Method for detecting integration site of HPV in host genome
A technology that integrates sites and genomes. It is applied in biochemical equipment and methods, and the determination/inspection of microorganisms. It can solve problems such as application limitations and user selection ability to correct differences in software results.
Active Publication Date: 2019-12-03
GENEIS TECH BEIJING CO LTD
View PDF3 Cites 5 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Although there are many error correction methods, such as correcting the third generation according to the sequencing results of the second generation and the third generation, or performing multiple self-corrections on the third generation results to reduce the error rate, due to the difference in the results of the correction software and the limitations of user selection capabilities , which greatly restricts its application
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment
[0056] This example is used to illustrate the method of the present invention.
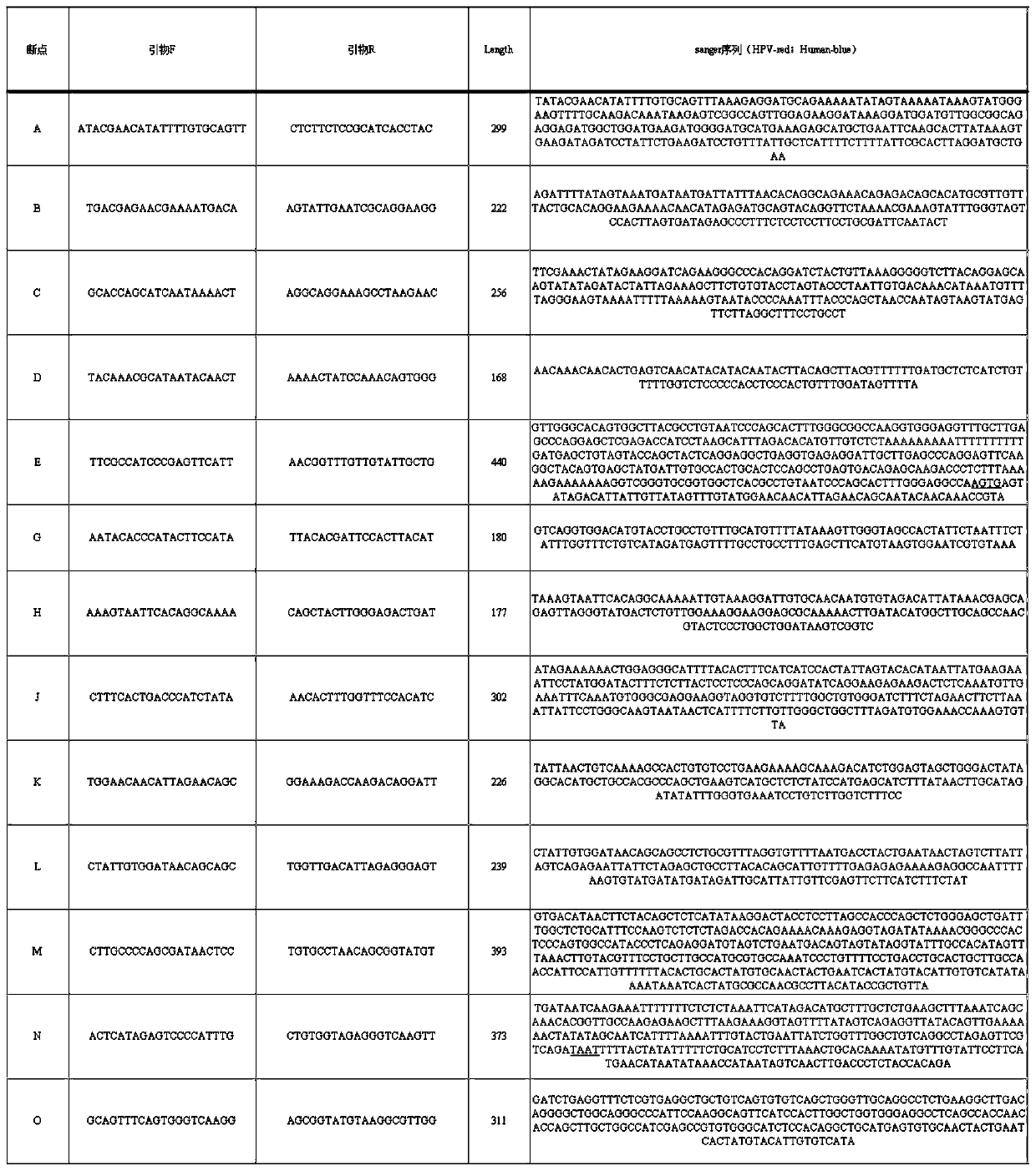
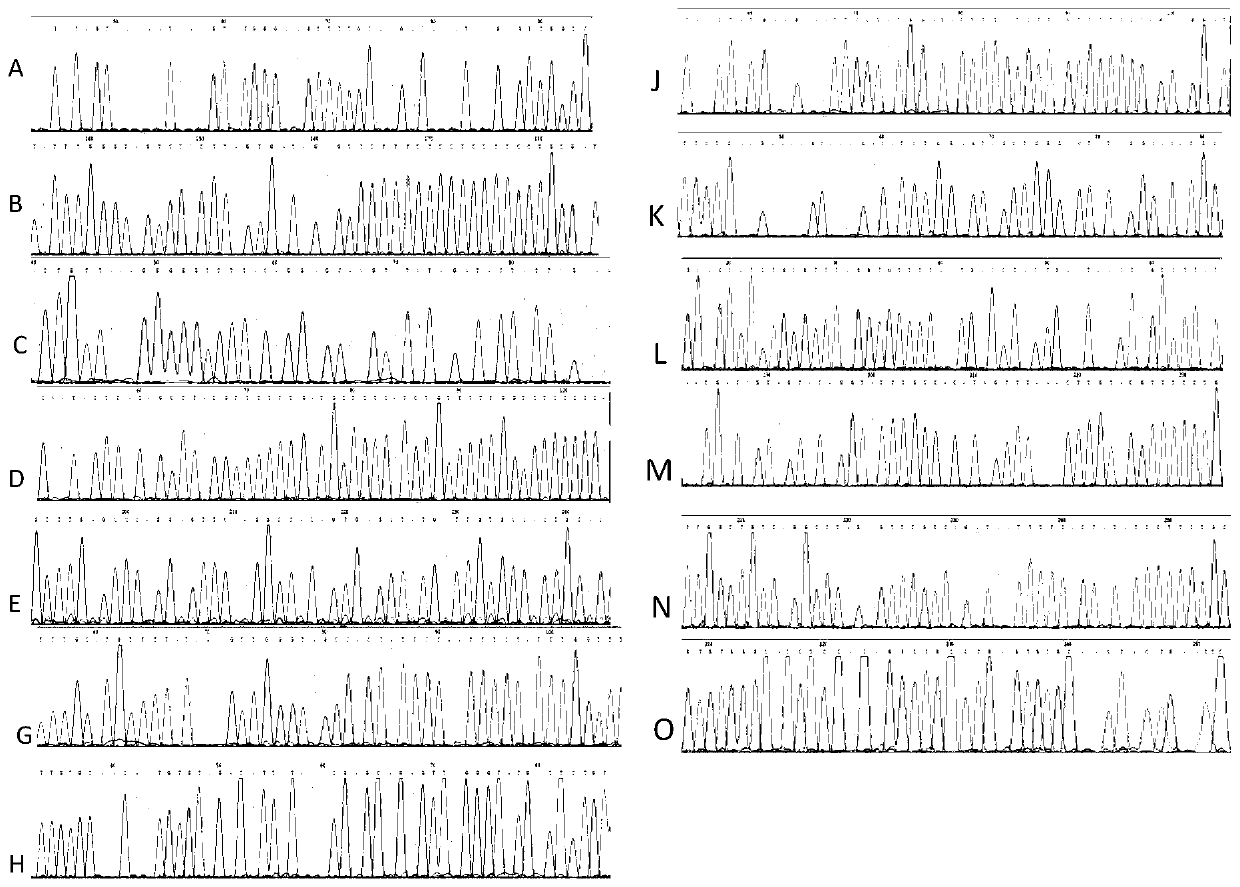
[0057] 1. Sample information
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
The invention discloses a method for detecting an integration site of HPV (human papillomavirus) in a host genome. The method comprises the following steps: extracting host genome DNA from a biological sample from a subject, and breaking the host genome DNA to fragments with a main peak of 800bp-4kb; performing a first detection on the obtained fragments by using a probe set to obtain a target sequence; preprocessing the target sequence to improve detection sensitivity; and performing a second detection on the target sequence without PCR amplification based on an electric signal sequencing technology so as to confirm the integration site of the HPV in the host genome. The method can quickly and timely analyze the state of the HPV virus existing in host cells, has obvious clinical application advantages, and can more effectively judge the integration information of the HPV without error correction analysis.
Description
technical field [0001] The invention relates to the detection of virus integration sites, in particular to a method for detecting HPV integration sites in host genomes. Background technique [0002] The integration of HPV virus into the human genome is an important process event that causes cancer. At present, the understanding of this process is still relatively limited. More and more studies have shown that HPV integration is a potential molecular marker for molecular diagnosis and individualized treatment, and it is also very important. valuable prognostic indicator. The research methods for HPV integration are currently obtained by using the probe capture method based on next-generation sequencing technology. For example, CN107739761A discloses a high-throughput sequencing detection method for HPV typing and integration. This method selects the genes of the current HPV subtypes, combined with the second-generation high-throughput sequencing technology, to detect the ty...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Applications(China)
IPC IPC(8): C12Q1/6869C12Q1/70
CPCC12Q1/6869C12Q1/708C12Q2525/191C12Q2565/631C12Q2537/165
Inventor 孟博田埂王伟伟董如一杨文娟
Owner GENEIS TECH BEIJING CO LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com