome in a site-specific manner.
Owner:CITY OF HOPE
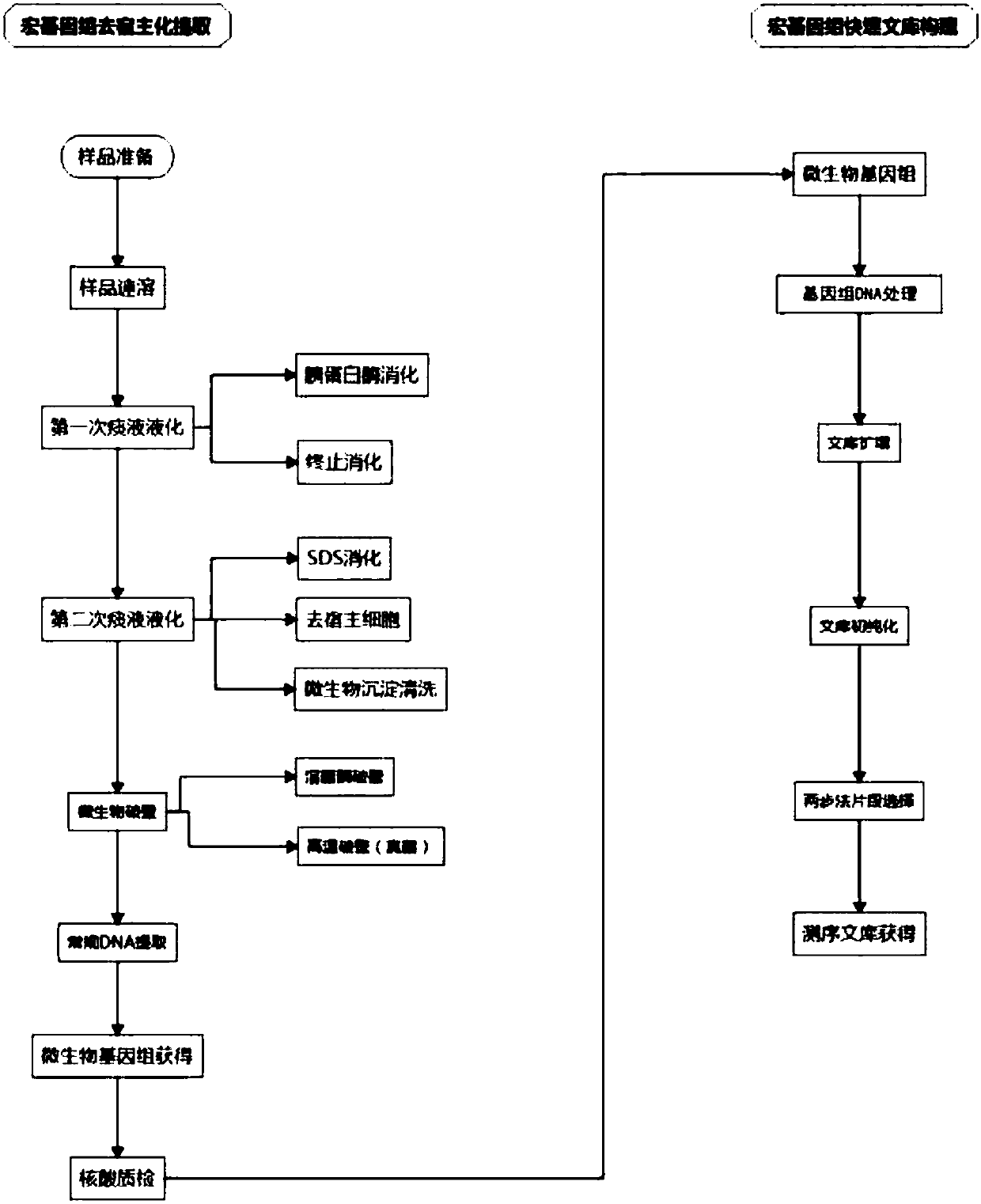
Host removal extraction and database building method for sputum microorganism metagenome
ActiveCN108048450AFully consider the physical characteristicsTaking biological differences into accountMicrobiological testing/measurementMicroorganism lysisCell wallDigestion
The invention belongs to the technical field of microorganism molecular biological detection, and specifically discloses a host removal extraction method for a sputum microorganism metagenome. The method comprises primary sputum liquefaction, secondary sputum liquefaction, microorganism wall breaking, DNA extraction and nucleic acid quality detection. The invention further discloses a database building method for a sputum microorganism metagenome, wherein the method comprises genome DNA extraction, genome DNA treatment, library amplification, library pre-purification, two-step fragment selection and computer detection. A mild digestion method combining trypsin with SDS is adopted to release a host genome to a solution, thereby obtaining microorganism with complete cell wall structure maintained and extracting nucleic acid from microorganisms with host removed. The provided extraction method and database building method are good in compatibility, and can be widely used to a mainstream sequencing platform. The methods are flexible and simple, efficient and fast, and cost-saving, and have bright popularization application prospects.
Owner:湖南赛哲智造科技有限公司
CRISPER-Cas9-system-mediated sheep MSTN (myostatin) gene knock-out and exogenous gene site-specific integration method
ActiveCN105671080AImprove integration efficiencyShorten the lengthNucleic acid vectorOxidoreductasesBiotechnologyMyostatin
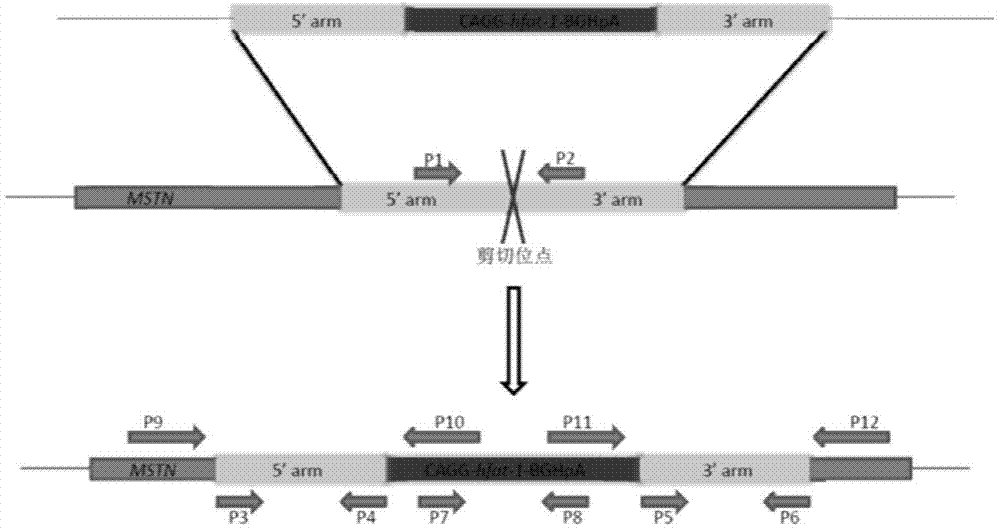
The invention relates to a CRISPER-Cas9-system-mediated sheep MSTN (myostatin) gene knock-out and exogenous gene site-specific integration method. The method comprises the following steps: establishing a CRISPER-Cas9-system-based gRNA (guide ribonucleic acid) expression vector according to a sheep MSTN gene sequence, establishing an exogenous-gene-containing donor plasmid capable of being integrated into a host genome according to the acting site of the gRNA, and transforming the optimized CRISPER-Cas9 vector, the gRNA expression vector and the linearized donor plasmid into sheep fibroblasts, thereby obtaining the sheep MSTN gene knock-out and exogenous gene site-specific integrated cells. The CRISPER-Cas9-mediated targeted vector provides a simple quick safe way for sheep MSTN gene knock-out and exogenous gene site-specific integration, thereby greatly enhancing the screening efficiency of the transgenic cell line. The method can screen the site-specific integrated exogenous gene cell line without adding any selection marker, thereby greatly enhancing the safety of the transgenic animals and having important value for sheep genetic breeding.
Owner:INNER MONGOLIA UNIVERSITY
Technology for identifying HBV (hepatitis B virus) gene integration sites and recurrently targeted genes in host genome
ActiveCN103725773AImprove accuracyGrowth inhibitionMicrobiological testing/measurementHost genomeHepatitis B virus
The invention discloses a novel MAPS (massive anchored parallel sequencing) technology which can be used for separating and sequencing integration sequences so as to identify HBV gene integration sites and recurrently targeted genes in a host genome. The invention further discloses a kit used for implementing the technology as well as six genes recurrently integrated by the HBV and related kits.
Owner:HANGZHOU PROPRIUM BIOTECH
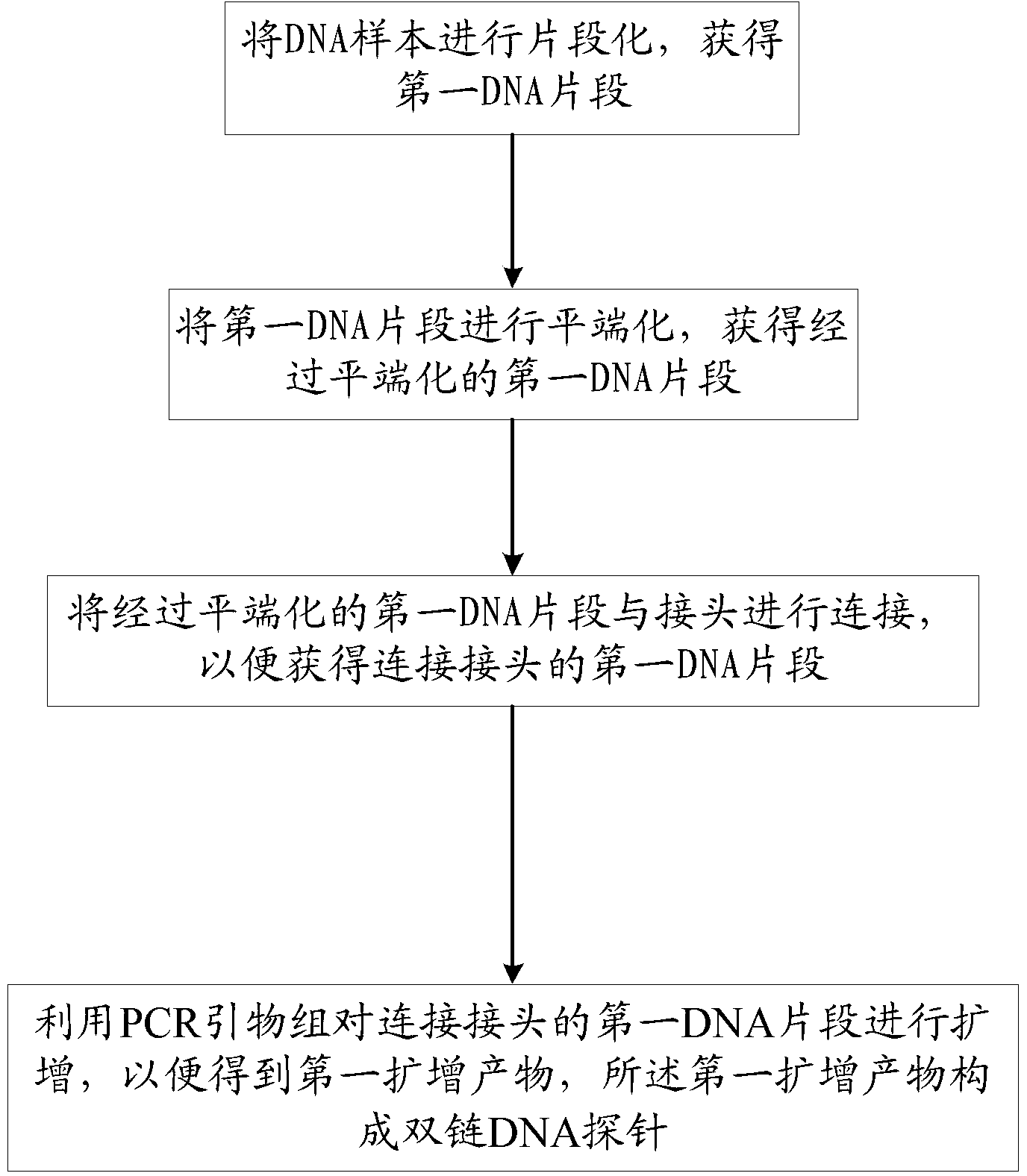
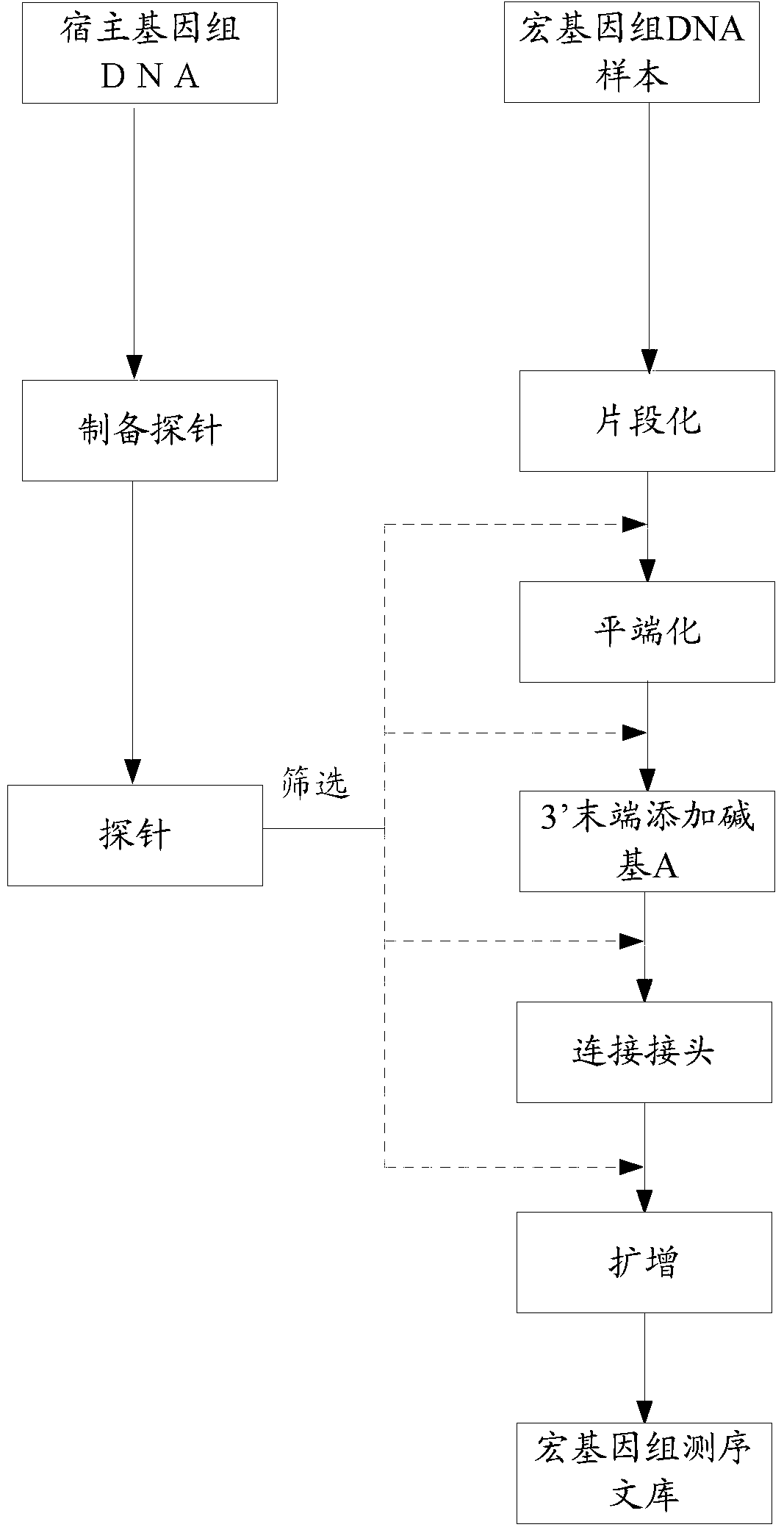
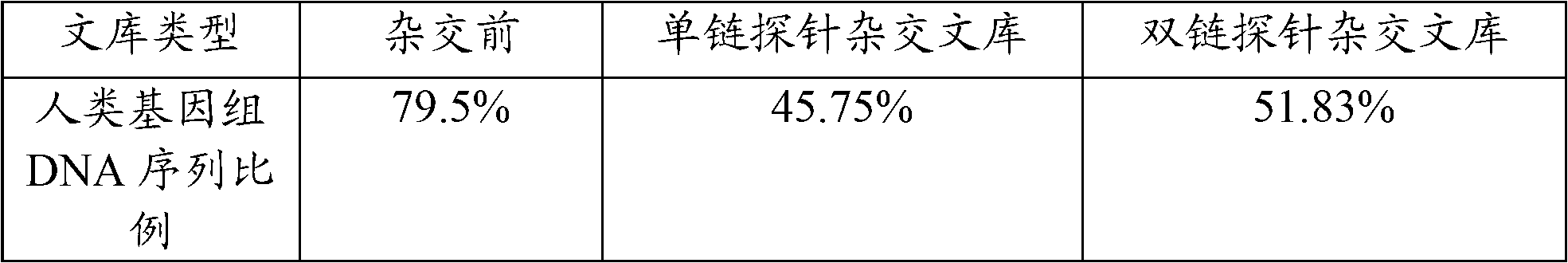
Nucleic acid probe, and preparation method and application thereof
InactiveCN102839168AEffective means of screeningNucleotide librariesMicrobiological testing/measurementNucleic Acid ProbesDNA fragmentation
The invention provides a probe, and a preparation method and an application thereof, wherein the method for preparing the probe comprises the steps of carrying out fragmentation on a DNA sample to obtain a first DNA fragment; blunting the first DNA fragment to obtain a blunted first DNA fragment; connecting the blunted first DNA fragment with a linker to obtain the first DNA fragment connected with the linker; carrying out amplification on the first DNA fragment connected with the linker by using PCR primers to obtain a first amplification product, wherein the PCR primers comprise a first PCR primer and a second PCR primer; the 5'-terminal bases of both the first PCR primer and the second PCR primer are marked by biotin; and the first amplification product forms a double-chain DNA probe. The probe obtained by the method can be effectively used for reducing pollutions of host genome DNA in a metagenome construction library.
Owner:SHENZHEN HUADA GENE INST
Novel lentiviral vectors for site-specific gene insertion
ActiveUS20050266565A1Easy to reorganizeGenetic material ingredientsDepsipeptidesTherapy TrialVector system
Murine leukemia virus (MLV) and lentivirus vectors have been used previously to deliver genes to hematopoietic stem cells (HSCs) in human gene therapy trials. However, these vectors integrate randomly into the host genome, leading to disruption or inactivation of vital host genes. The present invention discloses a novel lentiviral vector system that overcomes this problem by integrating into a host genome in a site-specific manner.
Owner:CITY OF HOPE
Retrons for gene targeting
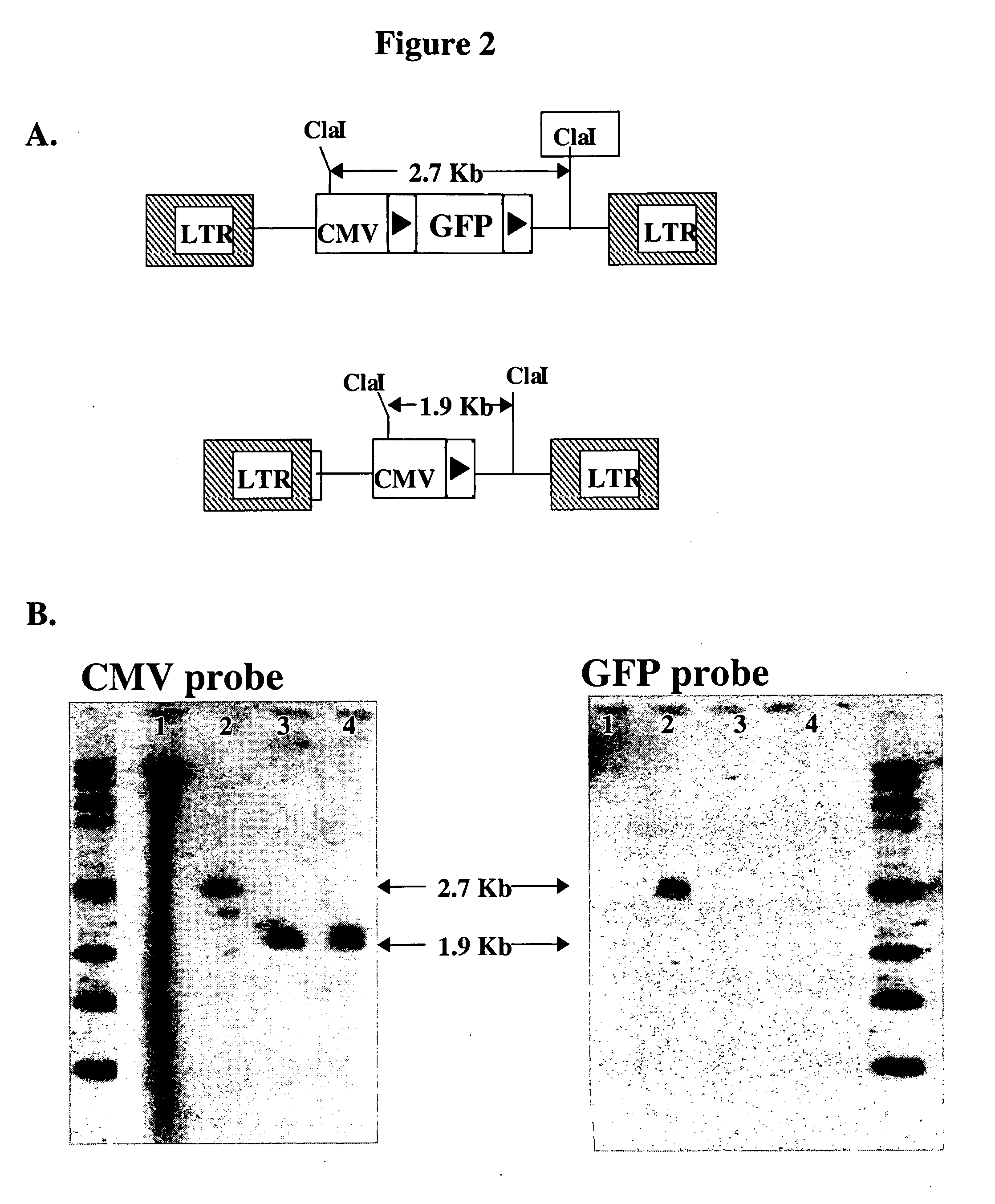
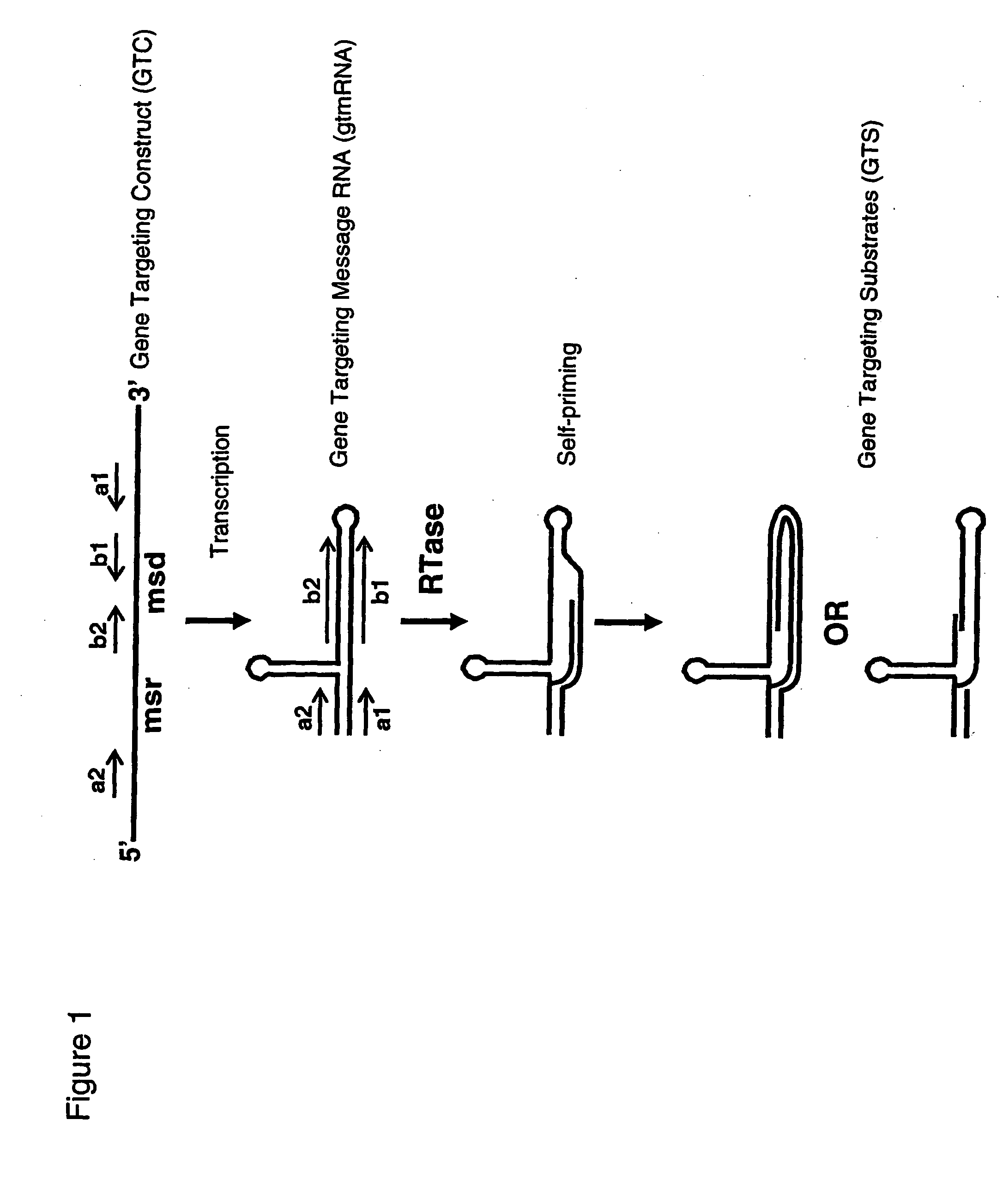
The invention provides methods and nucleic acid constructs that may be used to modify a nucleic acid of interest at a target locus within the genome of a host. In some aspects, the invention contemplates producing in vivo a gene targeting substrate (GTS), which may be comprised of both DNA and RNA components. The gene targeting substrate may comprise a gene targeting nucleotide sequence (GTNS), which is homologous to the target locus, but comprises a sequence modification compared to the target locus. The gene targeting substrate may be produced by reverse transcription of a gene targeting message RNA (gtmRNA). The gene targeting message RNA may be folded for self-priming for reverse transcription by a reverse transcriptase. The gene targeting message RNA may in turn be the product of transcription of a gene targeting construct (GTC) encoding the gene targeting message RNA. The gene targeting construct may for example be a DNA sequence integrated into the genome of the host, or integrated into an extrachromosomal element. Following expression of the gene targeting systems of the invention, hosts may for example be selected having genomic modifications at a target locus that correspond to the sequence modification present on the gene targeting nucleotide sequence. In some embodiments, the structure of retrons may be adapted for use in the gene targeting systems of the invention.
Owner:AGRI & AGRI FOOD
Method for performing genetic modification under a drug-free environment and components thereof
ActiveUS20110099649A1Minimize biasEffective targetingStable introduction of DNANucleic acid vectorGene targetsResistant genes
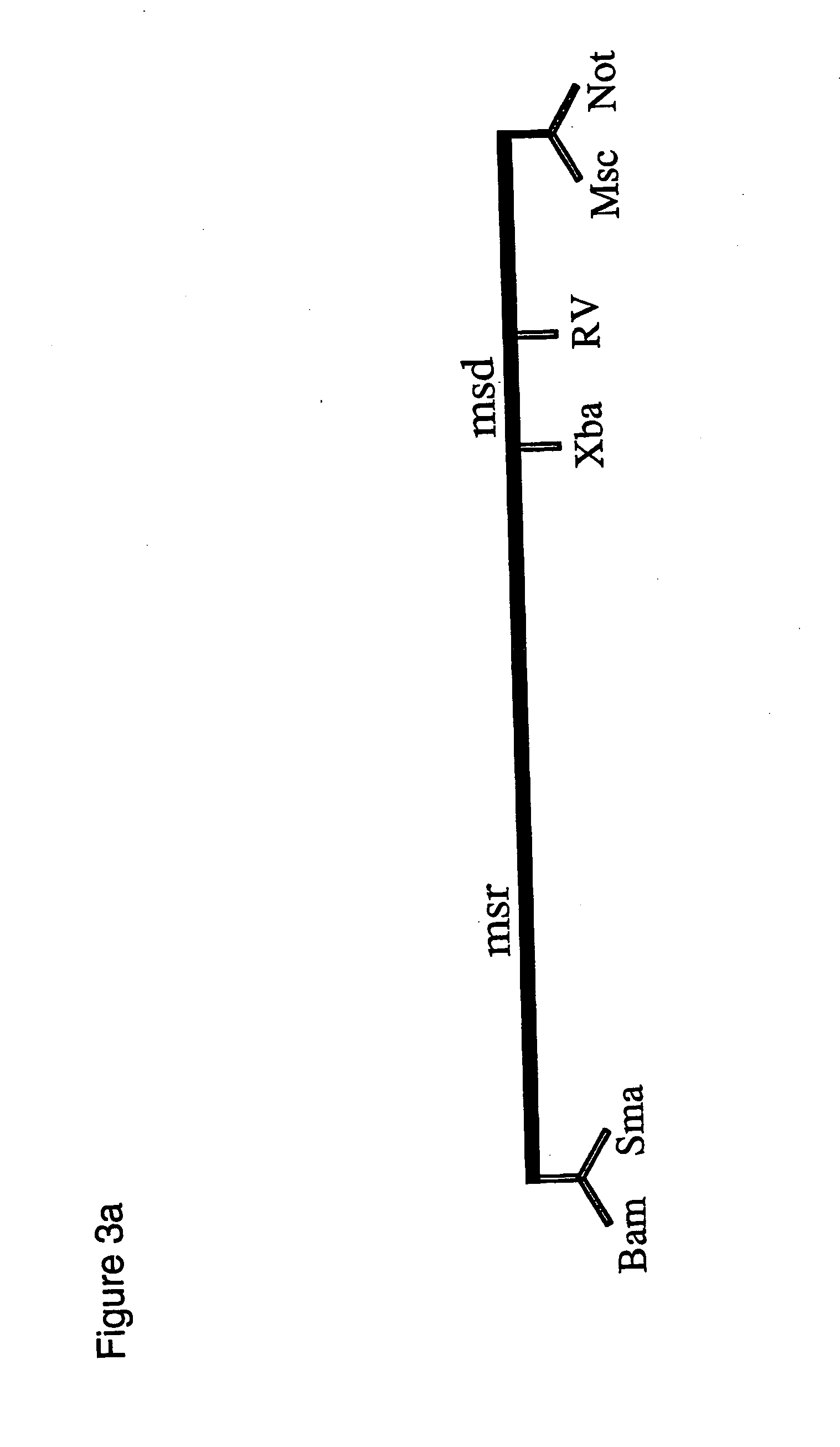
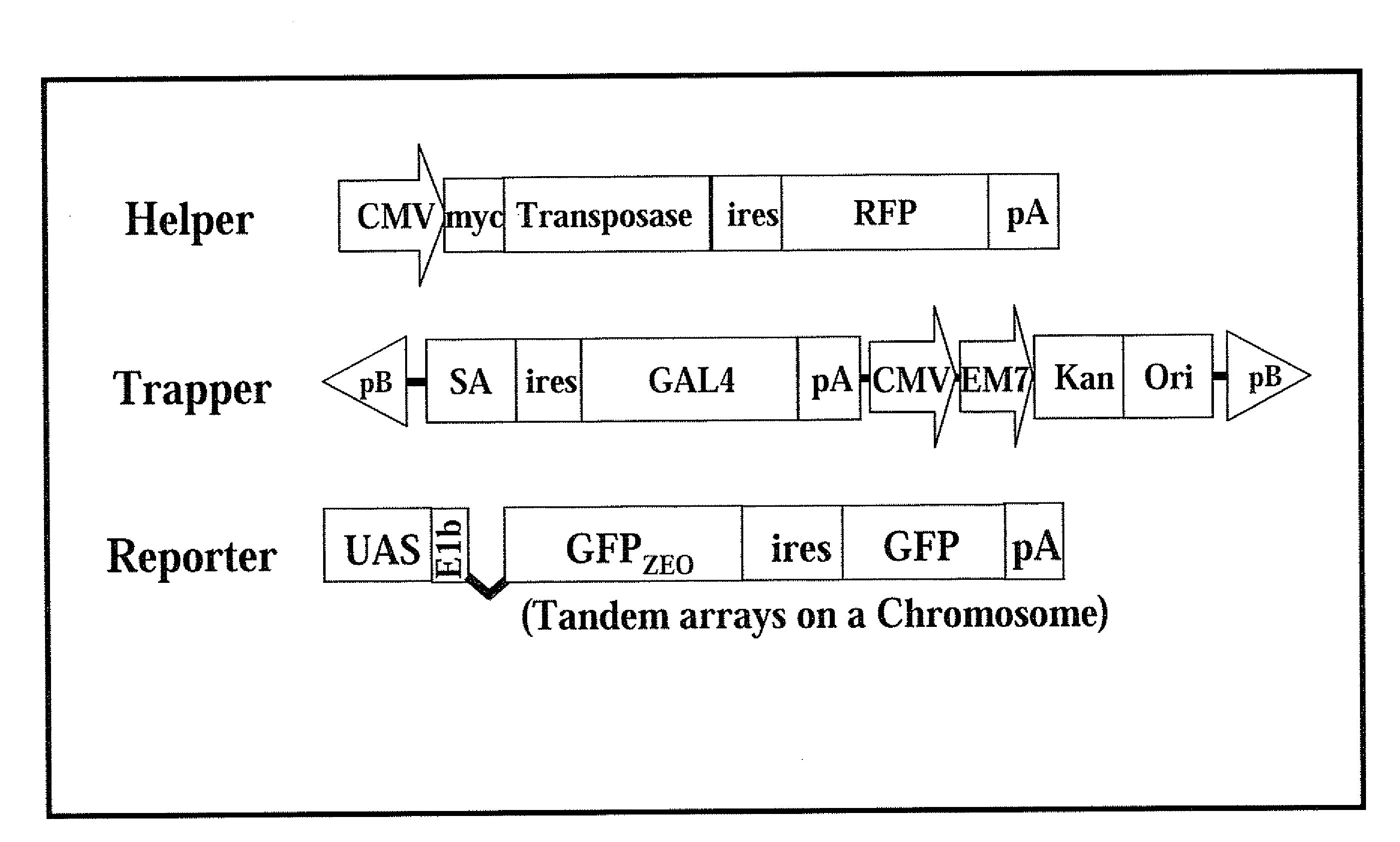
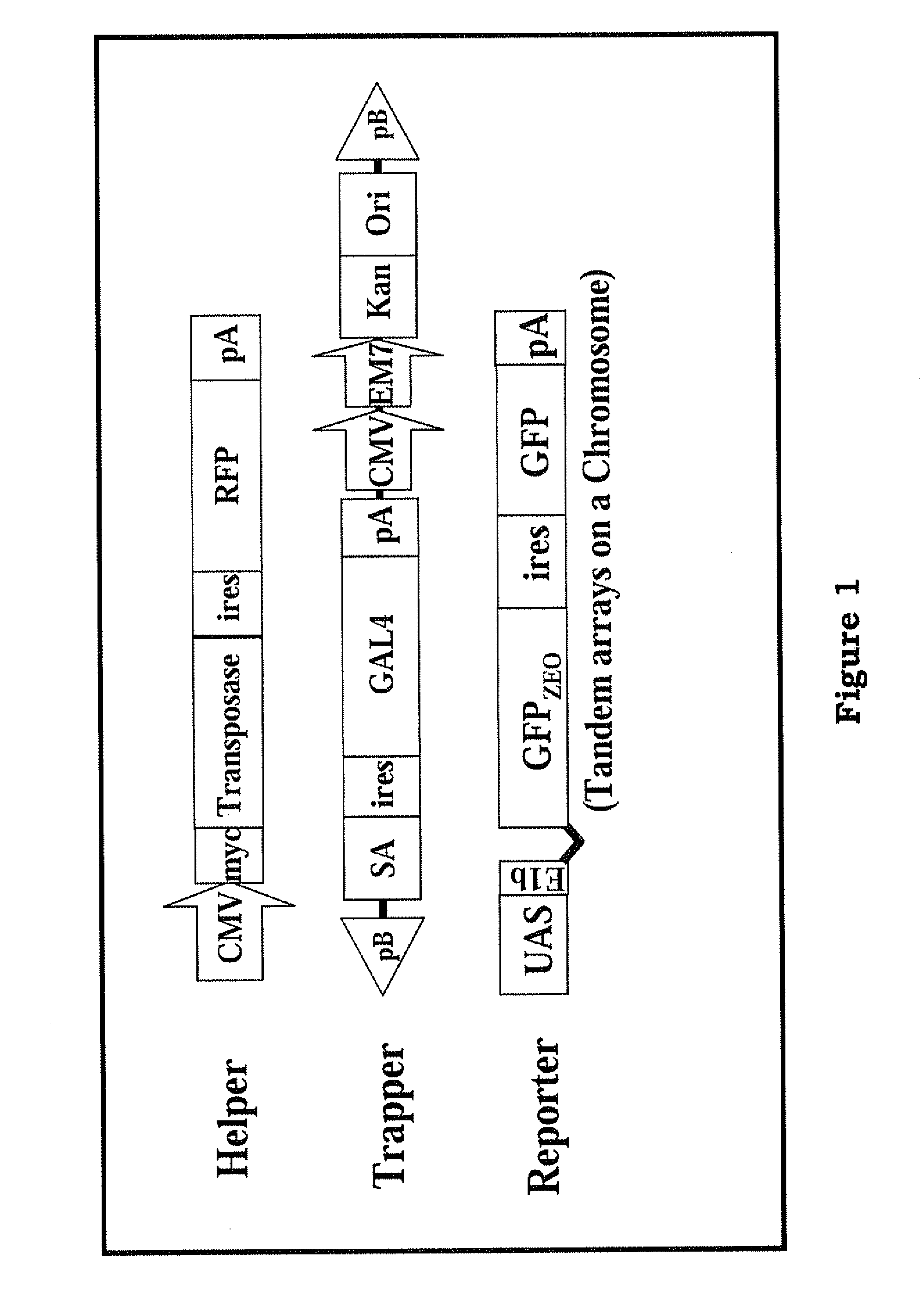
The present invention provides a method and components thereof of performing genetic modification under a drug-free environment. The method comprises the steps of generating a trapped mammalian cell library by trapper constructs (including the element of piggyBac terminal inverted repeats (TIRs)), reporter constructs, and helper constructs (including a sequence of an internal ribosomal entry site (IRES)). The present art allows: (1) to target & identify the silenced loci; (2) to separate genes with low-level expression at certain differentiation stages; (3) to evaluate the efficiency of gene targeting in the silent or repressed loci. The present invention avoids the biased gene targeting observed in the prior arts, and eliminates the needs of introducing antibiotic genes into the host genome which may lead to a potential threat of drifting antibiotic resistant genes into environment.
Owner:CHANG GUNG UNIVERSITY
Method and device for microbiological analysis of host sample
ActiveCN111009286AFast and accurate analysisFast and accurate detectionProteomicsGenomicsMicrobial GenomesMicroorganism
The invention relates to the field of microbiological detection, in particular to a method and a device for microbiological analysis of a host sample. The method comprises the following steps: (1) performing first filtering processing on a sequencing data set from a host sample by adopting a host genome database so as to remove sequencing data which can be compared with the host genome database from the sequencing data set; (2) performing second filtering processing on the sequencing data set by adopting a homologous database so as to remove the sequencing data which can be compared with the homologous database from the sequencing data set; and (3) comparing the sequencing data set subjected to the first filtering treatment and the second filtering treatment with a microbial genome database so as to determine microbial sequencing data from the microorganisms in the sequencing data set. By applying the method and the device provided by the invention, rapid and accurate analysis and detection of microorganisms in a host can be realized.
Owner:深圳华大因源医药科技有限公司 +1
Site-specific integration retroviral vector system and preparation thereof
InactiveCN101302537AAvoid toxicityOvercoming the Risk of Insertion MutationsGenetic engineeringFermentationTransgeneViral vector
Owner:北京康爱瑞浩细胞技术有限公司
Methods for targeted transgene-integration using custom site-specific DNA recombinases
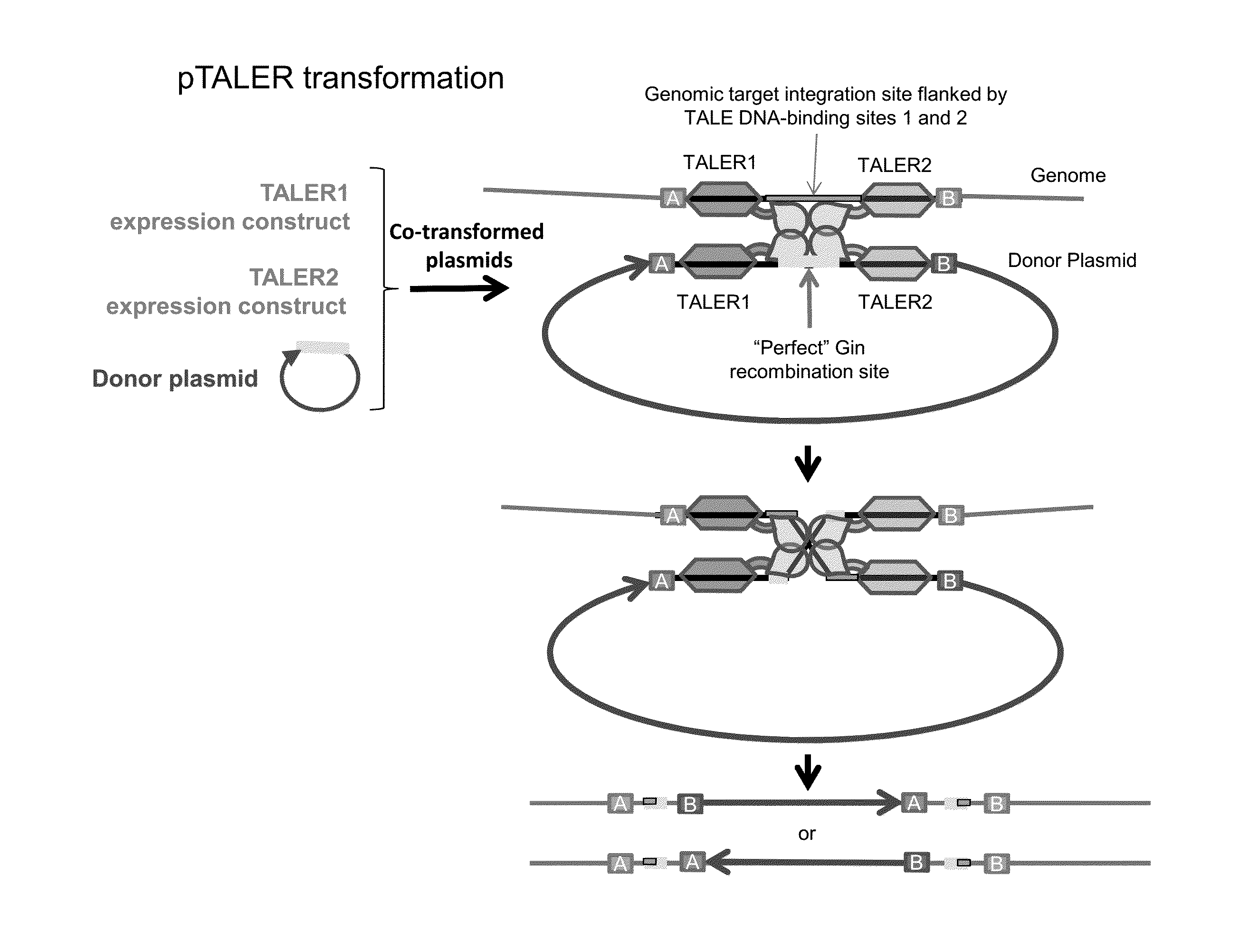
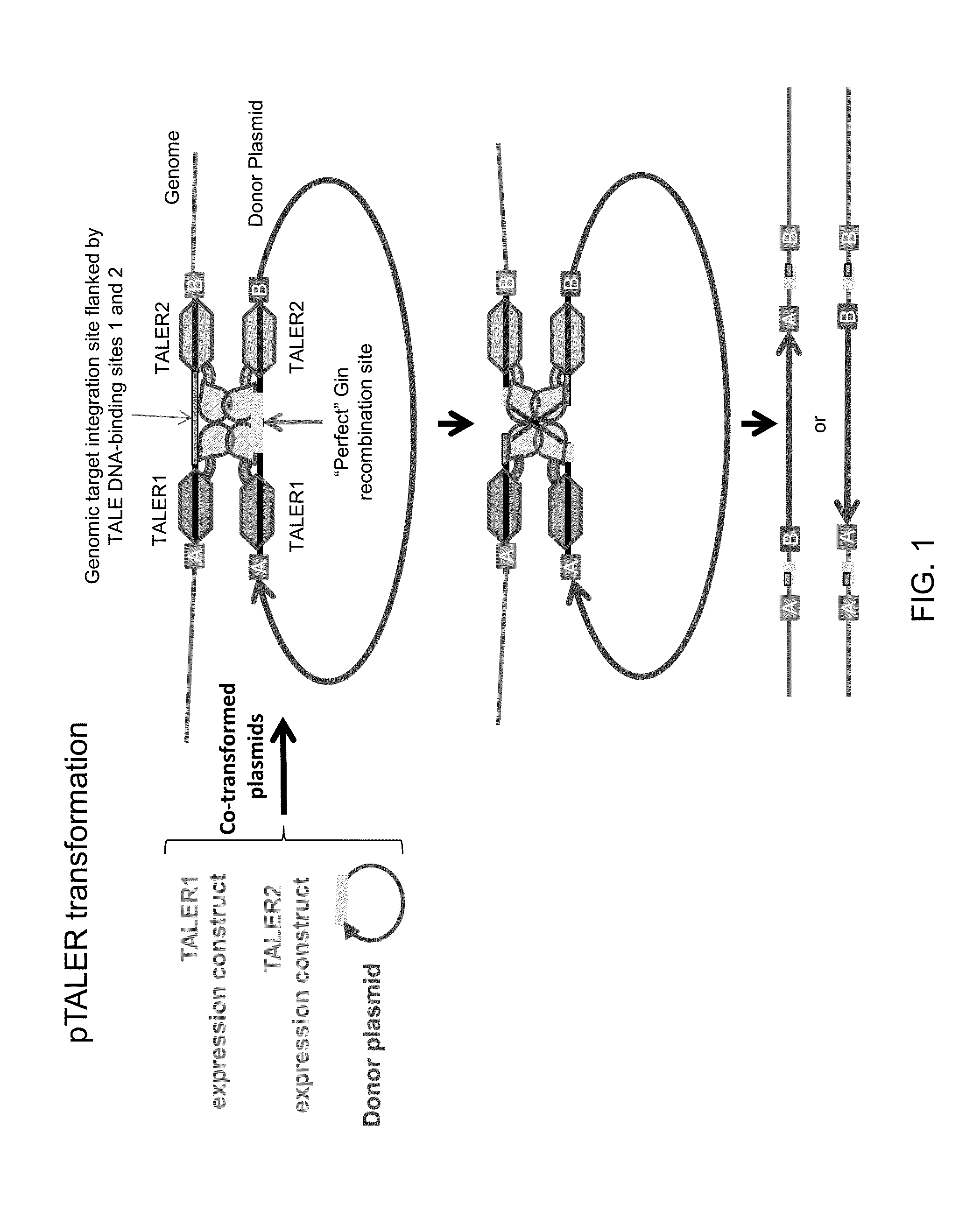
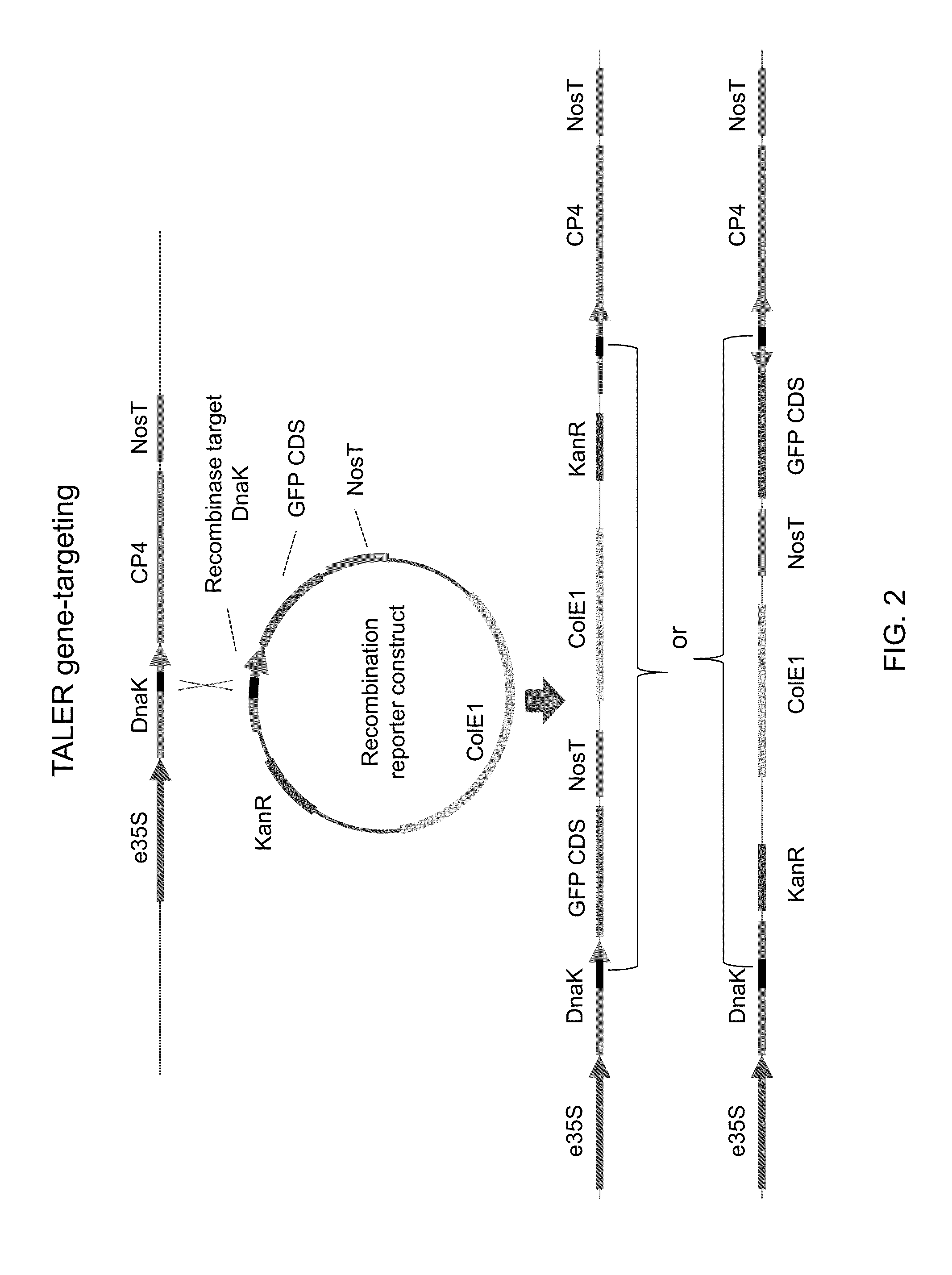
The invention relates to biotechnology and provides novel methods for sequence-specific or sequence-directed transcription activator-like effector recombinase-mediated integration of DNA sequences of interest into host genomes. The invention also provides methods of use for novel plant transformation vectors and expression cassettes, which include novel combinations of chimeric recombinases with plant expression and transformation elements. Methods for gene-targeting, DNA sequence removal, genome modification, and molecular breeding are also provided.
Owner:MONSANTO TECH LLC
Method for knocking out ZFNs (zinc finger nucleases)-mediated bovine MSTN (myostatin) gene and integrating exogenous gene at fixed point
InactiveCN103088046AHigh gene knockout efficiencyRealize fixed-point insertionFermentationVector-based foreign material introductionMyostatinZinc finger nuclease
The invention provides a method for knocking out a ZFNs (zinc finger nucleases)-mediated bovine MSTN (myostatin) gene and integrating an exogenous gene at the fixed point. The method comprises the following steps of: designing a specific locus expression vector of the ZFNs according to a bovine MSTN gene sequence; designing a target vector according to an acting site of the ZFNs, wherein the target vector contains the exogenous gene and can be integrated into a host genome; and transferring the expression vector and the target vector into a bovine fibroblast to obtain a cell for knocking out the bovine MSTN gene and integrating the exogenous gene at the fixed point. The DNA sequence acting on the designed specific ZFNs is located on the exon of the bovine MSTN gene. The constructed ZFNs-mediated target vector provides a way for easily and quickly knocking out the bovine MSTN gene and inserting the exogenous gene at the fixed point and has an important value to the genetic breeding of the new double-muscle bovine variety.
Owner:INNER MONGOLIA UNIVERSITY
Minimal plasmid vectors that provide for persistent and high level gene expression and methods for using the same
InactiveUS20040077576A1Genetic material ingredientsNucleic acid vectorBiological bodyHigh level expression
Methods are provided for the in vivo introduction of an expression cassette into a target cell of a vascularized organism, e.g., a mammal, in manner such that the encoded protein of the introduced expression cassette is persistently expressed at a high level in the target cell. In the subject methods, an aqueous formulation of a minimal plasmid vector that includes the expression cassette is administered into the vascular system of the organism. The minimal plasmid vector employed in the subject methods is one that provides for persistent and high level expression of an expression cassette that is present on the vector in a manner that is substantially expression cassette sequence and direction independent Also provided are the minimal plasmid vectors employed in the subject methods. The subject methods and compositions find use in a variety of different applications, including both research and therapeutic applications, and are particularly suited for use in the in vivo delivery of nucleic acids encoding protein products, particularly where persistent, high level protein expression is desired without integration of the vector into the host genome.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
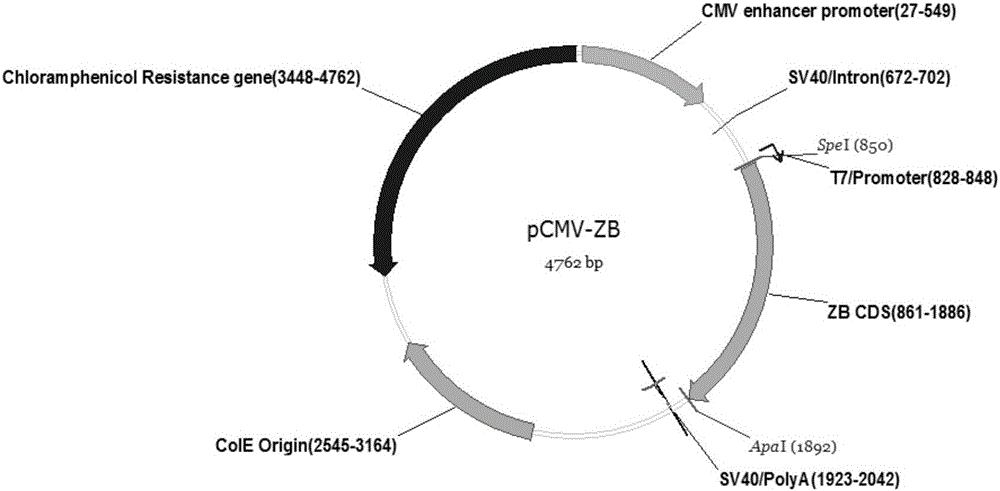
ZB (zebrafish) transposon system and gene transfer method mediated by same
ActiveCN105018523AEfficient transfer mediationImprove transfer efficiencyFermentationVector-based foreign material introductionCloning SiteBiology
The invention belongs to the field of animal genetic engineering, and relates to a ZB (zebrafish) transposon system of and a gene transfer method mediated by the ZB transposon system. The ZB transposon system is derived from zebra fish, and comprises two transgenic donor plasmids with terminal repetition sequences and capable of being inserted into a target gene box, and transposase auxiliary plasmids for providing activity of transposition; the transgenic donor plasmids comprise terminal repetition sequences and Msc1 insertion cloning sites at two sides of ZB transposon. The invention further discloses a gene transfer method based on the ZB transposon system, which is characterized that target genes are guided into a receptor genome through the ZB transposon system. The system and the method can be applied to multiple field of biotechnology: (1) the method can be used for effectively inserting the target gene box into a host cell genome, so that the gene transfer efficiency is improved; (2) by combining with the gene trapping technology, the researches on functions of animal genes can be effectively developed; (3) human gene therapy can be realized through mediation.
Owner:SHANGHAI CELL THERAPY GRP CO LTD
Antineoplastic ad virus preparation
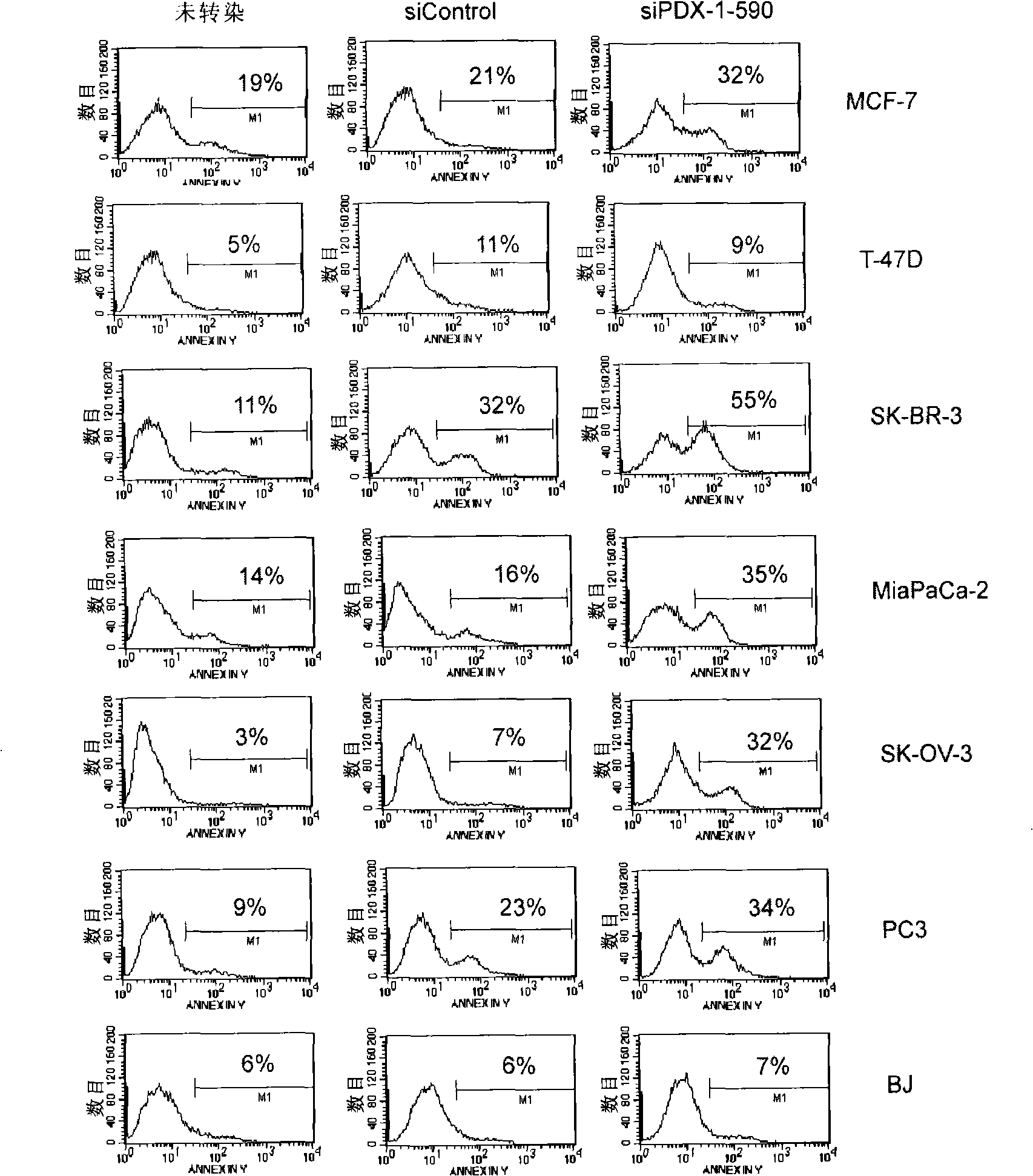
The invention discloses a siRNA which inhibits tumor through inhibiting the expression of PDX-1, and a recombinant adenovirus designed on the basis of the sequence of the siRNA. The invention also provides a drug composition containing the adenovirus. The adenovirus provided by the invention has the advantages of high titration, absence of integration into the genome of the host, and high safety.
Owner:ZHENGZHOU VIRIGE BIOTECH
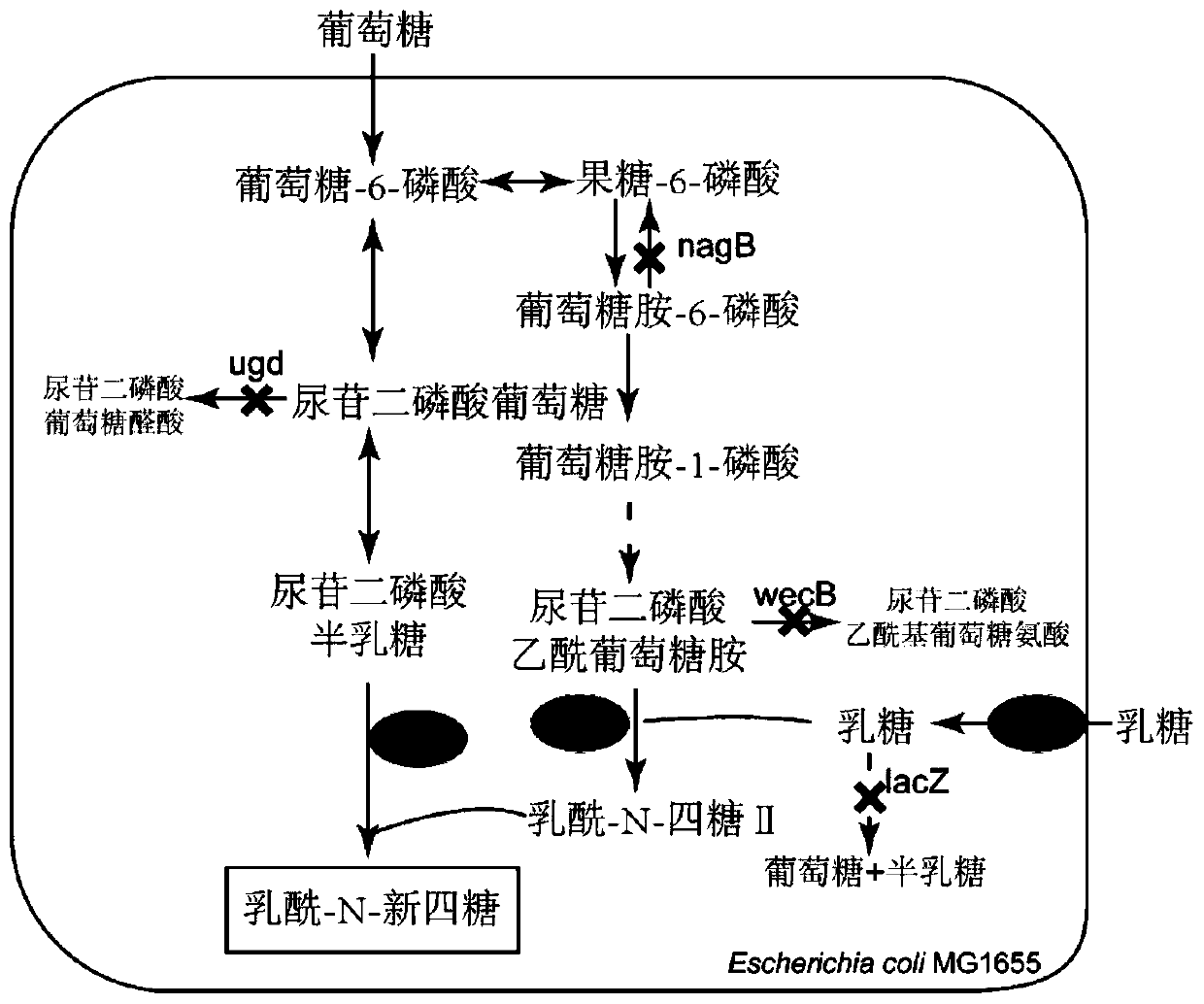
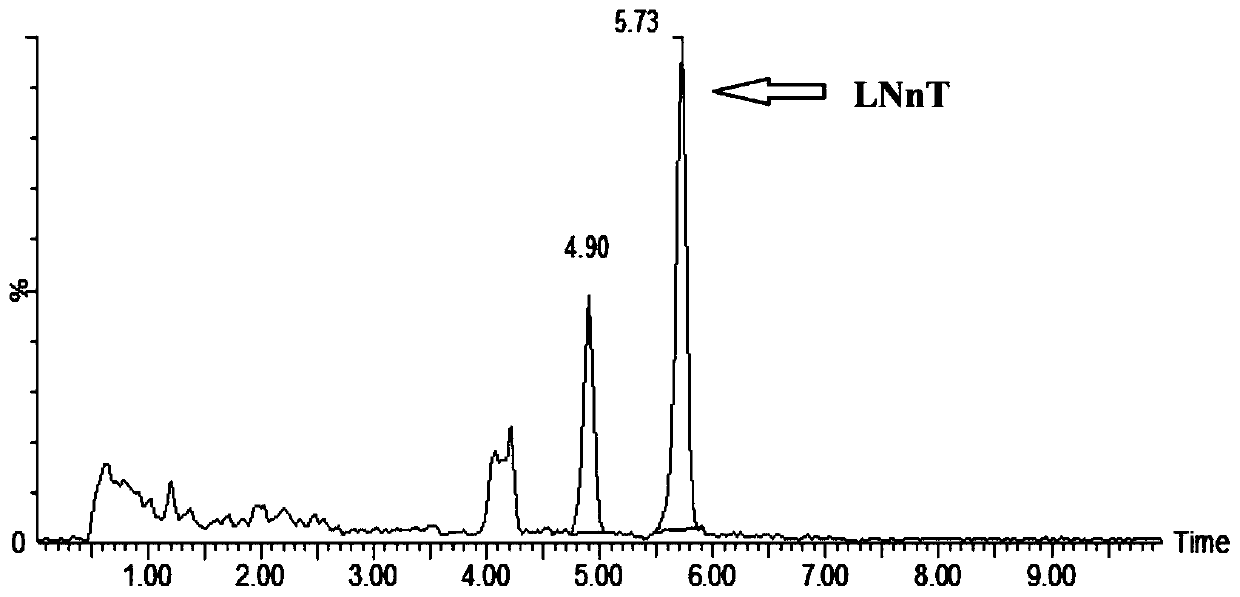
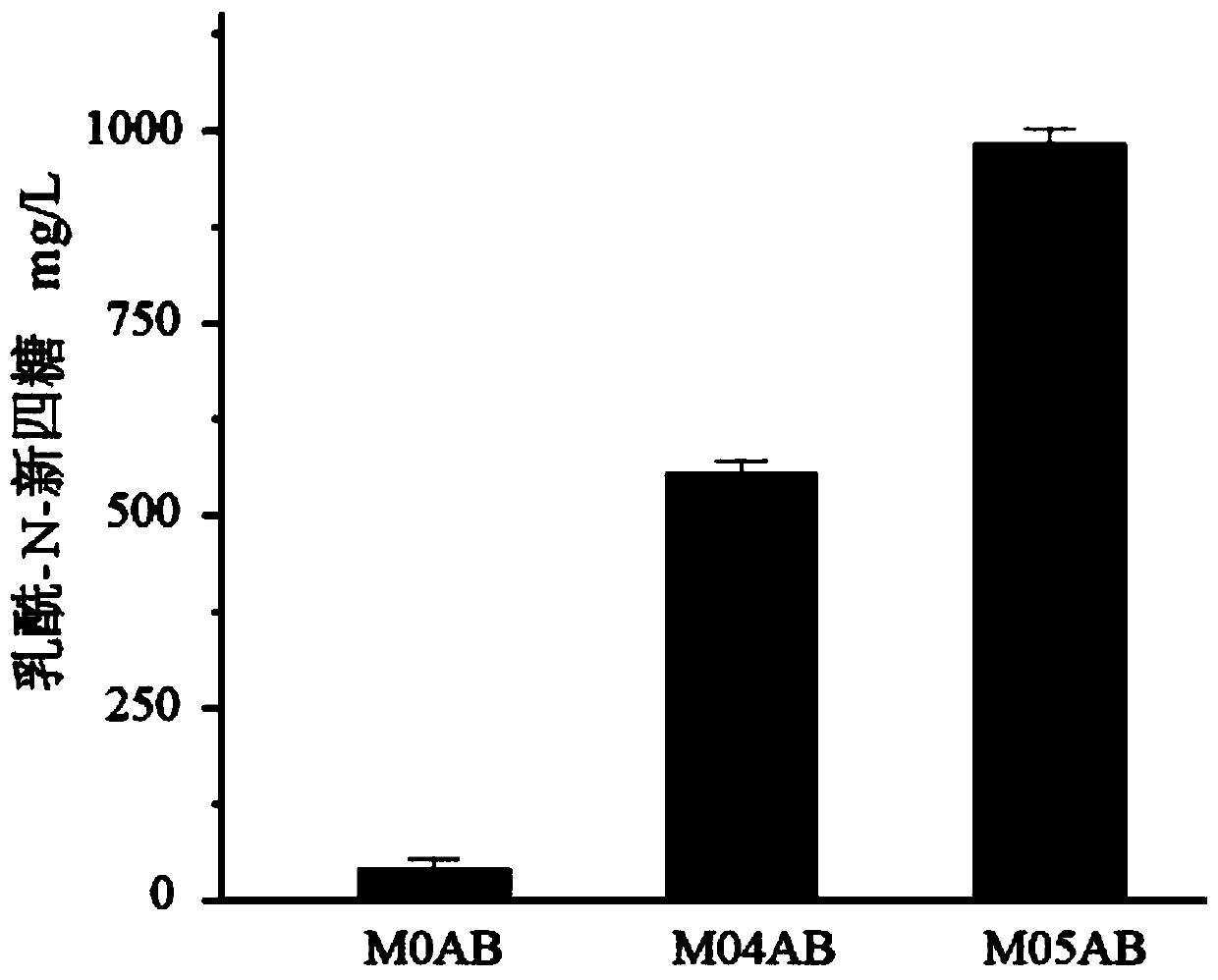
Recombinant escherichia coli for synthesizing lactoyl N-neotetraose and construction method and application of recombinant escherichia coli
The invention discloses recombinant escherichia coli for synthesizing lactoyl N-neotetraose and a construction method and application of the recombinant escherichia coli. The recombinant escherichia coli are obtained by knocking out a beta-galactosidase lacZ gene, a glucosamine-6-phosphodeaminase nagB gene, a UDP-acetylglucosamine epitope isomerase wecB gene and a UDP-glucose dehydrogenase ugd gene in the host genome of escherichia coli and overexpressing a lactose transportase lacY gene, beta-1, 3-N-glucosaminidase lgtA gene and a beta-1, 4 galactosyltransferase lgtB gene on the genome. The yield of lactoyl-N-neotetraose synthesized by recombinant escherichia coli reaches 985 mg / L, and a foundation is laid for further metabolic engineering transformation of escherichia coli to produce lactoyl-N-neotetraose.
Owner:JIANGNAN UNIV
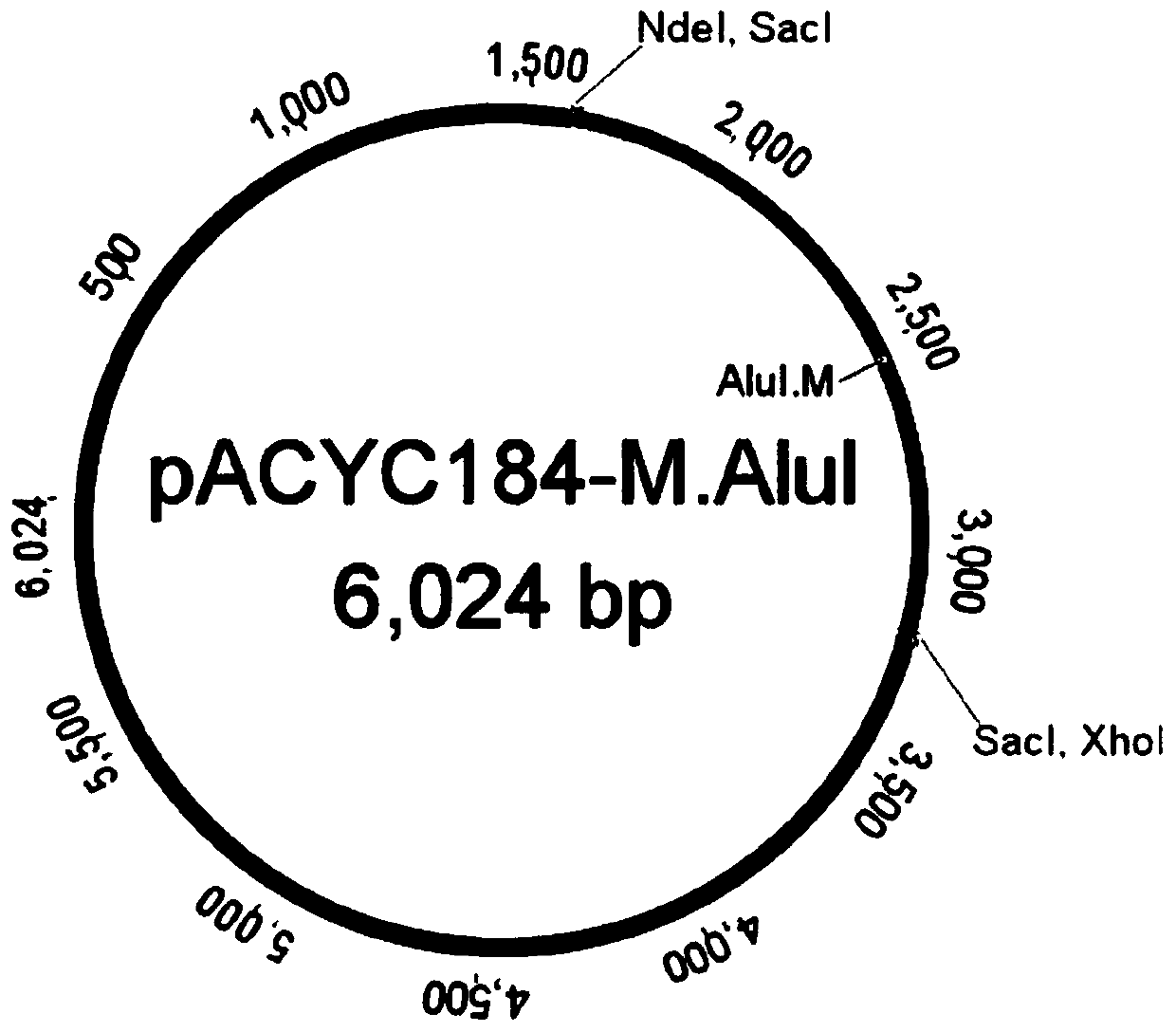
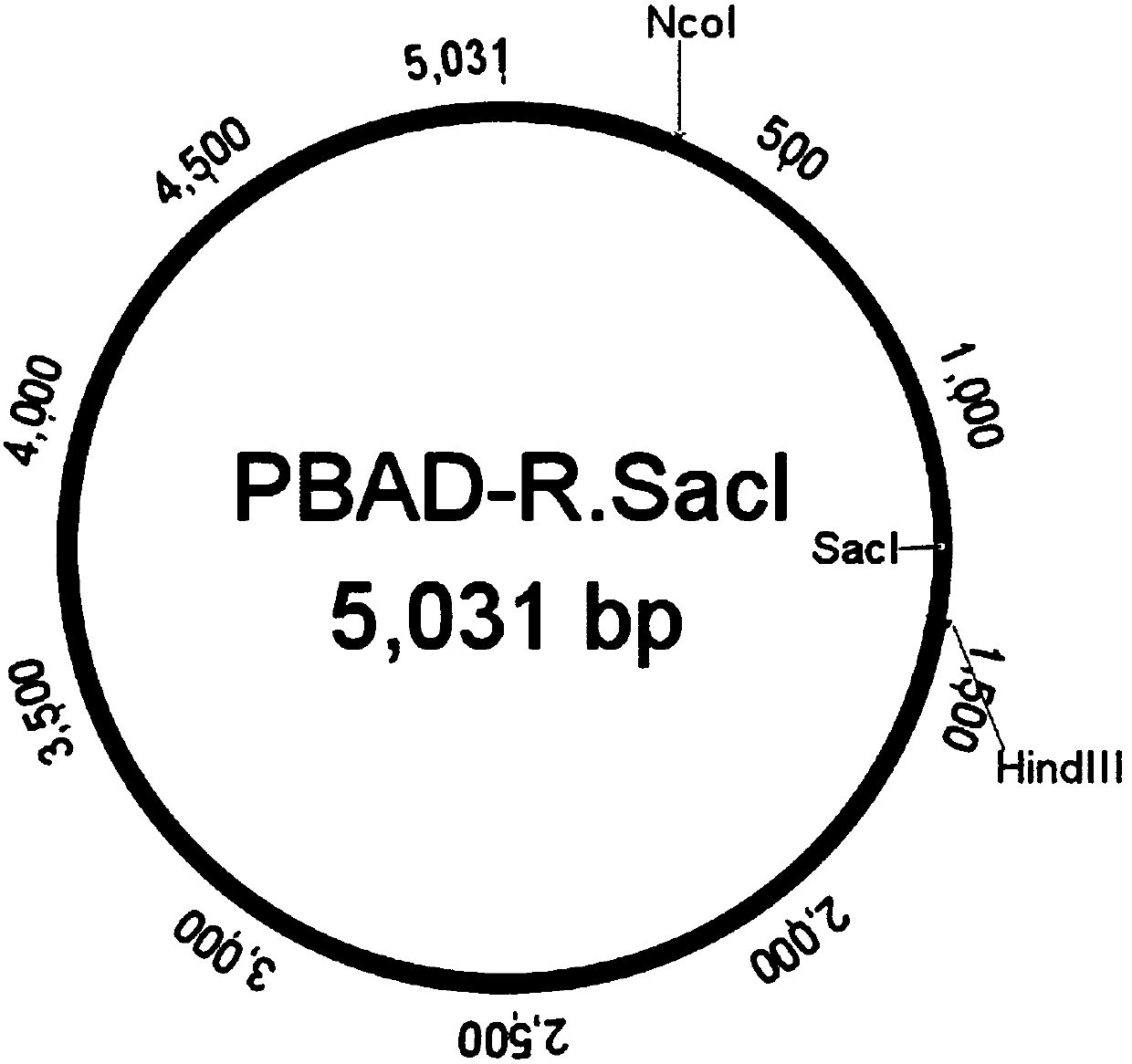
Screening method for methylated protective strain expressing restriction endonuclease SacI
InactiveCN107557372AEfficient recombinant expressionFermentationVector-based foreign material introductionHost geneOperon
The invention provides a screening method for methylated protective strain expressing restriction endonuclease SacI. The method comprises steps: screening and culturing recombinant expression plasmidpBAD-R. SacI transformed competence cells with correct sequencing to obtain a recombinant expression strain of restriction endonuclease SacI by establishment of recombinant expression strain of methyltransferase M. AluI, recombinant expression of restriction endonuclease SacI, and other steps; and performing enlarging cultivation on the strain, and inducing with arabinose to obtain the recombinantprotein of restriction endonuclease SacI. For the method disclosed by the invention, the efficient recombinant expression of the restriction endonuclease SacI can be achieved; the recombinant expression of the restriction endonuclease SacI is carried out by strict control of an arabinose operon expression system on background expression; the method is stable and efficient against digestion of therestriction endonuclease SacI on a host genome DNA; and an M. AluI methylated protection strain is also applicable to recombinant expression of other restriction endonucleases with similar recognition sites.
Owner:江苏愚公生命科技有限公司
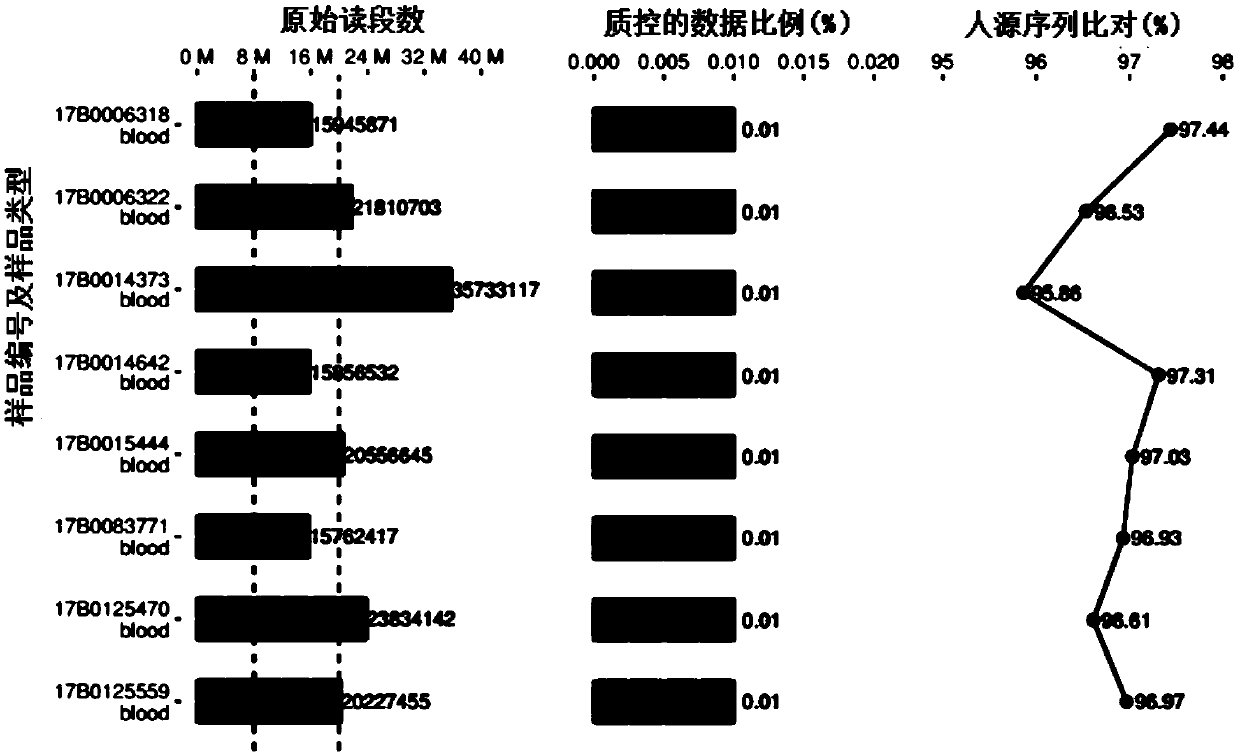
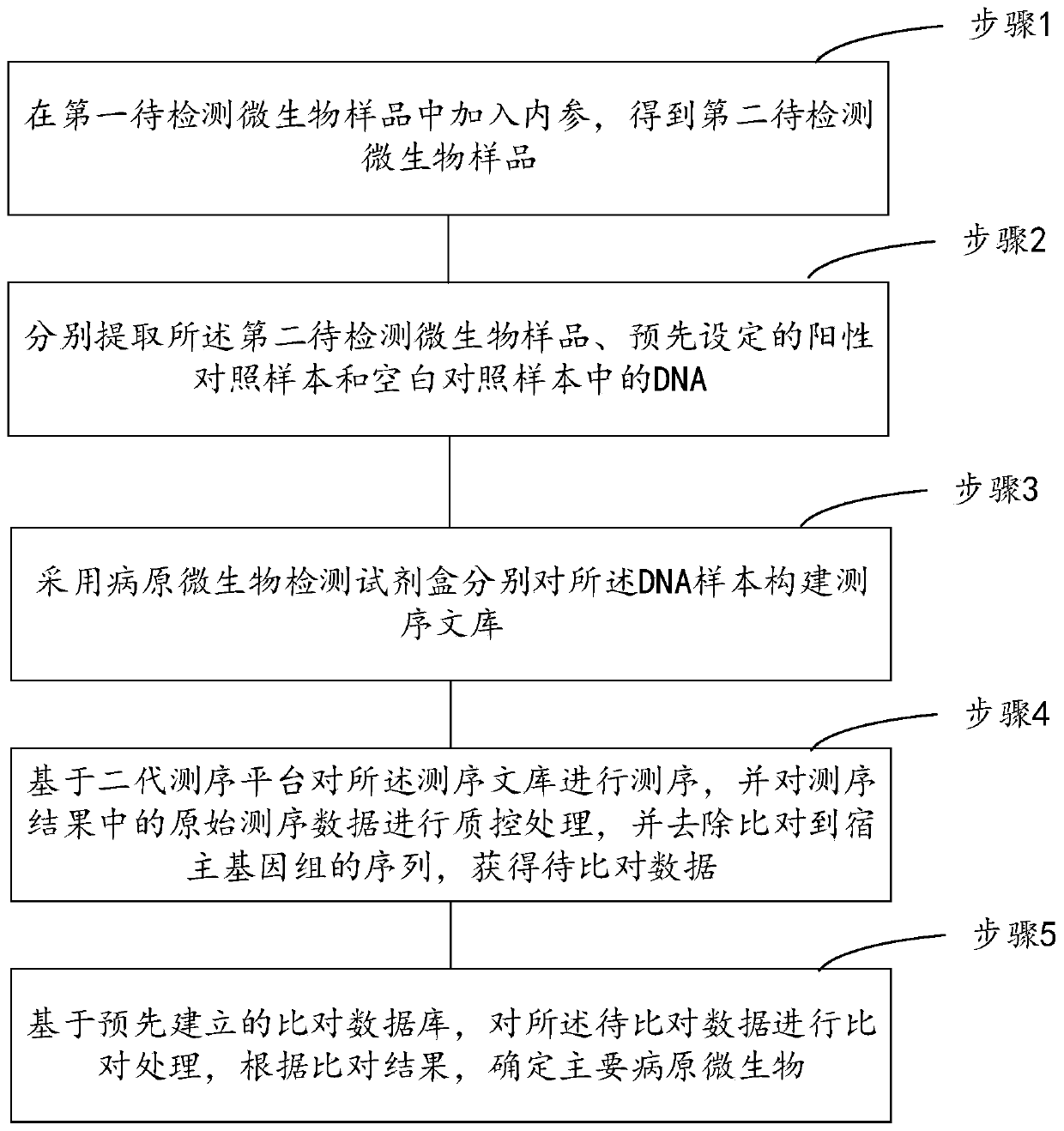
High-throughput sequencing detection method for pathogenic microorganisms with full-process quality control
ActiveCN111187813AImprove throughputRapid clinical testMicrobiological testing/measurementSequence analysisPathogenic microorganismHost genome
The invention provides a high-throughput sequencing detection method for pathogenic microorganisms with full-process quality control. The method comprises the following steps: adding an internal reference into a first to-be-detected microorganism sample to obtain a second to-be-detected microorganism sample; extracting DNA in the second to-be-detected microorganism sample, a preset positive control sample and a blank control sample respectively; constructing a sequencing library for the extracted DNA samples by adopting a pathogenic microorganism detection kit respectively; sequencing the sequencing library based on a second-generation sequencing platform, performing quality control processing on original sequencing data in a sequencing result, and removing a sequence compared to a host genome to obtain to-be-compared data; and comparing the to-be-compared data based on a pre-established comparison database, and determining main pathogenic microorganisms according to a comparison result. Rapid clinical detection of the pathogenic microorganisms which do not need to be predicted, do not have preference and have high throughput for infectious diseases is realized, meanwhile, full-process quality control of a system is realized, and false positive is reduced.
Owner:HUGOBIOTECH BEIJING CO LTD +2
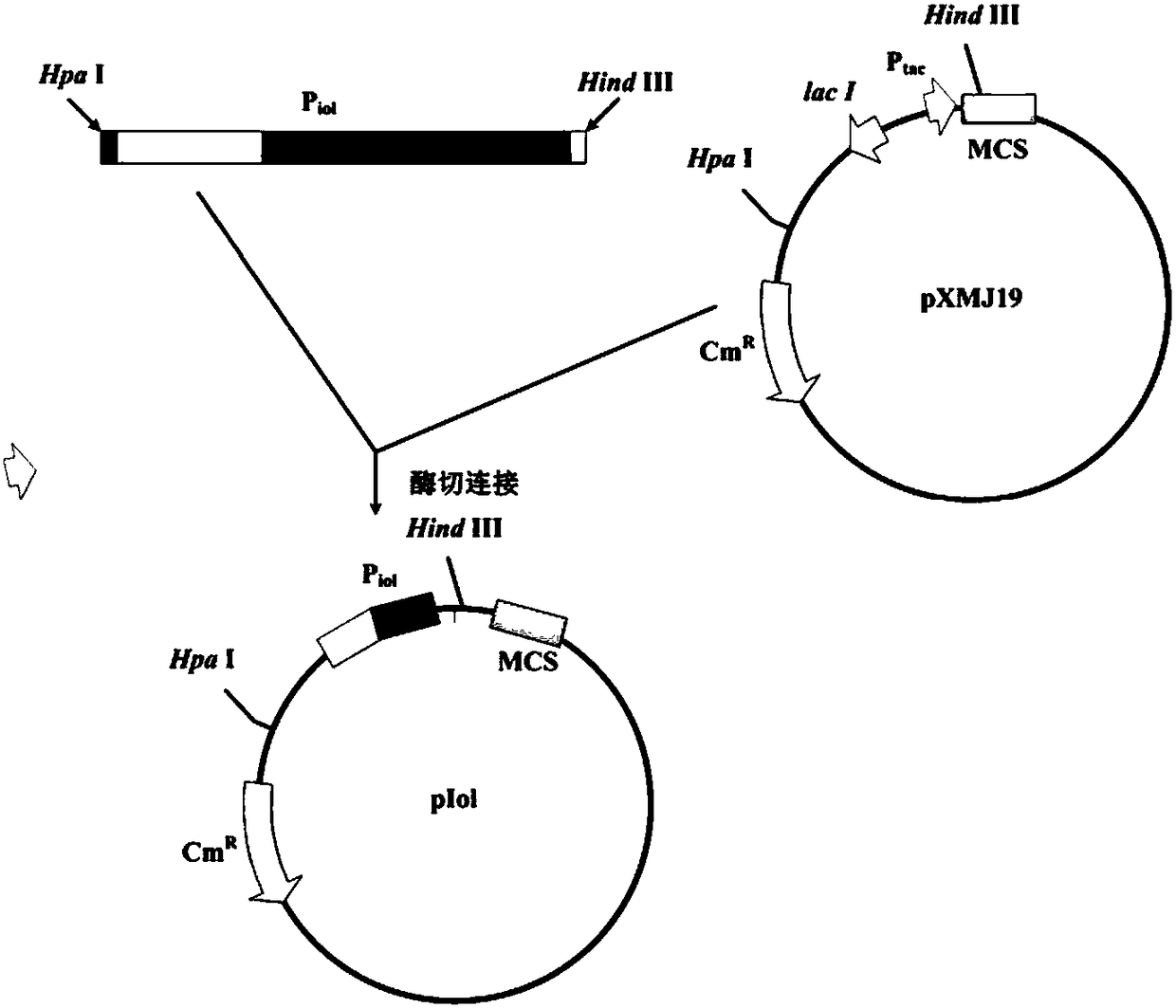
Corynebacteria inducible promoter, expression vector comprising same, and application thereof
The invention relates to a Corynebacteria inducible promoter, an expression vector comprising same, and an application thereof, wherein the expression vector is converted to corynebacterium glutamicumto obtain a 5-aminolevulinic acid producing bacterial strain, by that 5-aminolevulinic acid can be produced through a fermentation method. The expression vector constructed from the promoter, throughcontrollable expression of gene through inositol induction, can overcome problems of inhibition effect on growth of bacterial cells with and high production cost due to high price of an inducing agent, (isopropyl[beta]-D-thiogalactoside, IPTG). The expression vector has characters such as stability. By integrating the promoter with genome of a host or converting the expression vector into the host, corynebacterium glutamicum, the 5-aminolevulinic acid producing bacterial strain is acquired, yield of the 5-aminolevulinic acid reaching 24.2 g / L.
Owner:TIANJIN UNIVERSITY OF SCIENCE AND TECHNOLOGY
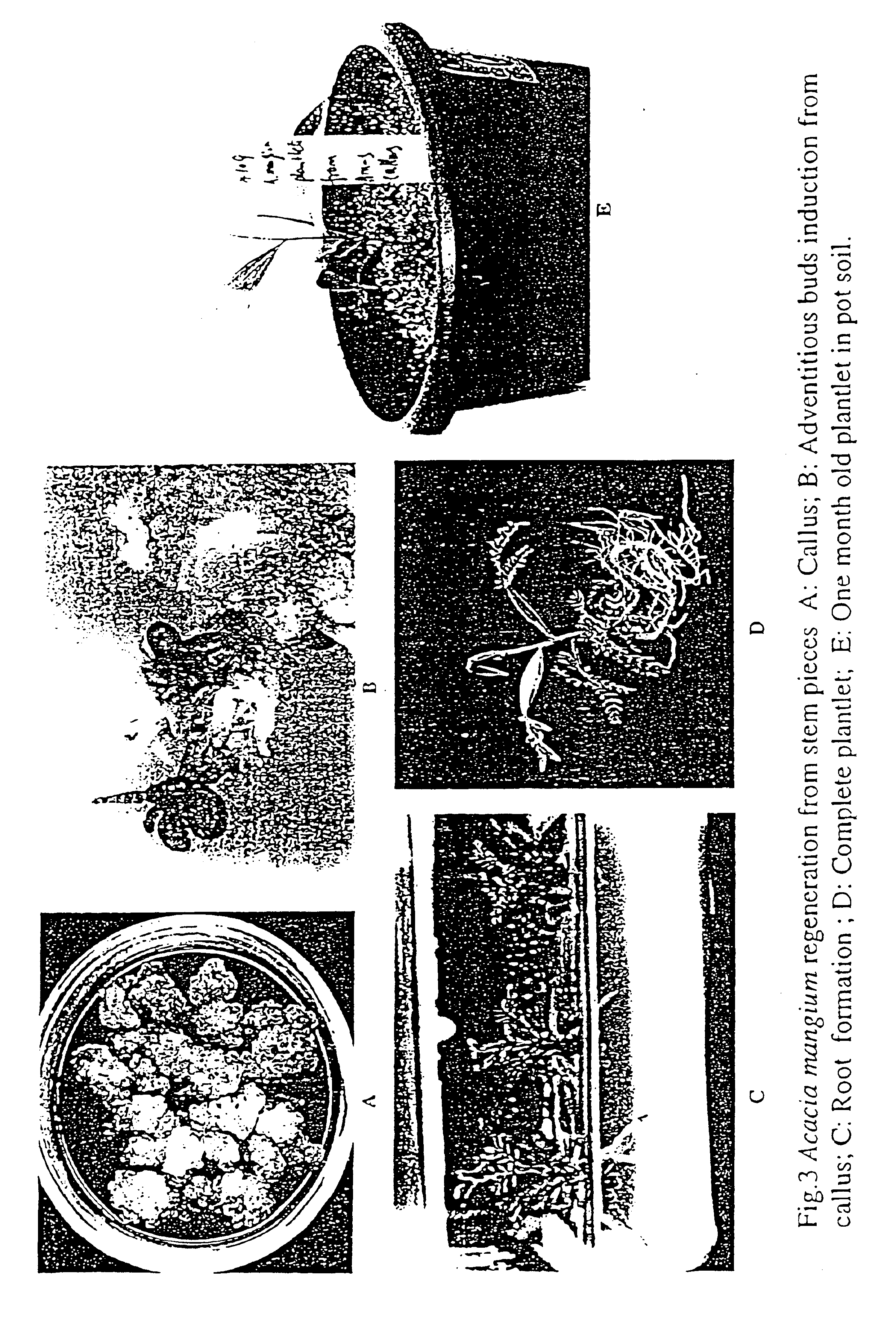
Regeneration and genetic transformation of Acacia mangium
InactiveUS6846971B1Other foreign material introduction processesFermentationCauliflower mosaic virusOrganogenesis
The present invention is directed to a method of Acacia mangium regeneration through organogenesis and a method of genetic transformation of Acacia mangium. The method of regeneration comprises inducing callus from different parts of seedlings and vegetatively micropropagated plantlets as explants; adventitious bud induction followed by pinnate leaf and bud elongation and eventually, elongated shoots were induced to root. Based on the regeneration system, a marker gene GUS under cauliflower mosaic virus promoter was introduced to Acacia mangium via Agrobacterium infection. GUS staining in the regenerated plants and Southern blot hybridization prove the incorporation of the foreign gene into the host genome and expression of the foreign gene.
Owner:TEMASEK LIFE SCIENCES LABORATORY
Adenovirus vector for gene therapy on hemophilia B and application thereof
The invention relates to an adenovirus vector for gene therapy on hemophilia B and application thereof, and the adenovirus vector can simultaneously express hFIX and red fluorescent protein; both sides of a tandem gene cassette of the hFIX and the red fluorescent protein are respectively provided with a loxp site of a Cre integrase action site in the same direction, and an action site attB of site-specific integrase PhiC31 is arranged in the gene cassette. The invention obtains a recombinant adenovirus vector which can be cyclized by DNA marked by the red fluorescent protein, and lays the foundation for the recombinant adenovirus vector to be further integrated into a main genome through attb sequences to exert the lasting function for treating hemophilia B. The adenovirus vector or adenovirus particles packaged outside the adenovirus vector is applied in the treatment of hemophilia B.
Owner:INST OF HEMATOLOGY & BLOOD HOSPITAL CHINESE ACAD OF MEDICAL SCI
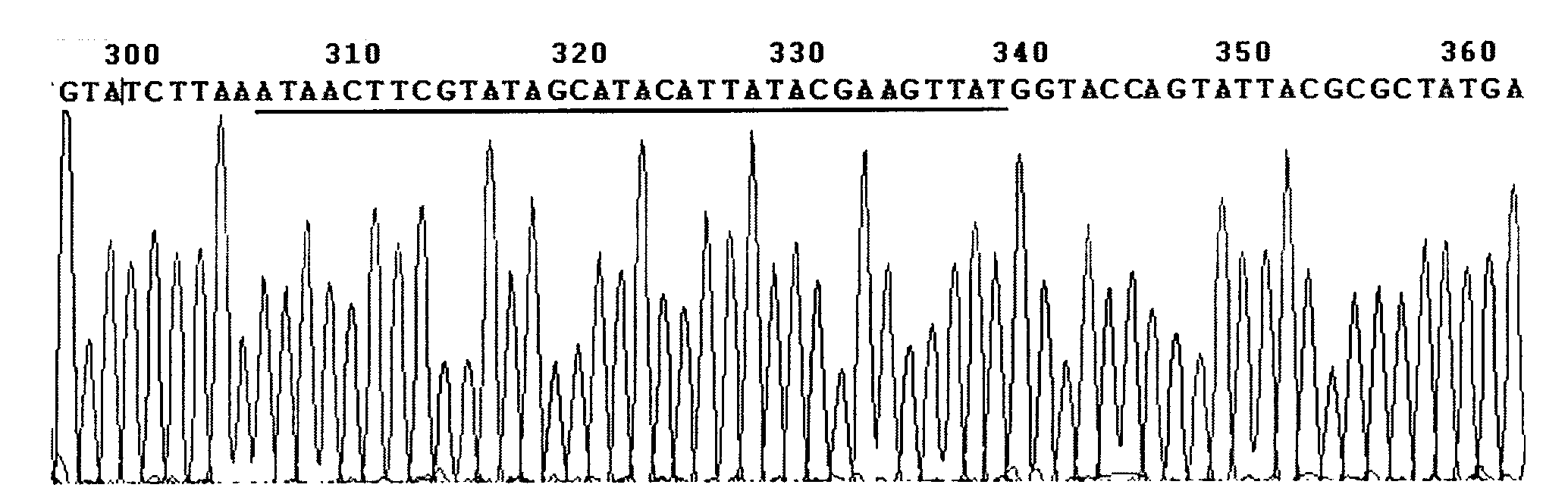
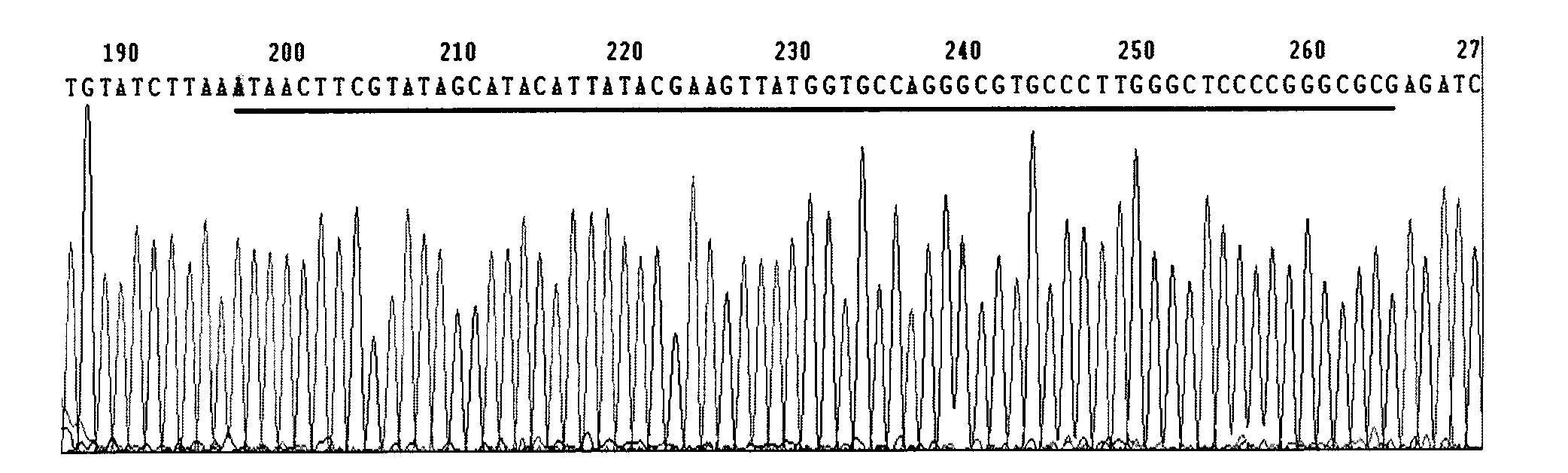
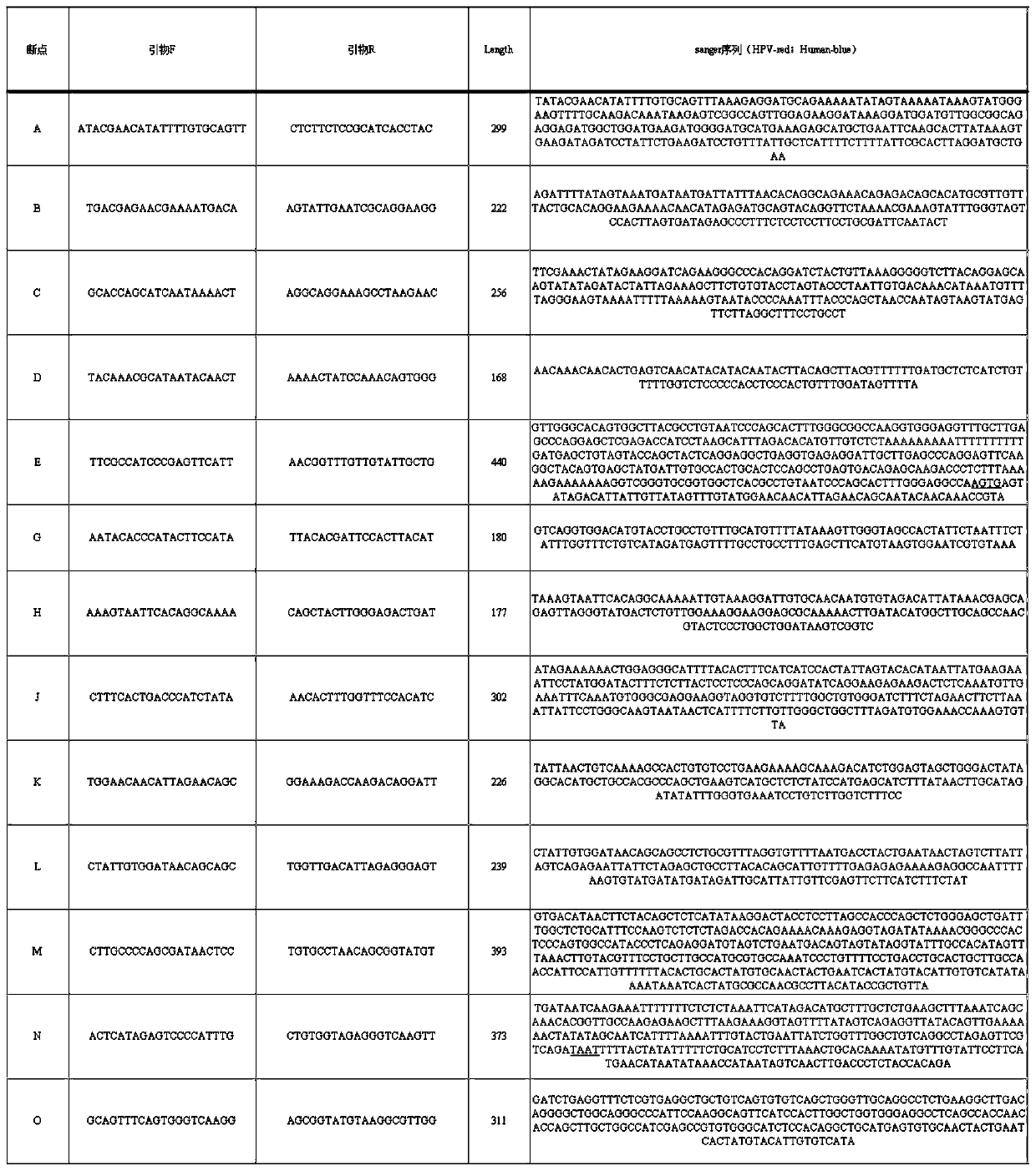
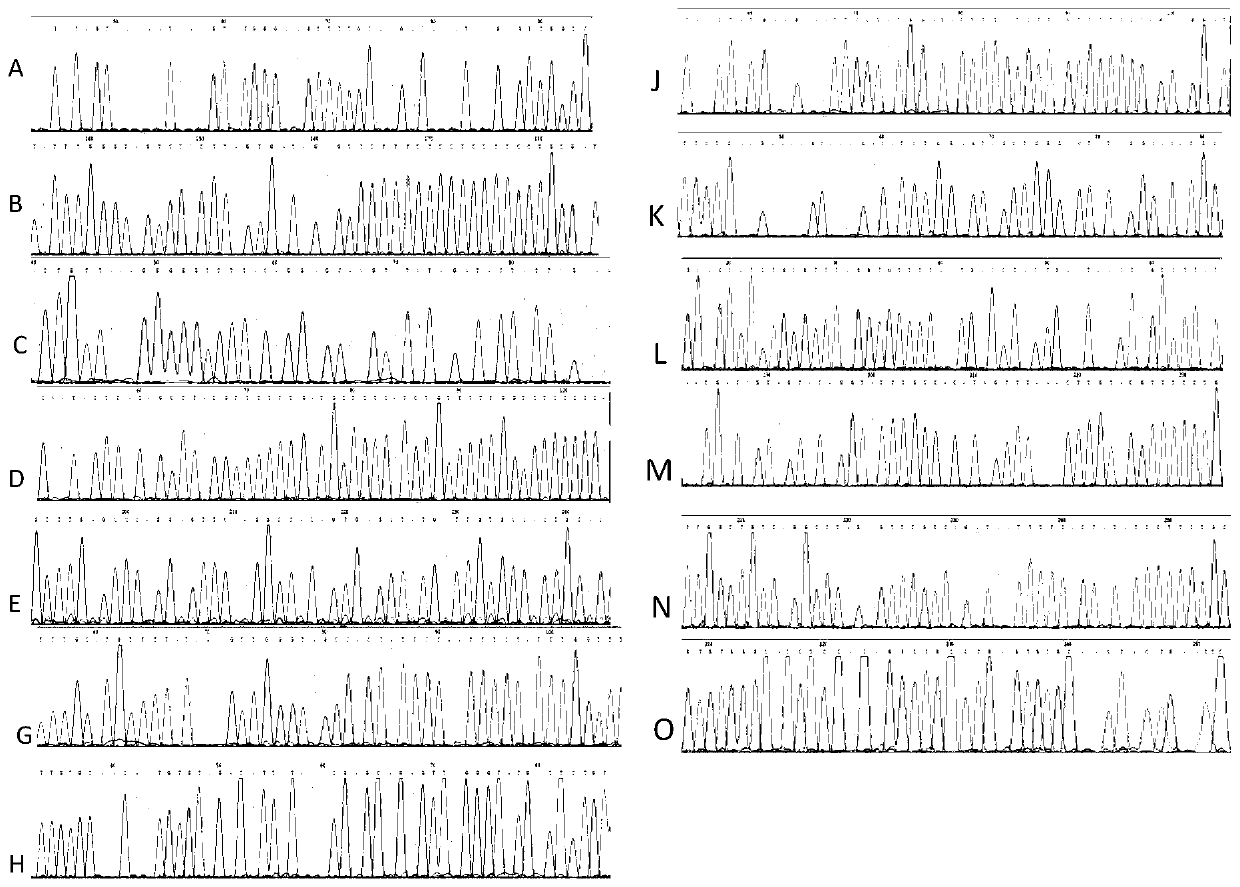
Method for detecting integration site of HPV in host genome
The invention discloses a method for detecting an integration site of HPV (human papillomavirus) in a host genome. The method comprises the following steps: extracting host genome DNA from a biological sample from a subject, and breaking the host genome DNA to fragments with a main peak of 800bp-4kb; performing a first detection on the obtained fragments by using a probe set to obtain a target sequence; preprocessing the target sequence to improve detection sensitivity; and performing a second detection on the target sequence without PCR amplification based on an electric signal sequencing technology so as to confirm the integration site of the HPV in the host genome. The method can quickly and timely analyze the state of the HPV virus existing in host cells, has obvious clinical application advantages, and can more effectively judge the integration information of the HPV without error correction analysis.
Owner:GENEIS TECH BEIJING CO LTD
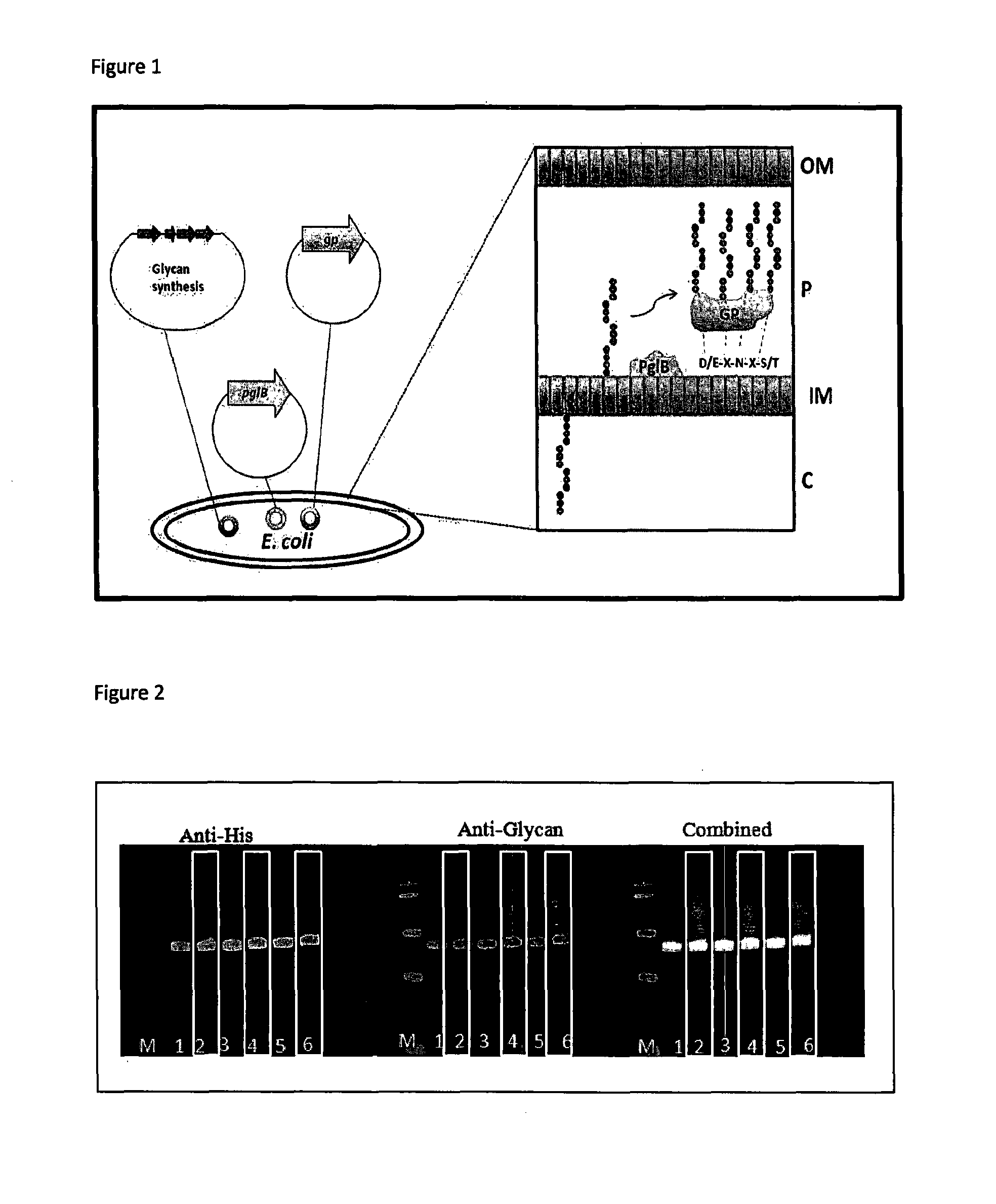
Glycosylation method
ActiveUS20150344928A1Maintain stabilityIncrease typeBacteriaMicrobiological testing/measurementBiotechnologyHost genome
The invention relates to microbial host cells engineered to produce glycoconjugate vaccines by stable integration of an acceptor protein and an oligosaccharyltransferase into the host's genome, wherein expression of the oligosaccharyltransferase is regulated.
Owner:LONDON SCHOOL OF HYGIENE & TROPICAL MEDICINE
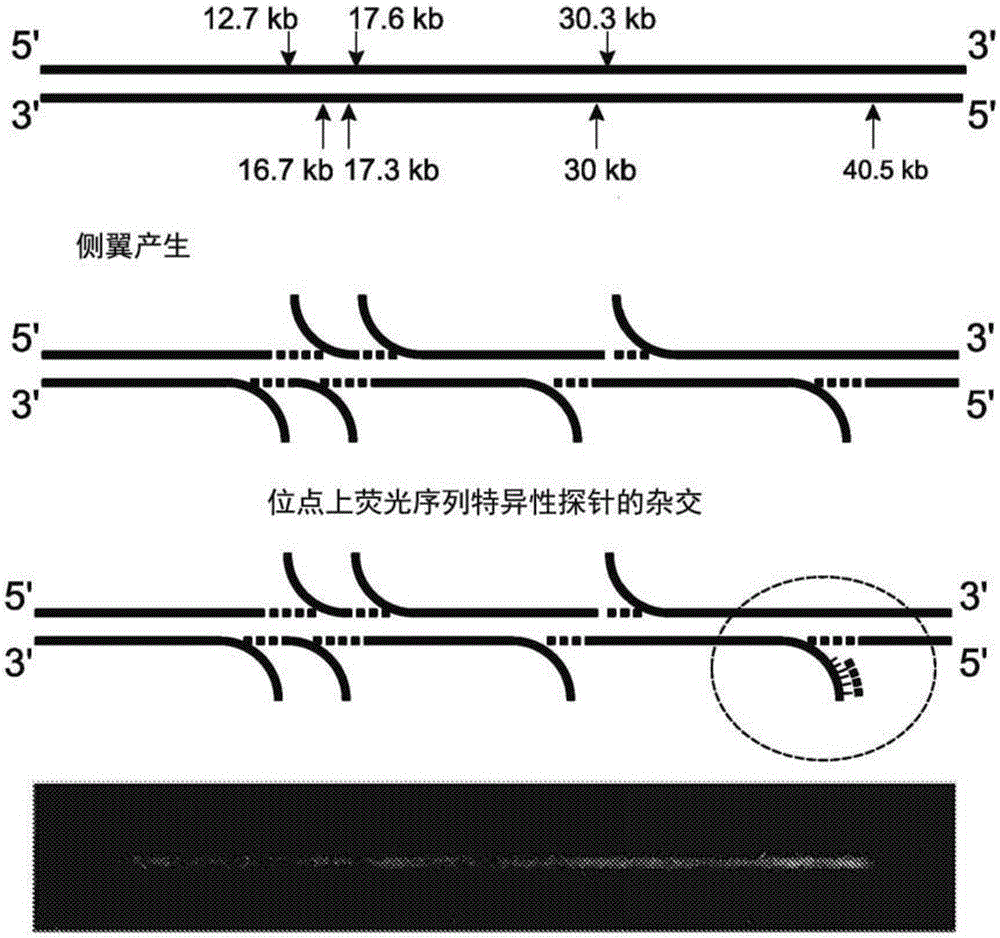
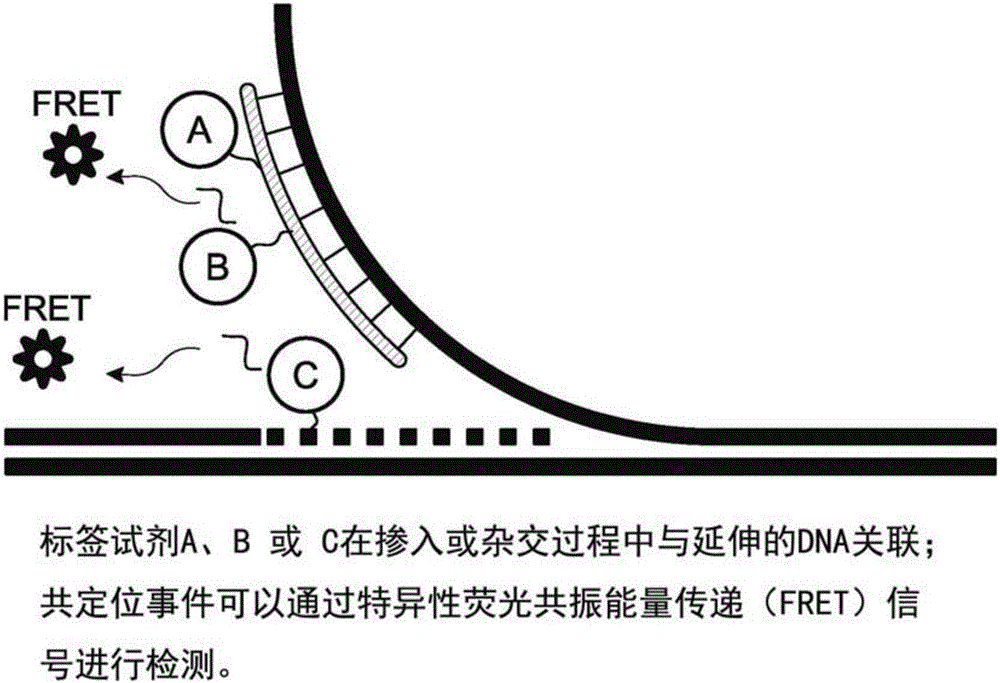
Analysis of polynucleotides
According to some embodiments herein, methods and kits for labeling and analyzing nucleic acids are provided, In some embodiments, sequence-specific labeling is performed on polynucleotide sequences associated with a host genome, and the presence or absence of patterns characteristic of extragenomic sequences are determined.
Owner:BIONANO GENOMICS
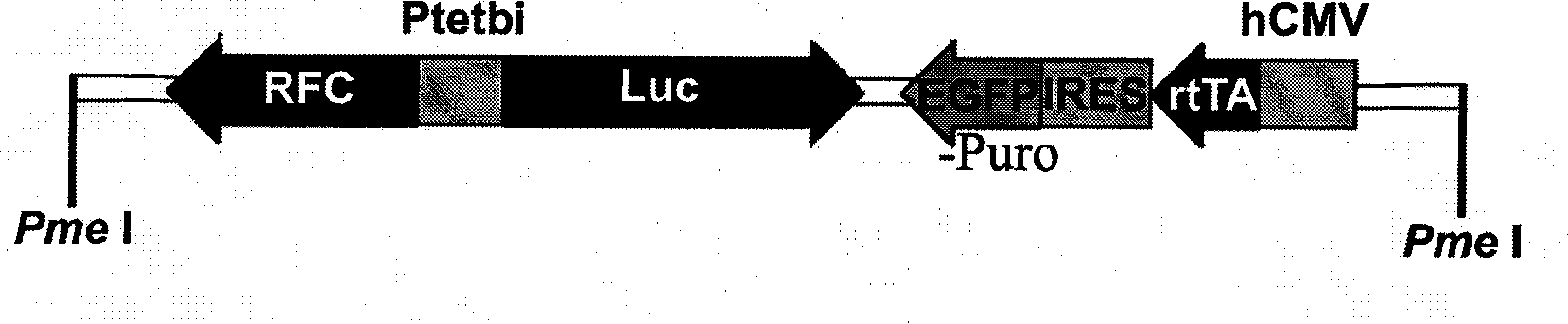
Tetracycline stringent controlled eucaryon expression vector
InactiveCN101481704ARepress transcriptionTranscription releaseVector-based foreign material introductionFluorescenceHost genome
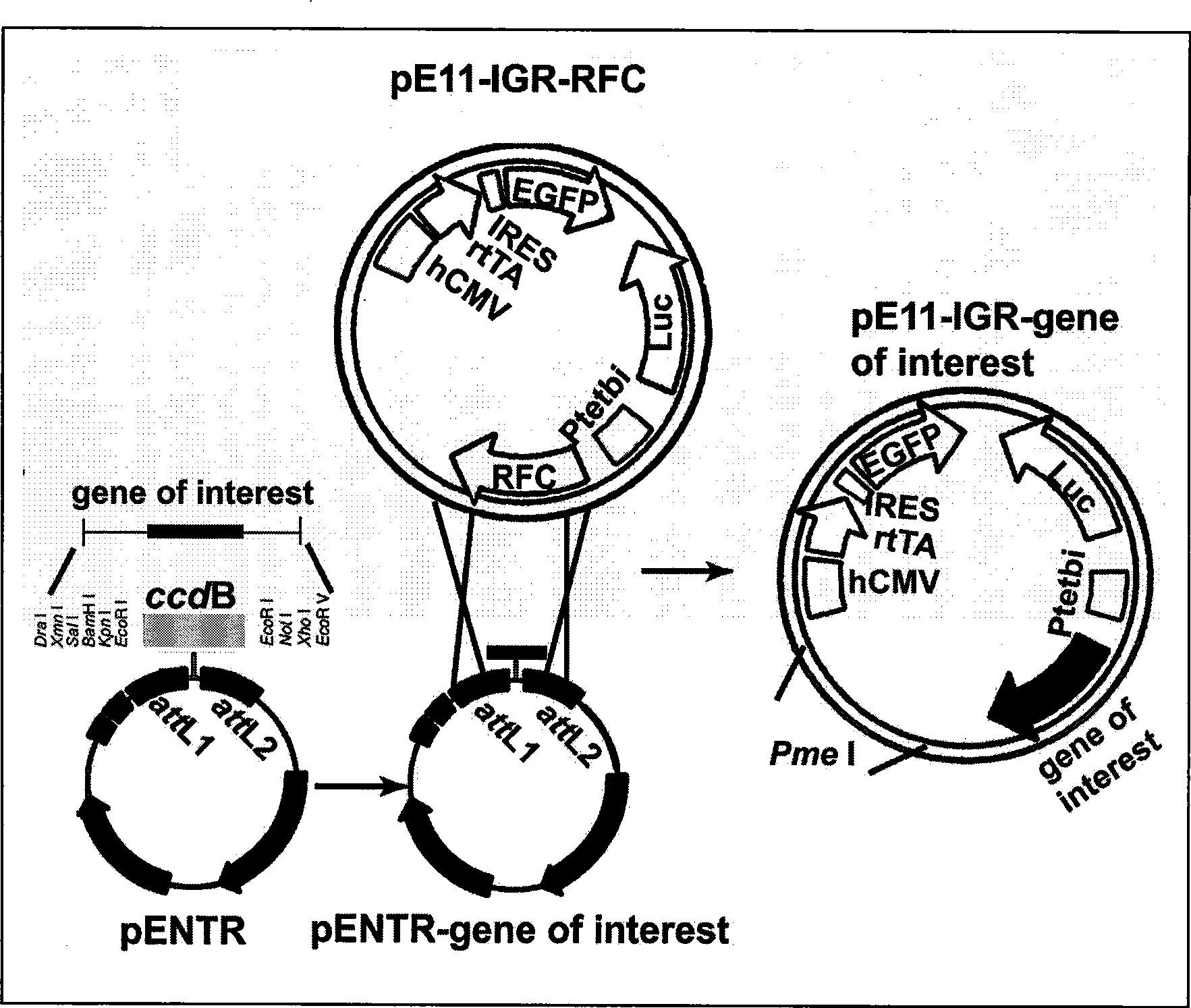
The invention provides a tetracycline strictly controlled eukaryotic expression vector. Following DNA elements including hCMV, rtTA2-M2, IRES, EGFP-Puro, element (Luc) of coding luciferase and Ptetbi-RFC are respectively introduced and are used for the expression and screening of exogenous interest genes in eukaryotic cells. The vector is strictly controlled by Tet or Dox after being transfected. And the expression level of the genes is quantitatively regulated through the amount of the added Tet or Dox. The expression level of the interest genes can be indirectly detected through the detection of the activity of Luc luciferase. The transfection efficiency of the vector can be completed through the detection of the proportion of green fluorescyte. The stably transfected cells are sorted by a flow cytometry. Thus, the expression of the interest genes is ensured to be simple and convenient. And in a stable cell line obtained through screening, the interest genes are integrated with a host genome. The tetracycline strictly controlled eukaryotic expression vector has long stable expression characteristics and advantages.
Owner:FOURTH MILITARY MEDICAL UNIVERSITY
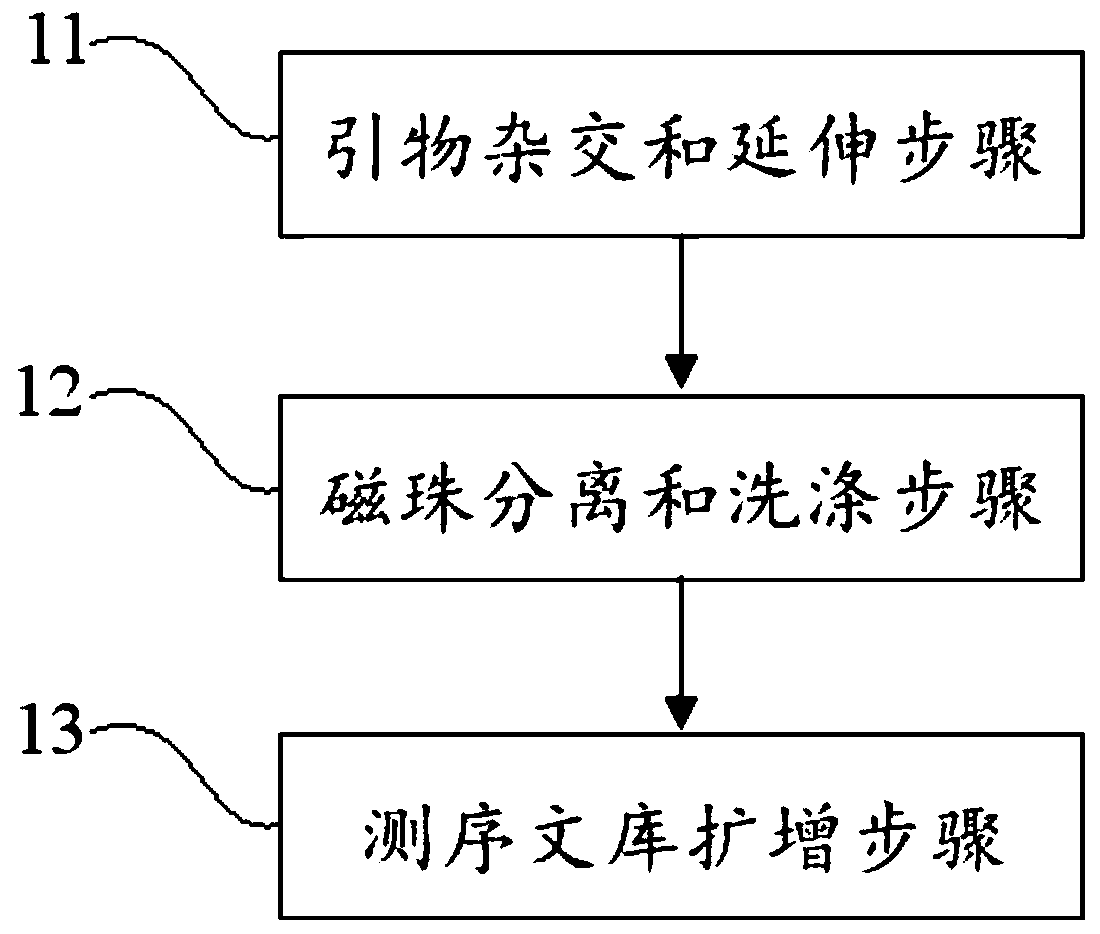
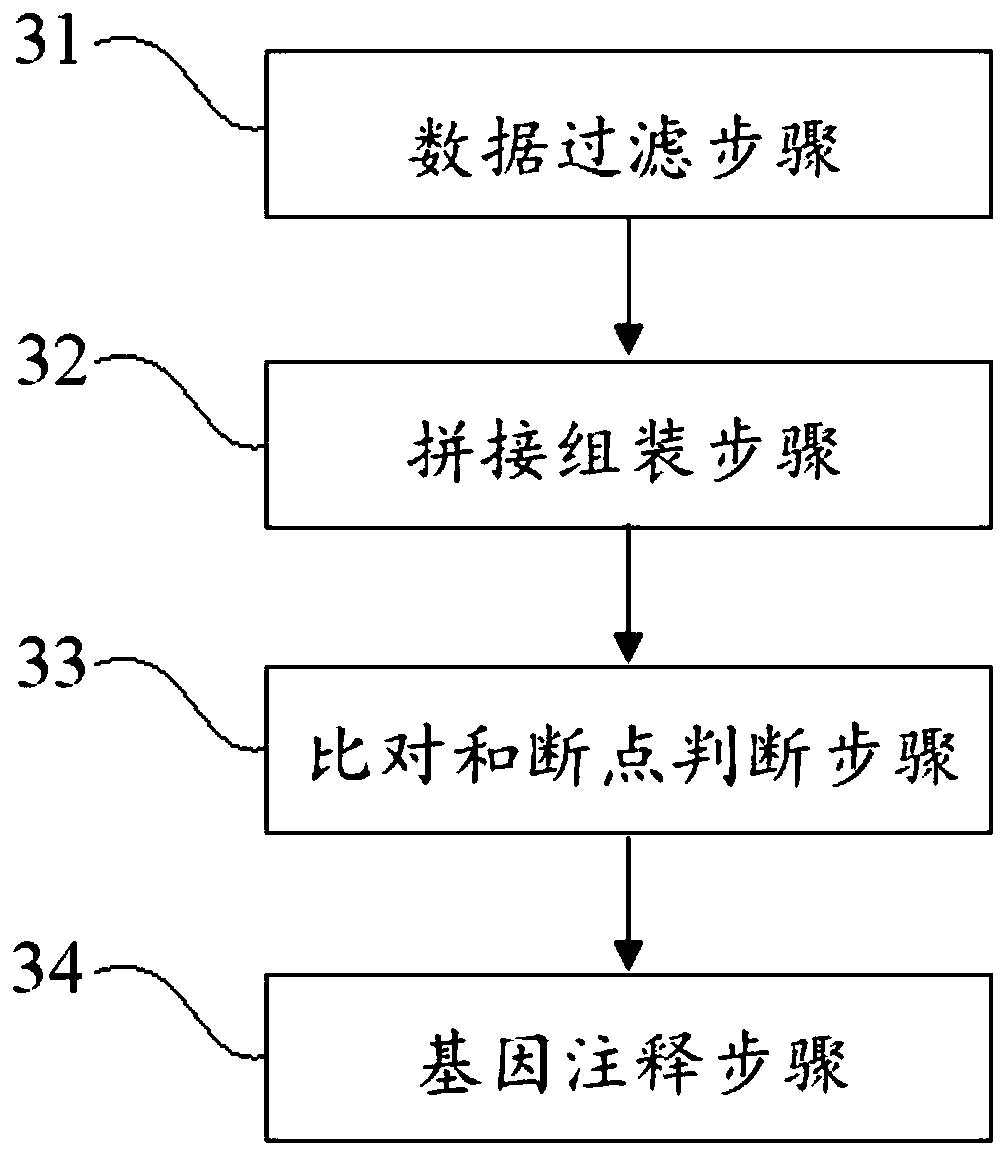
Virus integrated DNA enrichment method, sequencing data analyzing method and device
PendingCN110273028AReduce data volumeIncreased sequencing depthMicrobiological testing/measurementMicroorganism based processesMagnetic beadEnrichment methods
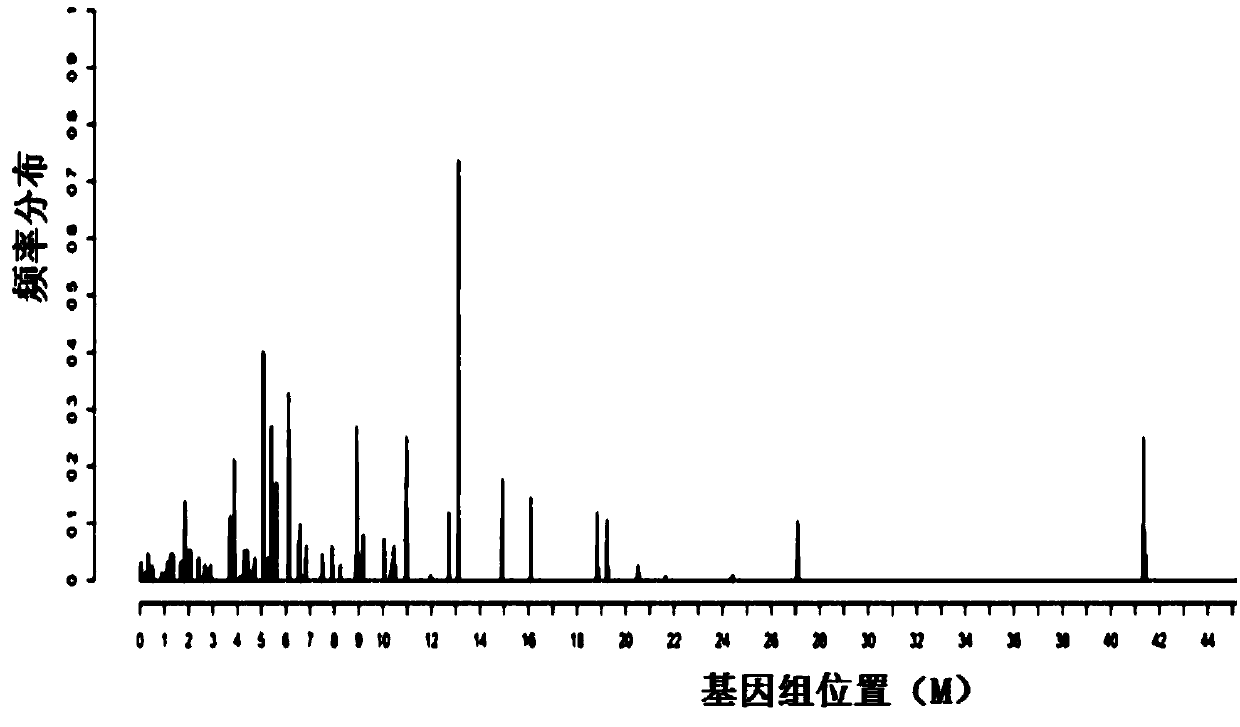
The application discloses a virus integrated DNA enrichment method, a sequencing data analyzing method and a device. The enrichment method of the application comprises the steps of primer hybridizing and extending, magnetic bead separating and washing. The sequencing data analyzing method disclosed by the application comprises the steps of data filtering, splicing and assembling, comparing and breakpoint judging and gene noting. According to the enrichment method disclosed by the application, specific primers are adopted for extending virus genome target genes so as to replace a long probe, so that the cost is low, the efficiency is high, the specificity is high, and the enrichment time is shortened. Compared with a complete genome, the sequencing data analyzing method disclosed by the application can save data amount, can obtain higher sequencing depth, can obtain more virus DNA breakpoint information and integration position information on the host genome, and can obtain clonal integration events supported by more reads, and besides, can detect non-clonal integration supported by single reads, so as to construct a comprehensive prospect integrated on the host genome.
Owner:SHENZHEN HAPLOX BIOTECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com