ARG1, ARG2, ARG3, HIS1, HIS2, HIS5, HIS6 genes and methods for stable genetic integration
a technology of stable genetic integration and arg2, which is applied in the field of new genes, can solve the problems of auxotrophic strains of i>p. pastoris /i>suffering the disadvantage of gene reversion
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
General Materials and Methods
[0114]Escherichia coli strain DH5α (Invitrogen, Carlsbad, Calif.) was used for recombinant DNA work. P. pastoris strain YJN165 (ura5) (Nett and Gerngross, Yeast 20: 1279-1290 (2003)) was used for construction of yeast strains. PCR reactions were performed according to supplier recommendations using ExTaq (TaKaRa, Madison, Wis.), Taq Poly (Promega, Madison, Wis.) or Pfu Turbo® (Stratagene, Cedar Creek, Tex.). Restriction and modification enzymes were from New England Biolabs (Beverly, Mass.).
[0115]Yeast strains were grown in YPD (1% yeast extract, 2% peptone, 2% dextrose and 1.5% agar) or synthetic defined medium (1.4% yeast nitrogen base, 2% dextrose, 4×10−5% biotin and 1.5% agar) supplemented as appropriate. Plasmid transformations were performed using chemically competent cells according to the method of Hanahan (Hanahan et al., Methods Enzymol. 204: 63-113 (1991)). Yeast transformations were performed by electroporation according to a modified procedu...
example 2
Cloning of P. pastoris ARG1, ARG2, ARG3, HIS1, HIS2, HIS5, and HIS6 Genes
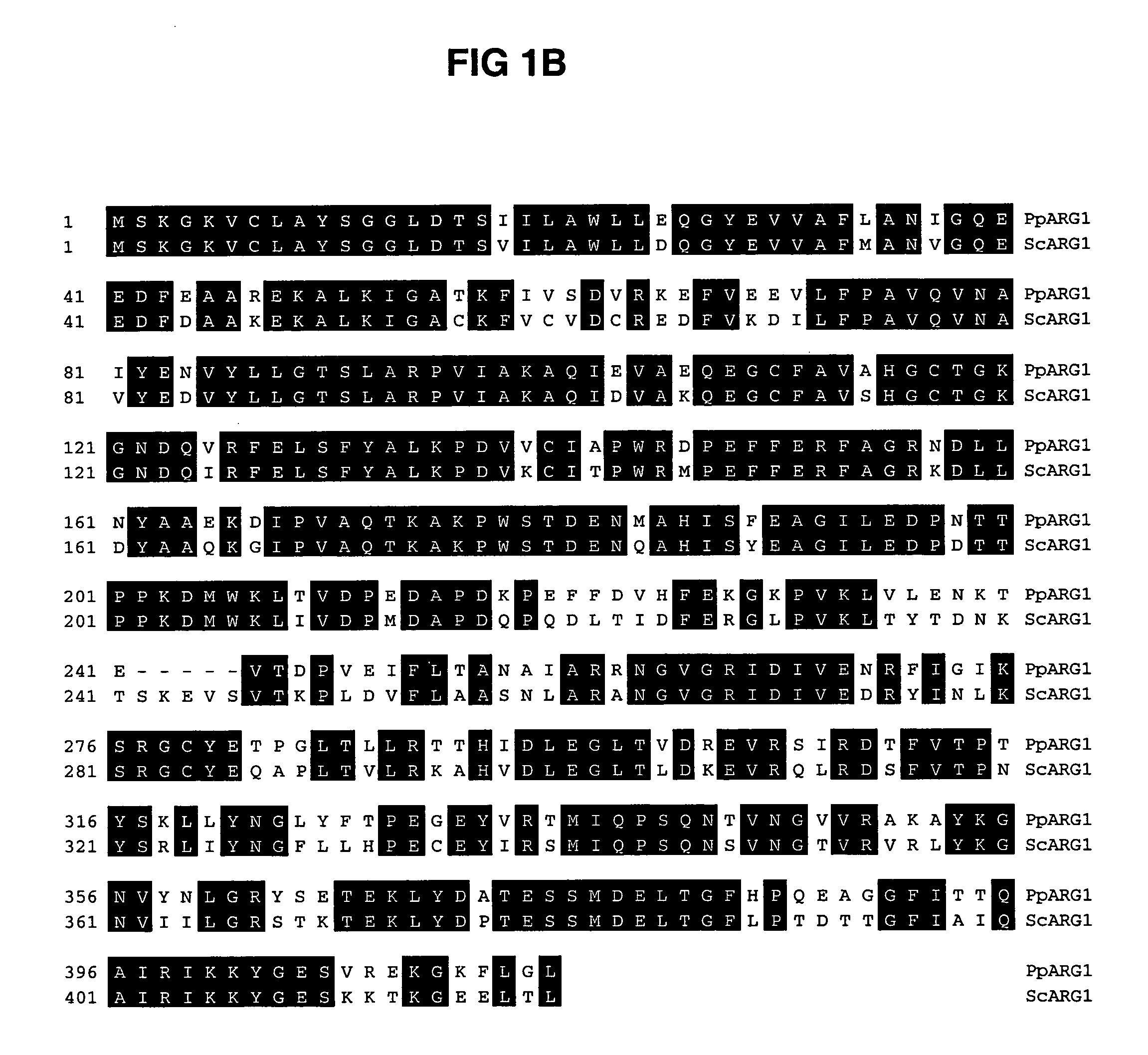
[0117]The P. pastoris orthologues to the S. cerevisiae ARG1, ARG2, ARG3, HIS1, HIS2, HIS5 and HIS6 genes were cloned by comparison of the translations of the respective S. cerevisiae open reading frames to a partial P. pastoris genomic sequence provided by Integrated Genomics Inc. using the BLAST program (Altschul et al., J. Mol. Biol. 215: 403-410 (1990)). Using the S. cerevisiae ARG1 ORF (SEQ ID NO: 3) as bait we identified a P. pastoris ORF of 416 amino acids (SEQ ID NO: 2) with 72.9% identity (FIG. 1). The ARG2 orthologue encoded an ORF of 590 amino acids (SEQ ID NO: 5) with 29.8% identity to ScARG2 (SEQ ID NO: 6) (FIG. 2). The ARG3 orthologue encoded an ORF of 342 amino acids (SEQ ID NO: 8) with 57.3% identity to ScARG3 (SEQ ID NO: 9)(FIG. 3). Using the ORF encoded by ScHIS1 (SEQ ID NO: 12) as bait we identified a P. pastoris gene sequence consisting of two exons of 51 and 843 nucleotides separated by an i...
example 3
Construction of Disruption Vectors and Strains
[0118]All disruption vectors were derived from plasmid pJN653 (FIG. 8), which was constructed using a method previously described for pJN267 (Nett and Gerngross, Yeast 20: 1279-1290 (2003)). For amplification of the PpGAPDH promoter and the PpCYC1 transcriptional terminator, the oligos GAP5 clean (SEQ ID NO:22), GAP3 clean (SEQ ID NO:23), CYC5 clean (SEQ ID NO:24) and CYC3 clean (SEQ ID NO:25) were used to amplify genomic DNA from P. pastoris strain NRRL Y-11430 for the GAPDH promoter and S. cerevisiae strain W303 for the CYC1 terminator. The plasmid consists of two fragments of the 5′ and 3′ regions of the PpKEX1 gene, flanking the GAPDH / CYC1 expression cassette and a ScURA3-auxotrophic marker cassette which was isolated as a BamHI / BglII fragment from pNKY51 (ATCC). The restriction sites flanking all segments allow for the convenient replacement to generate disruption vectors for the gene of choice. To generate disruption vectors for th...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Tm | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
| resistance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com