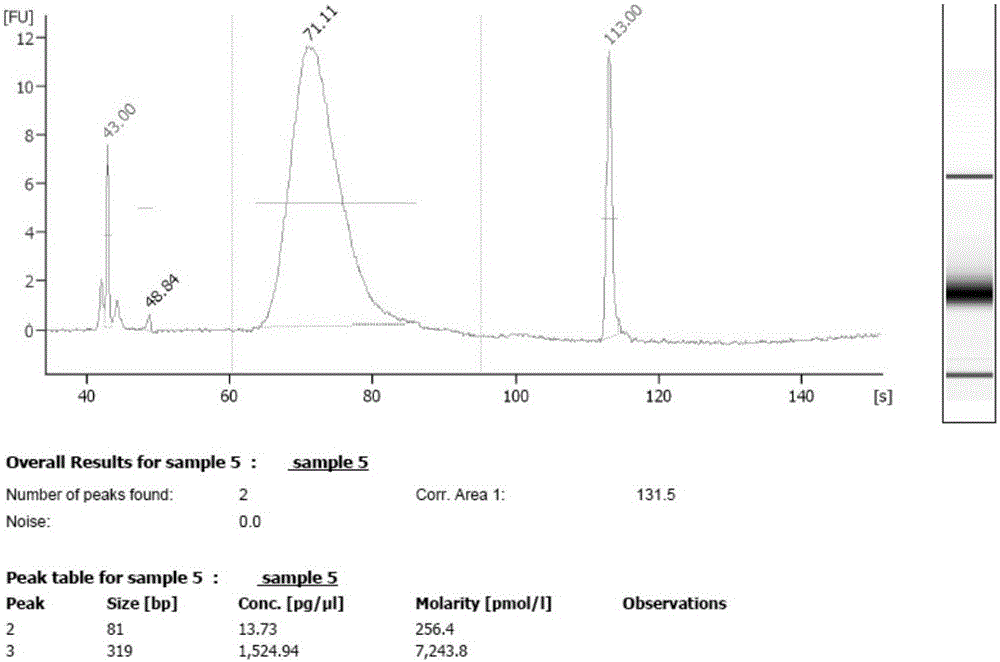
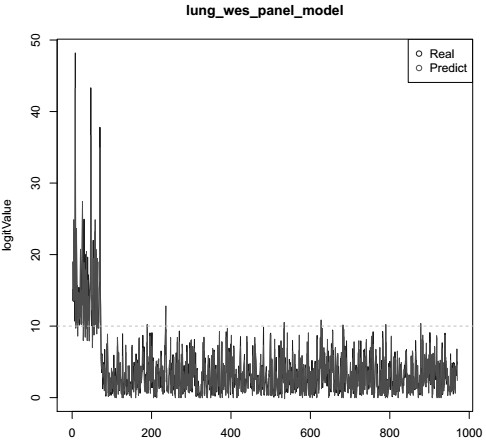
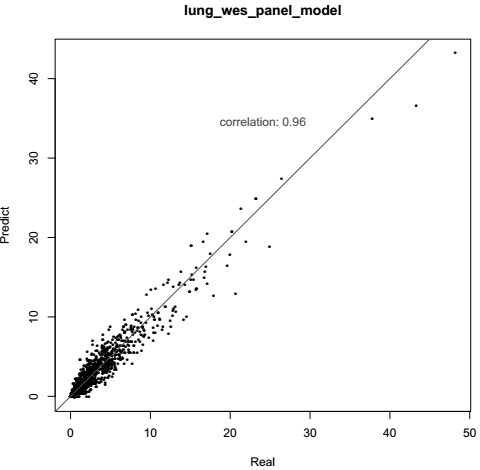
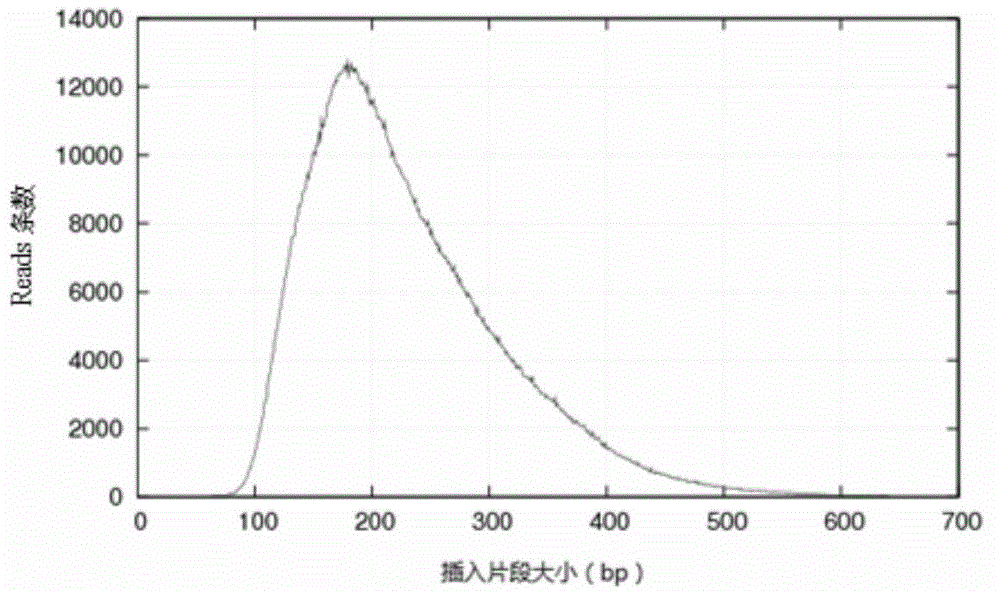
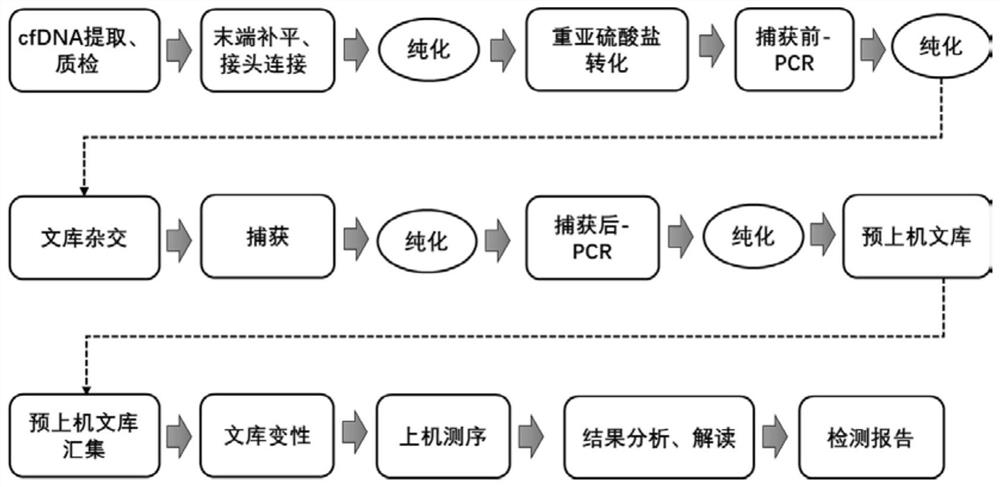
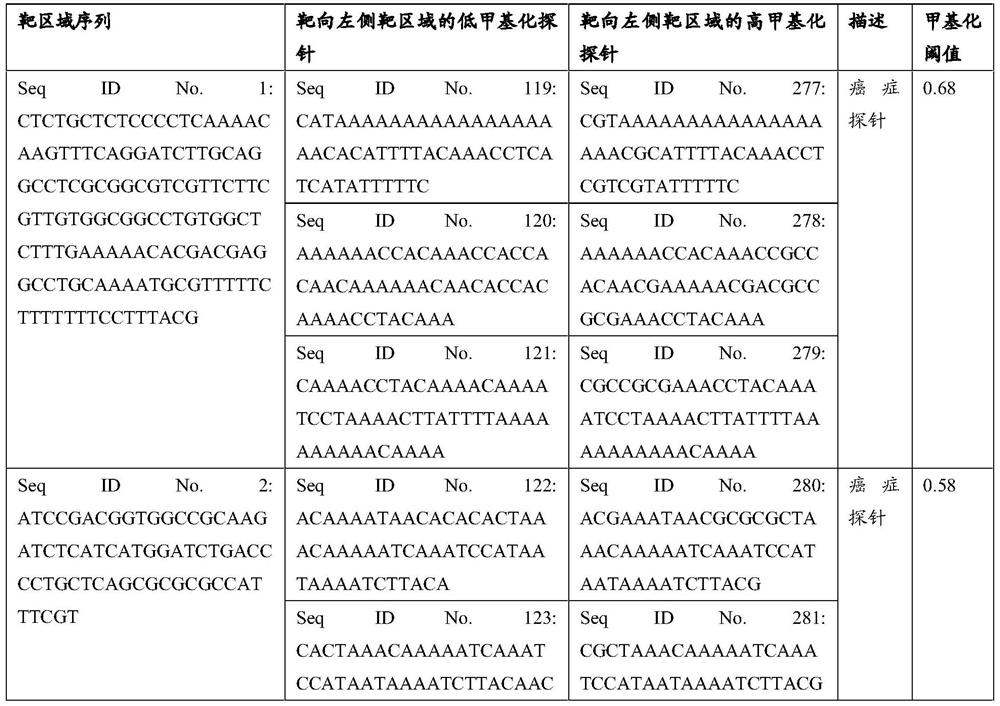
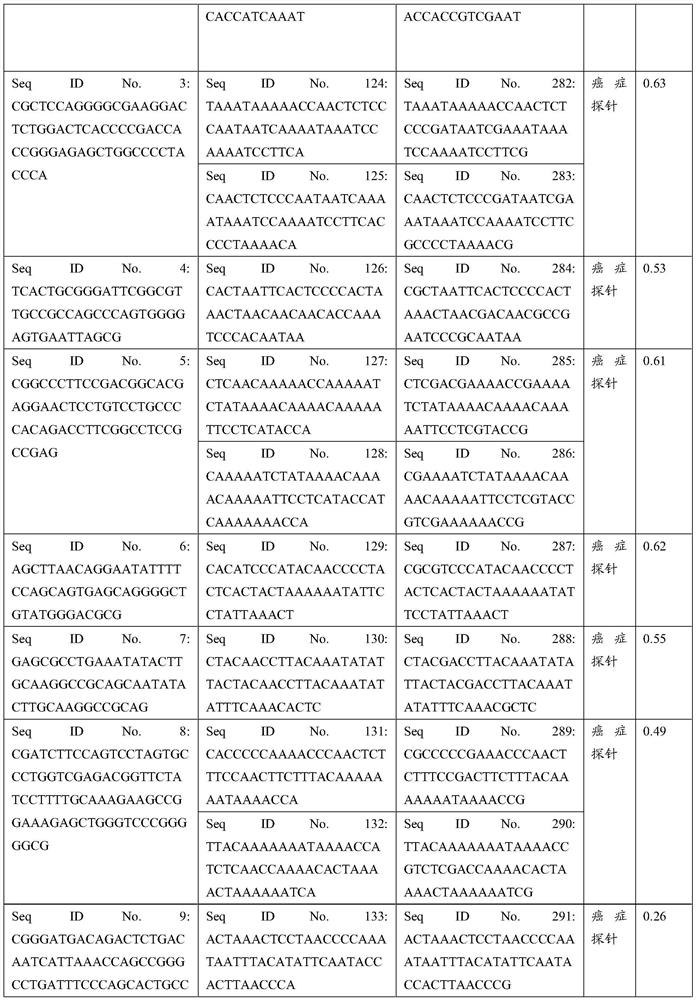
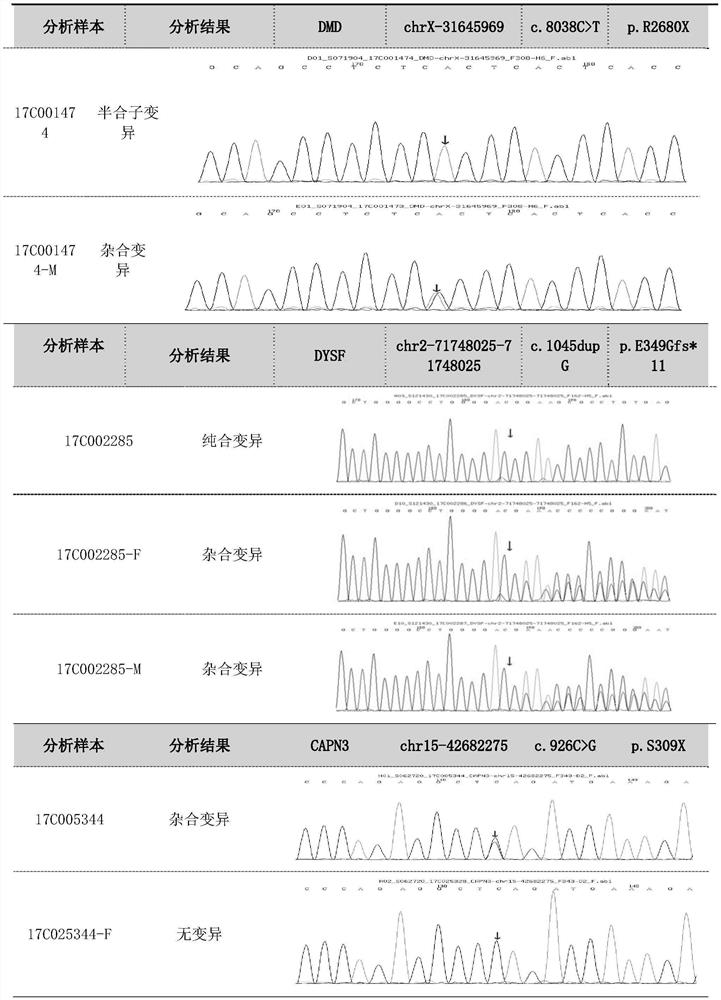
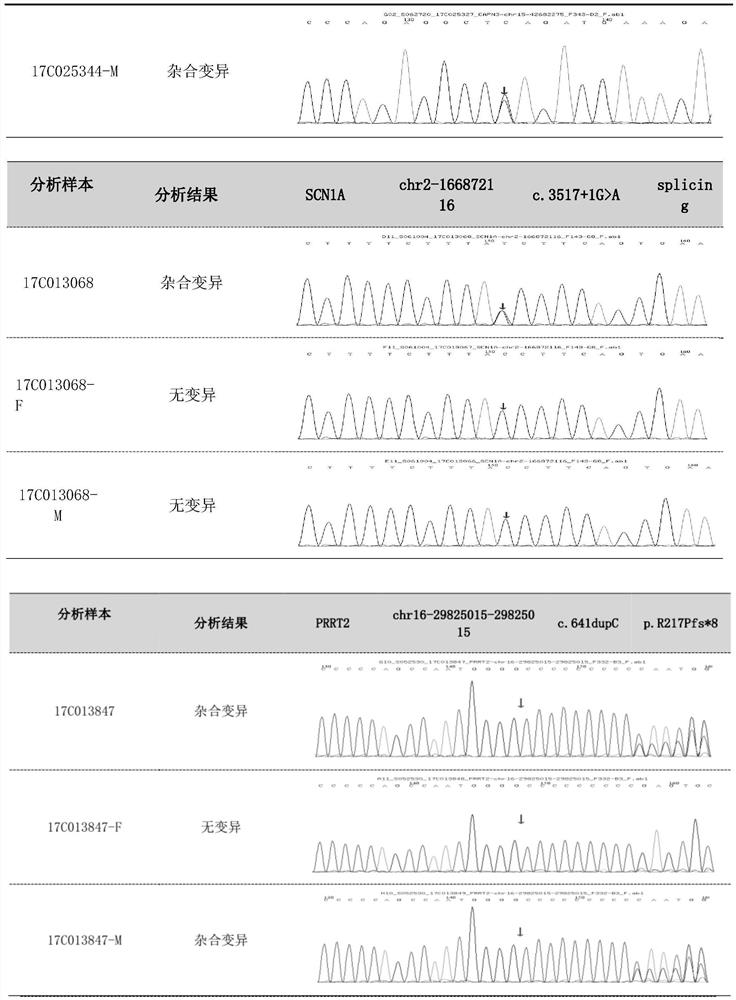
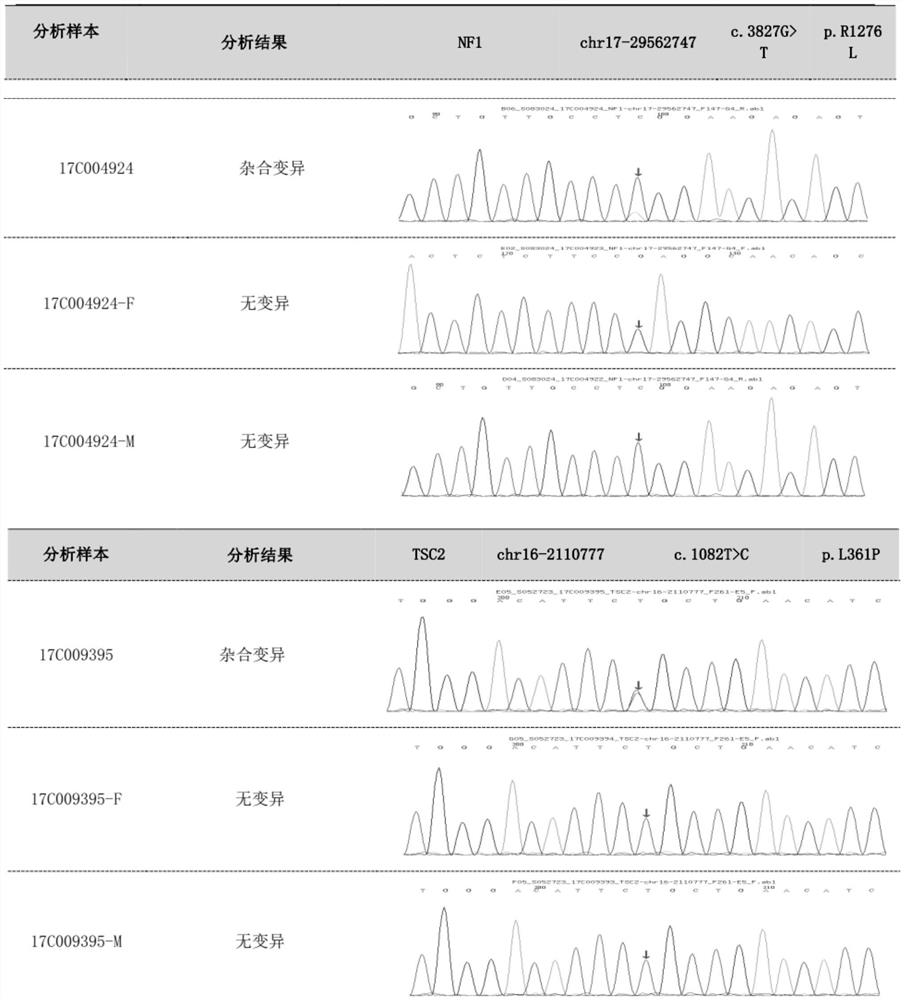
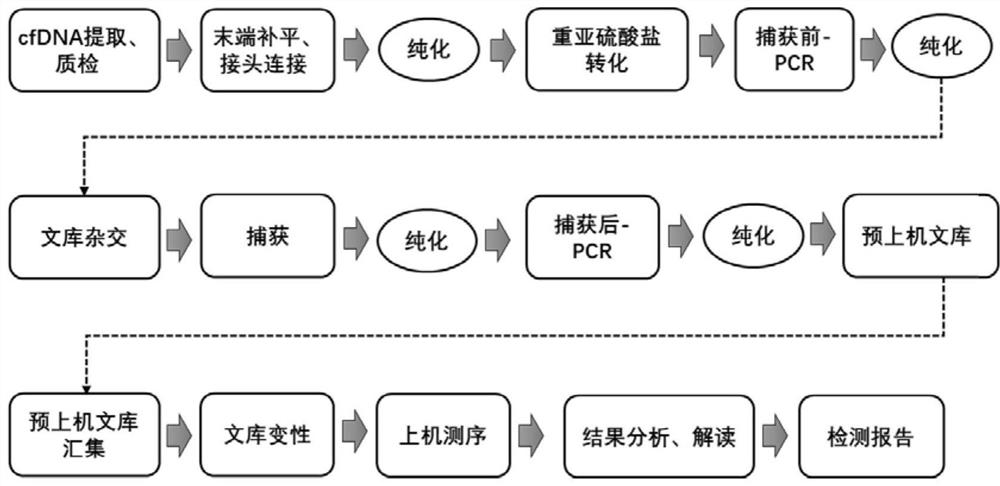
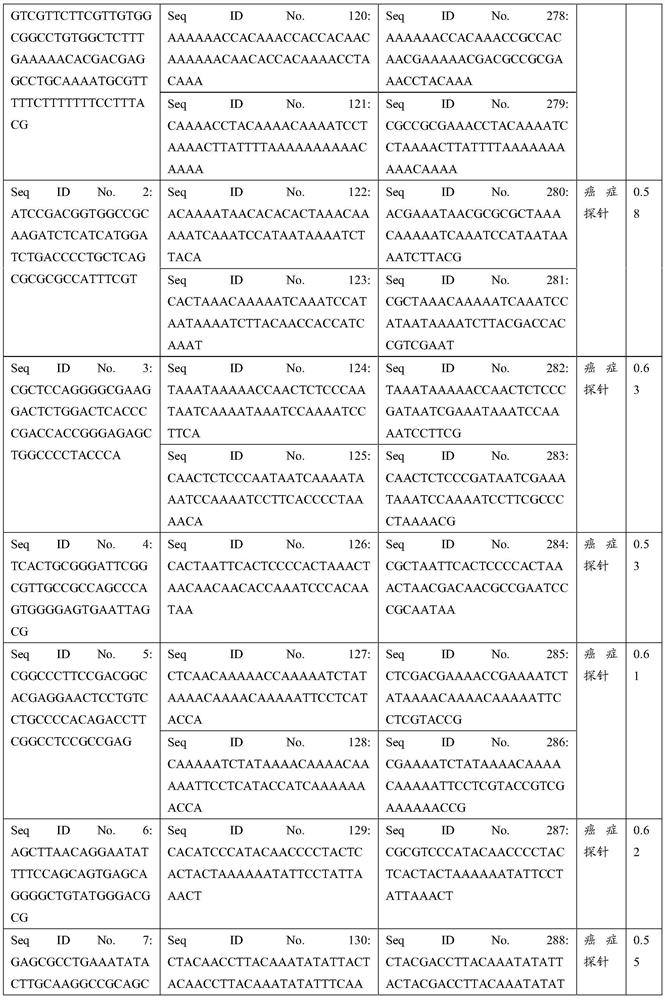
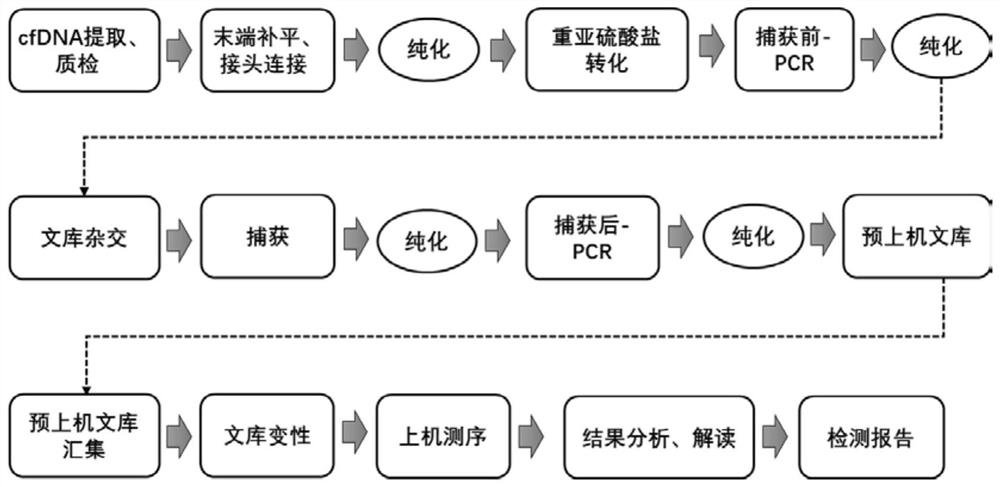
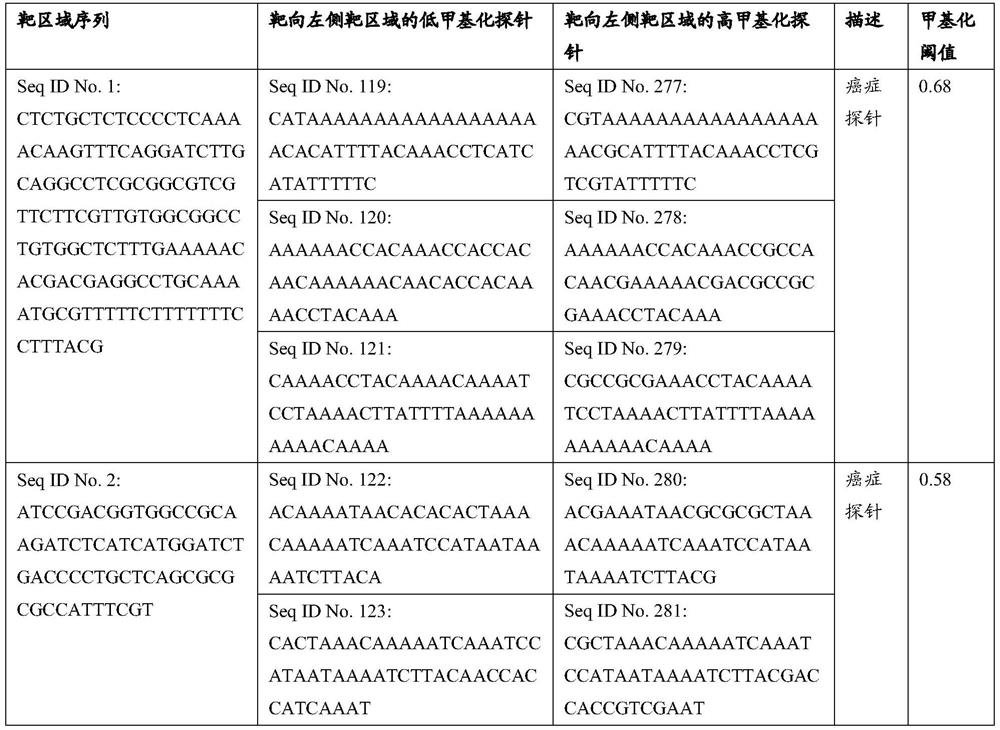
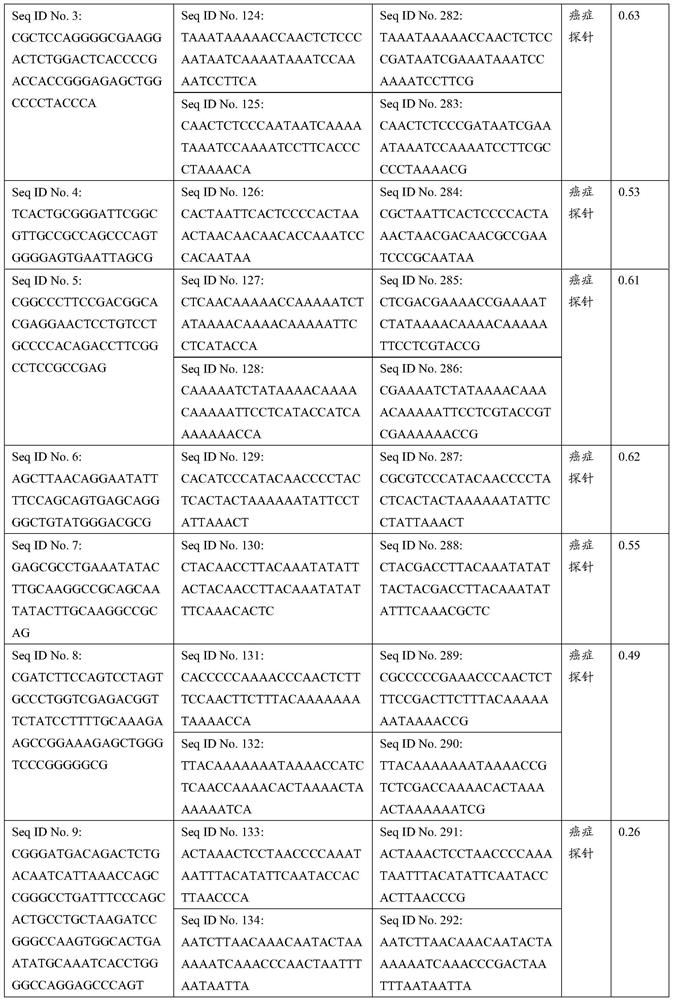
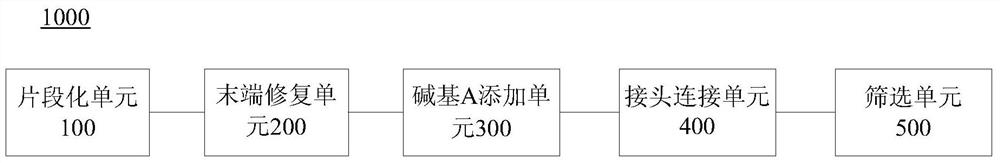
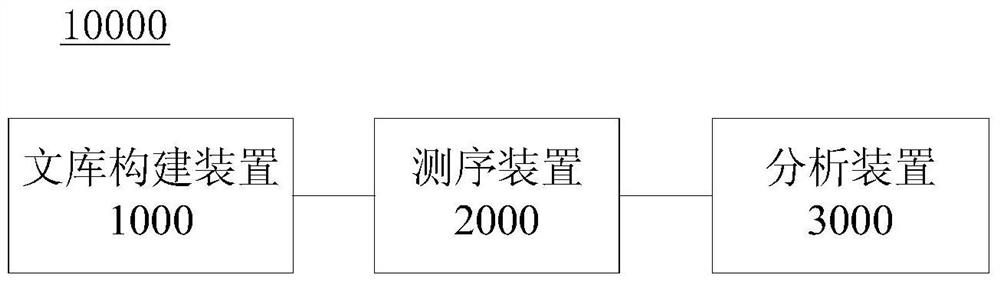
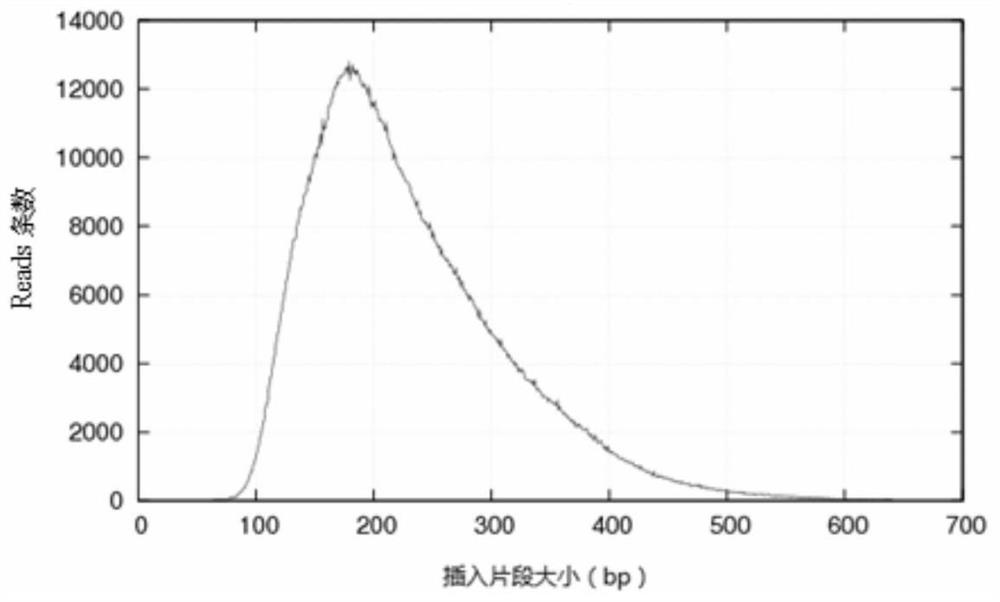
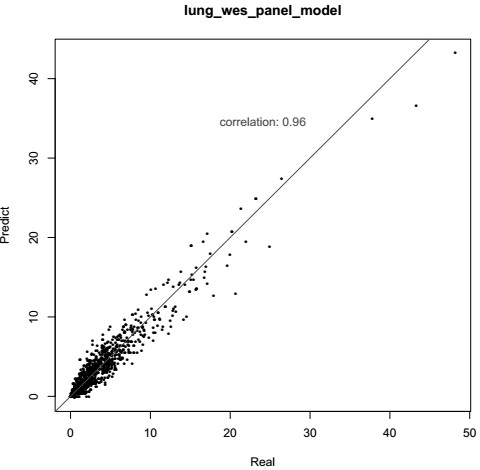
Patents
Literature
39results about How to "Increased sequencing depth" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Method and device for detecting tumor mutational load by using high-throughput sequencing data
InactiveCN108588194AStrong specificityIncreased sensitivityBioreactor/fermenter combinationsBiological substance pretreatmentsMutation detectionA-DNA
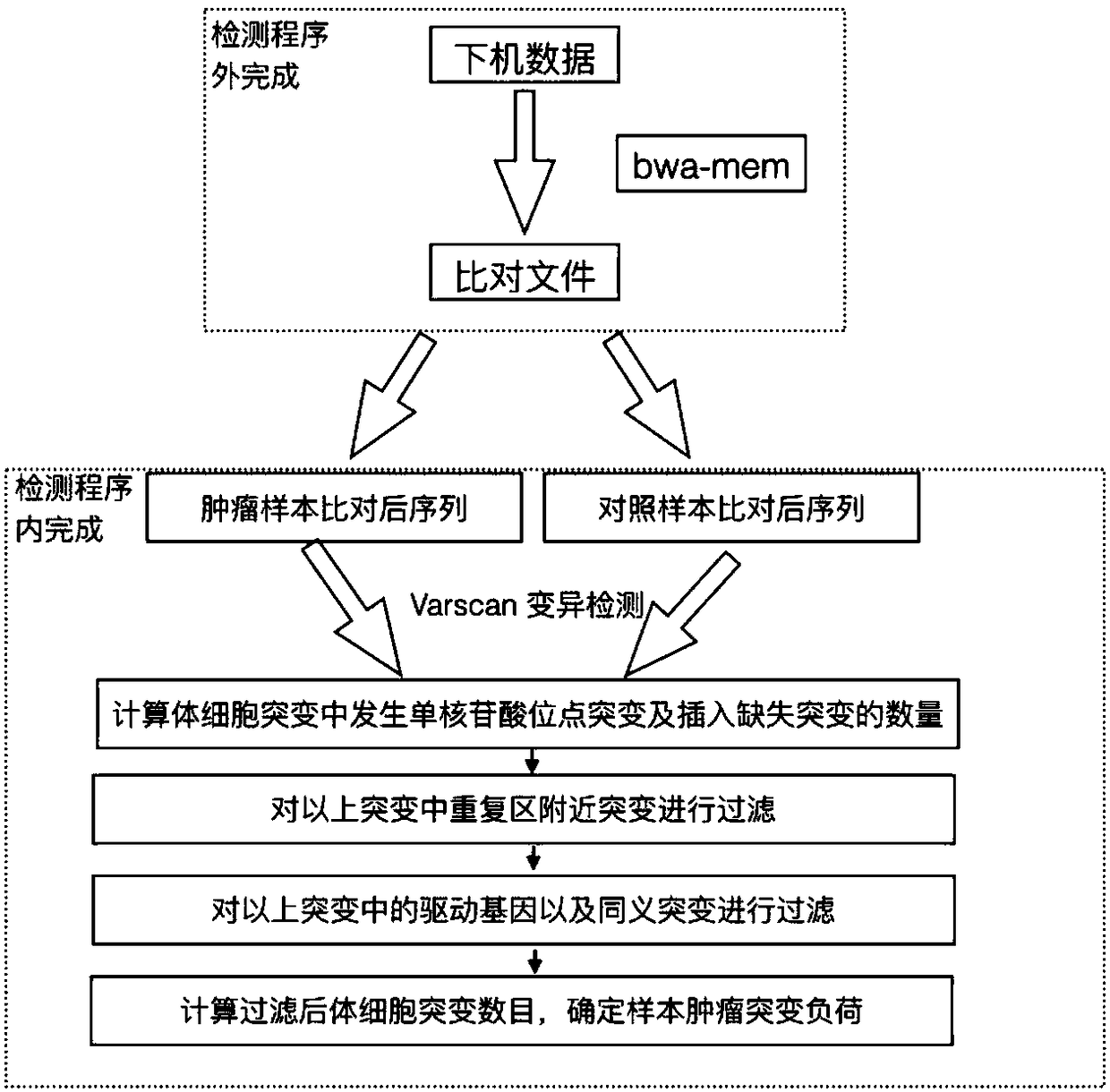
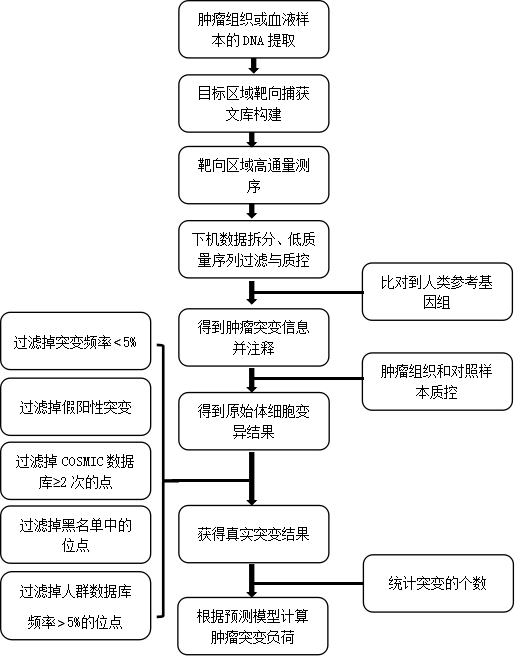
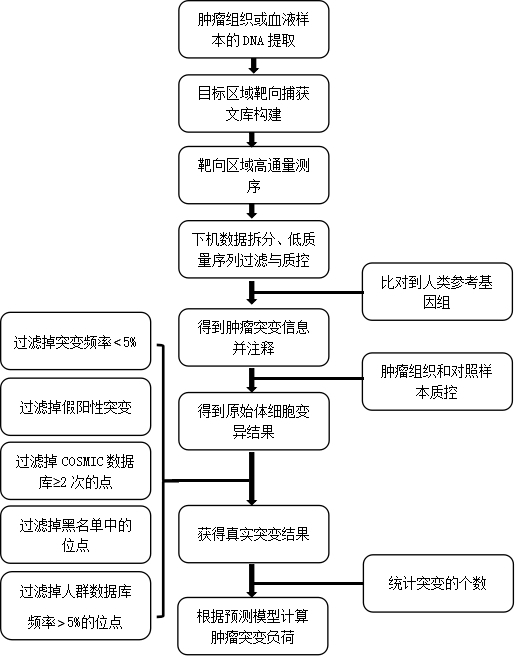
The invention discloses a method and device for detecting a tumor mutational load by using high-throughput sequencing data. The method comprises the steps that a to-be-detected sample is pretreated and a DNA is extracted; according to a target area capture principle, a probe is used for capturing tumor related genes; sequencing is carried out through a high-throughput method to obtain sequencing information; low-quality sequences are filtered out, a checkout process is used for detection, the checkout process comprises that a high-throughput sequencing sequence is compared to a reference genome by using a comparison software, a non-compared sequence forms a soft truncation, according to the compared position, sorting is carried out, and an index is established; mutation detection is carried out on a compared sequence, the detected mutation is annotated by using a RMSK database, a repetitive area is removed on the annotation, driver genes and same sense mutation are filtered, and the number of the filtered somatic cell mutations is calculated to determine the sample tumor mutational load. By applying the technical scheme, the detection precision is higher, and the effect is better.
Owner:天津诺禾致源生物信息科技有限公司
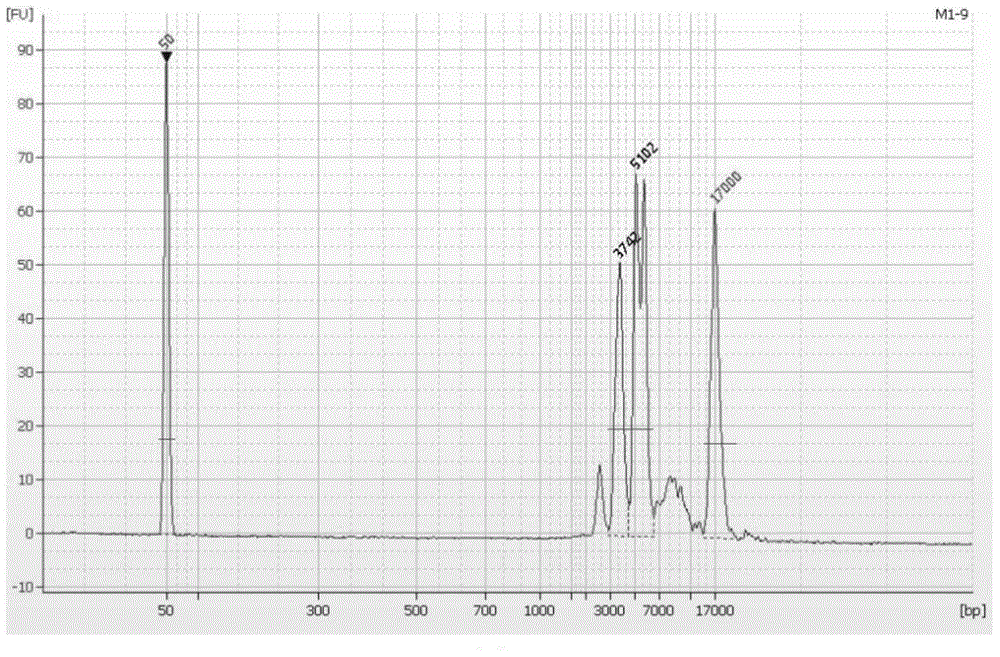
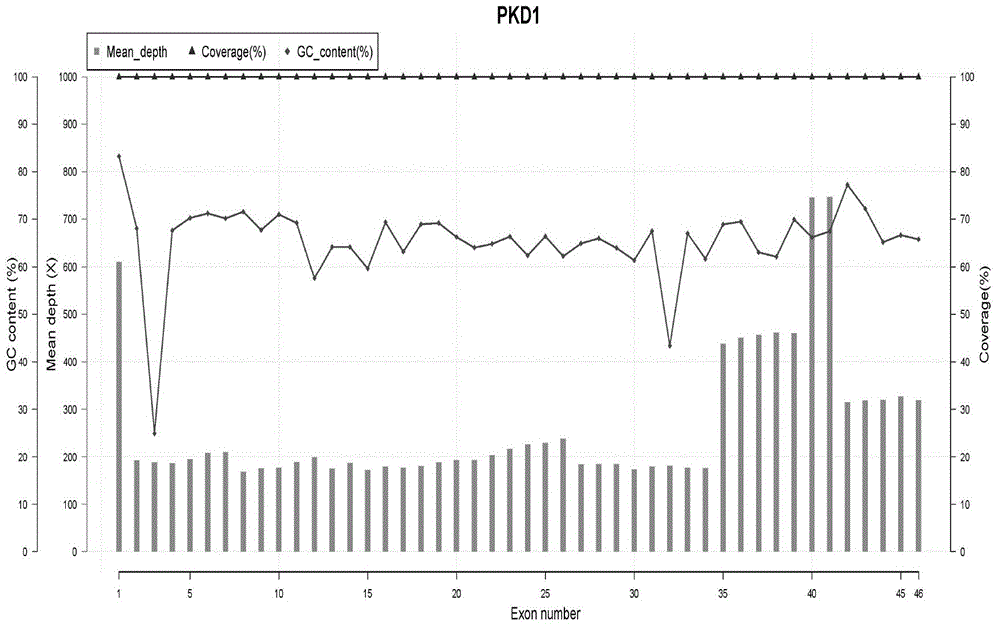
Amplimers, kit and method for detecting PKD1 gene mutation
ActiveCN104975081AAchieve separate markingAchieving Parallel SequencingMicrobiological testing/measurementDNA/RNA fragmentationPKD1Pcr ctpp
The present invention relates to a method for detecting PKD1 gene mutation. The method utilizes sequence information of the PKD1 gene, designs nine pairs of PCR tag primers, and uses a long-chain PCR technology for targeting amplification of PKD1 gene; while amplification products are labeled, PCR products are purified and mixed for direct construction of a single SMRTBell library; a PacBio RSII sequencer is employed for sequencing; and bioinformatic analysis is carried out to obtain PKD1 gene mutation information. The invention has the beneficial effects that a simplified pair of primers is designed and combined with tag primer technology to realize respective marking of PCR products of multiple DNA samples; the amplified PCR products do not need to perform knockout and can be directly used for the library construction, and the library construction does not need extra PCR, so that a plurality of samples are mixed into one library and processed simultaneously by a PacBio RSII sequencing library construction link, and the experimental operation is greatly simplified.
Owner:NANJING MATERNITY & CHILD HEALTH CARE HOSPITAL
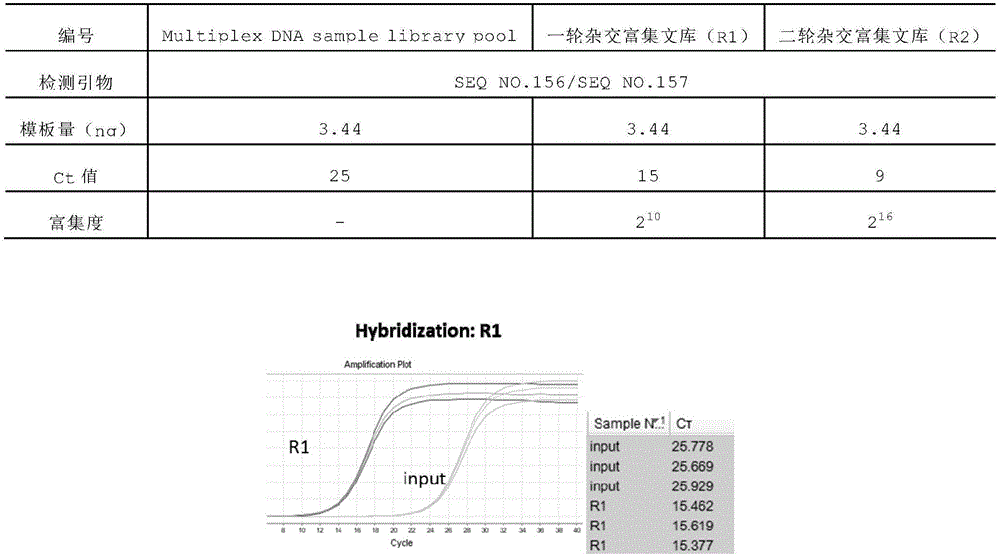
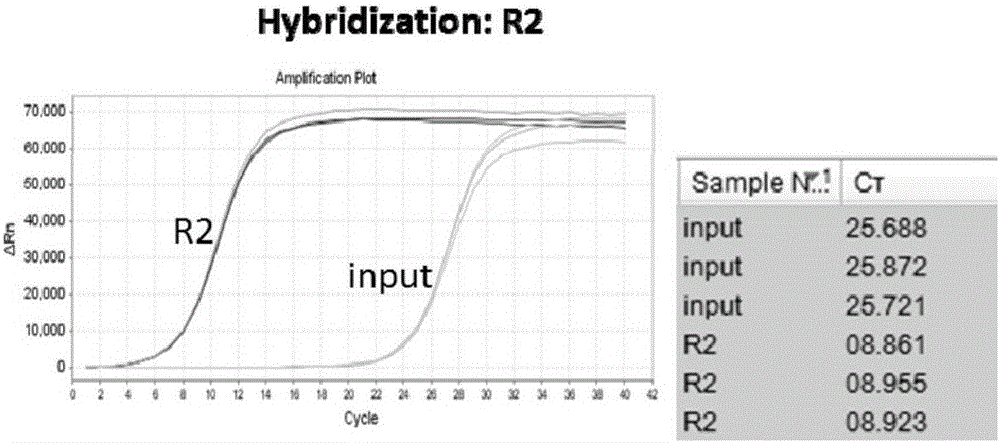
Hybrid capture kit and method for detecting mutation of breast cancer susceptibility genes BRCA1 and BRCA2
ActiveCN106676169AImplement mutation detectionLow costMicrobiological testing/measurementBreast cancer susceptibility genesMutation detection
The invention discloses a hybrid capture kit for detecting mutation of breast cancer susceptibility genes BRCA1 and BRCA2. The hybrid capture kit comprises a hybridization reagent, a PCR amplification reagent and an enrichment degree detection reagent, wherein the hybridization reagent comprises probe mixtures from SEQ NO.1 to SEQ NO. 173, and the mole ratio is SEQ NO.1: SEQ NO. (2-172): SEQ NO. 173=1: 1: 1. During detection, the use amount of each of the probe mixtures with the concentration of 2nM is 1 [mu]l. The kit disclosed by the invention can be used for high-sensitivity, high-flux, low-cost and easy-operation full-explicit mutation detection for the genes BRCA1 and BRCA2, and can assist a clinical doctor in screening out individuals with the breast cancer susceptibility risk to fulfill the aim of preventing occurrence of breast cancers.
Owner:SHANGHAI PERSONAL BIOTECH
Mitochondrial genome library based on high-throughput sequencing and building method thereof
ActiveCN105907748AGuaranteed Accurate AmplificationSensitive and accurate mutation detectionNucleotide librariesMicrobiological testing/measurementHuman DNA sequencingHomologous sequence
The invention relates to a mitochondrial genome library based on high-throughput sequencing and a building method thereof. The building method comprises the following steps of S1, performing one-step PCR (polymerase chain reaction) amplification of mitochondria full-length DNA (deoxyribonucleic acid) in total DNA; S2, crushing the mitochondria full-length DNA to obtain DNA fragments; S3, performing tail end restoration on the DNA fragments to obtain flat tail end DNA fragments; S4, adding A to the end 3' of the flat tail end DNA fragments to obtain the A-added DNA fragments; S5, adding a connector to the end 3' of the A-added DNA fragments to obtain connector-added DNA fragments; S6, performing front LM-PCR on the connector-added DNA fragments; S7, performing post PCR on the LM-PCR product. The building method of the mitochondrial genome library has the advantages that the mitochondrial full-length DNA is obtained through further PCR amplification from the total DNA, so that the precise amplification of the human mitochondrial genome can be guaranteed; the pollution mixing by the homologous sequence of the human genome is avoided, so that the mutation detection is more sensitive and accurate.
Owner:GUANGZHOU JIAJIAN MEDICAL TESTING CO LTD
Combination tag, combination tag joint, and application ofcombination tagand combination tag joint
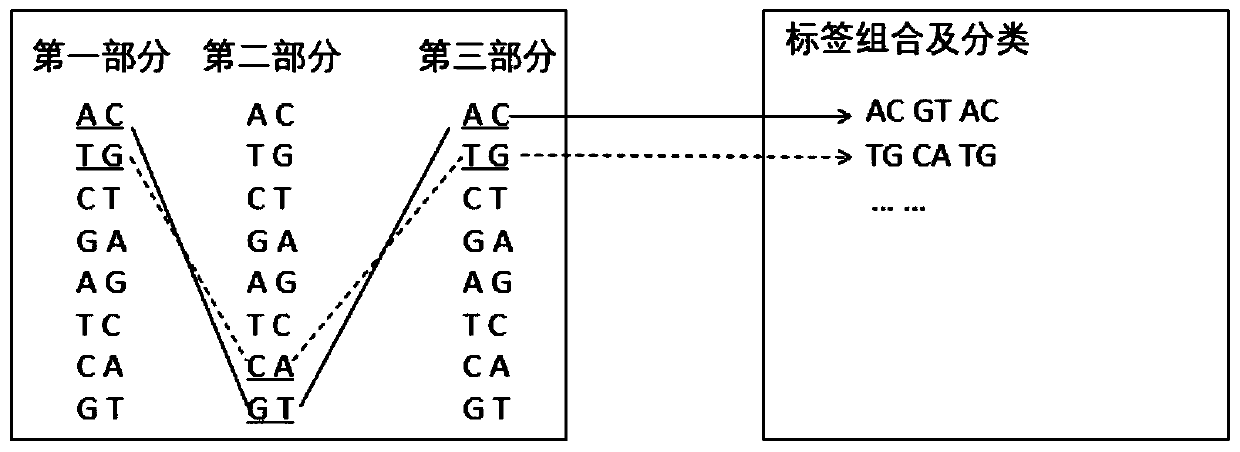
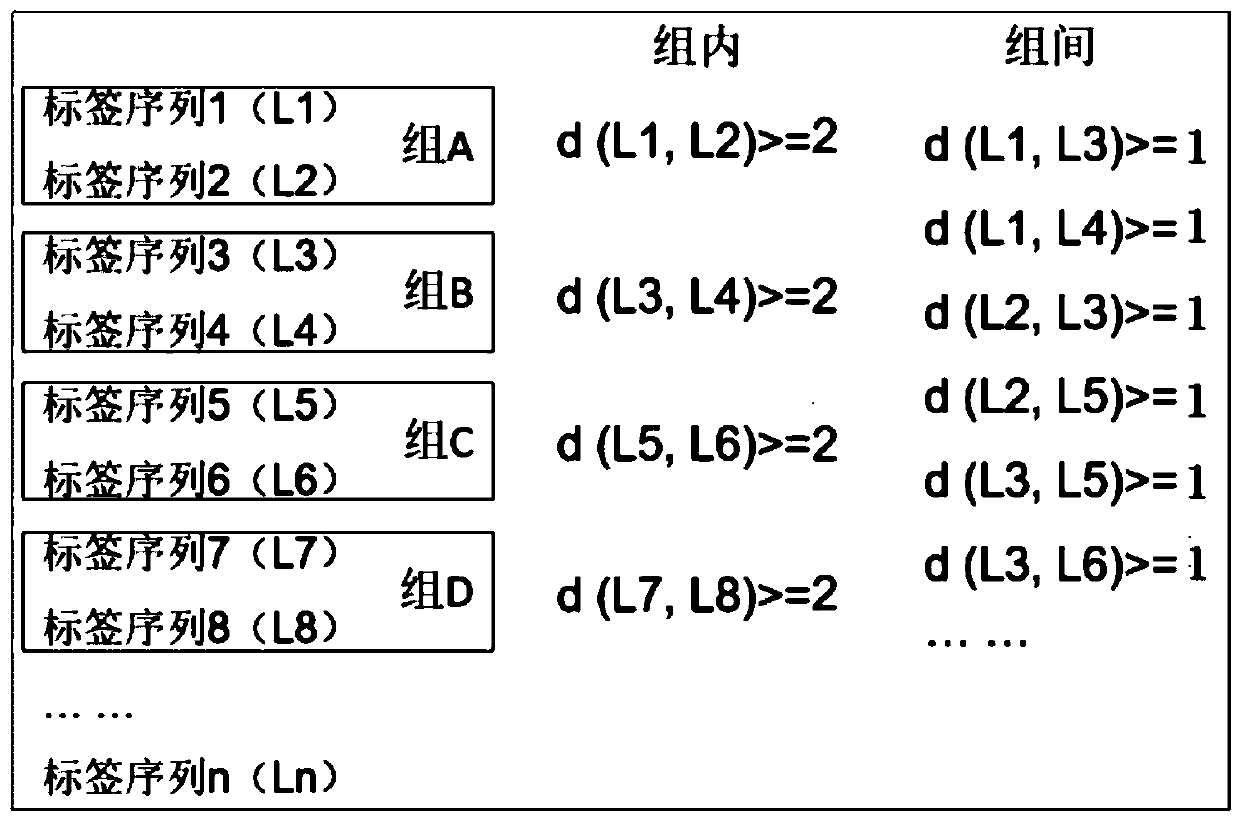
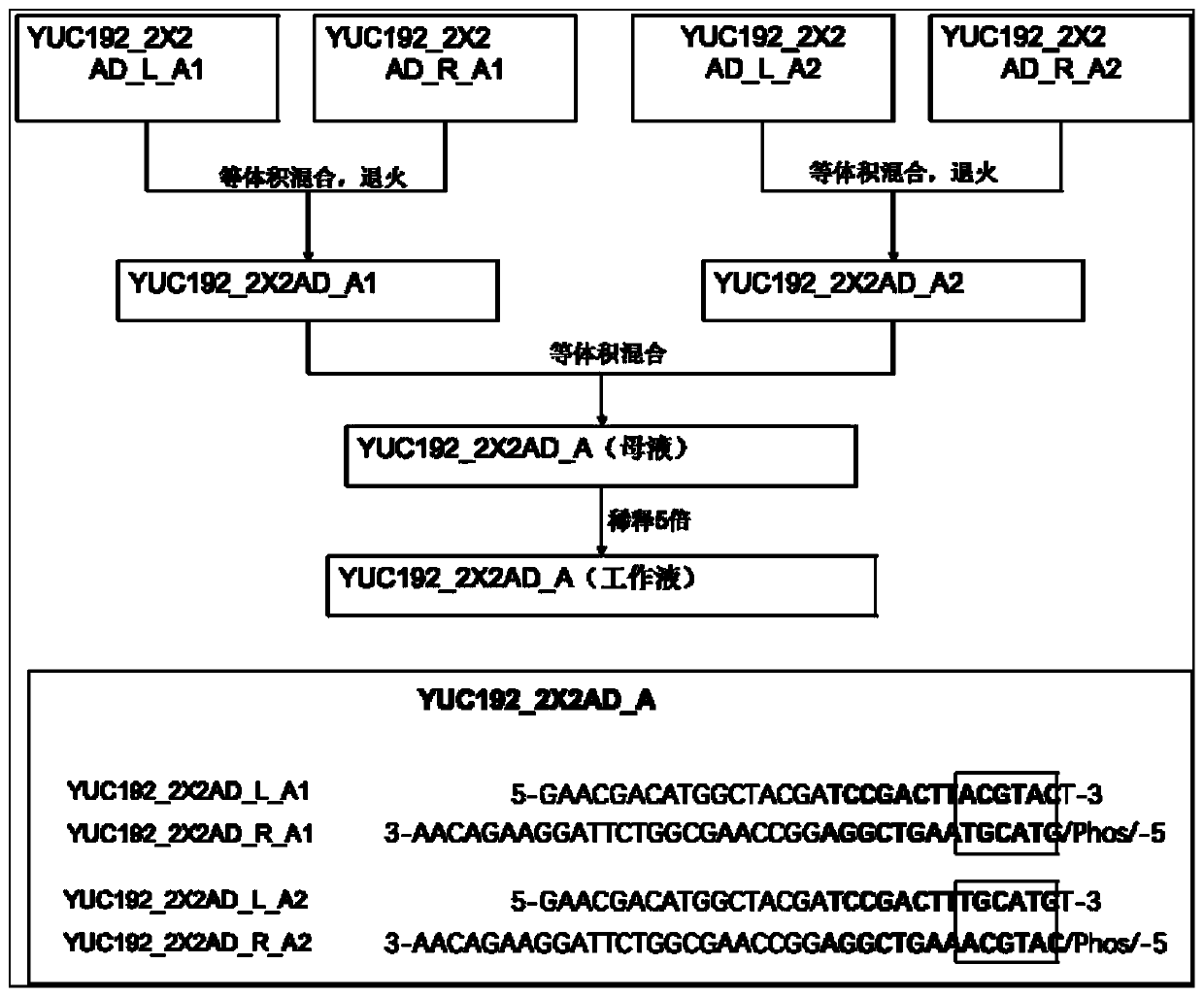
PendingCN110331187AReduce pollutionIncreased sequencing depthLibrary tagsMicrobiological testing/measurementHamming distanceComputer science
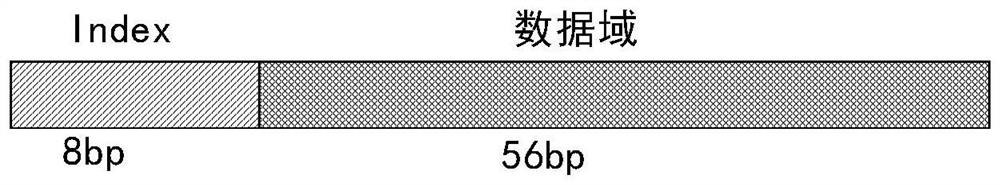
The invention relates to the field of gene sequencing, in particular to a combination tag, a combination tagjoint, and application ofthe combination tagandthe combination tag joint. The combination tagcomprises two or more sets of tags, each set of the two or more sets of tags contains two or more tagsequences, the Hamming distance between any tag sequences in each of the two or more sets of tagsis greater than or equal to 2, the Hamming distance between any of the tag sequences between each of the two or more sets of tags is greater than or equal to 1. The combination tag joint contains a joint sequence and the combination tag. The combination tag or the combination tag jointis used for gene library construction, sequencing and the like, contamination between samples can be effectively avoided,and the sequencing depth is increased.
Owner:TIANJIN MEDICAL LAB BGI +1
Method and prediction model for detecting tumour mutation burden
ActiveCN111826447AStrong specificitySensitive highMedical simulationMicrobiological testing/measurementTumour tissueTarget capture
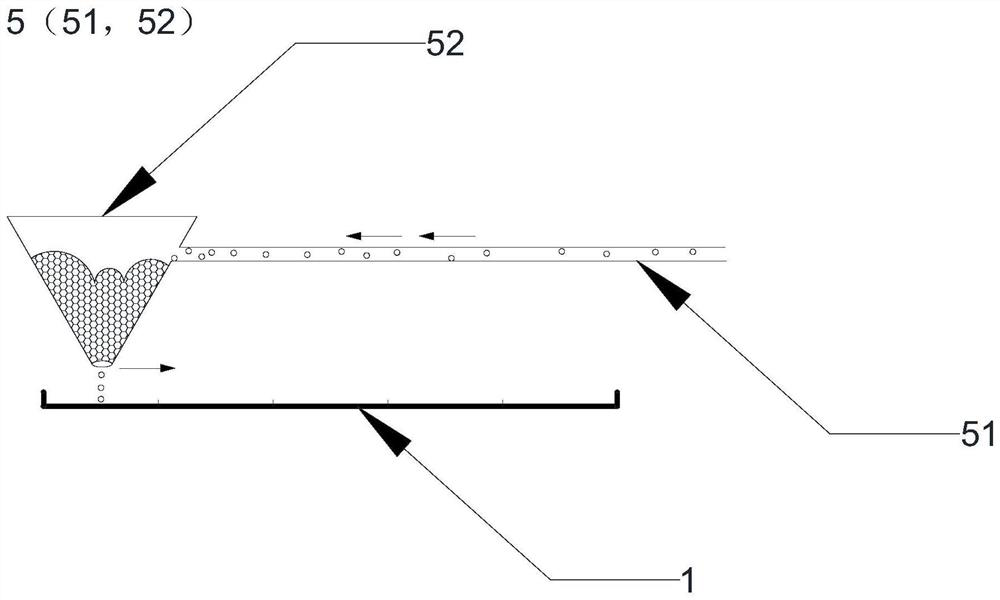
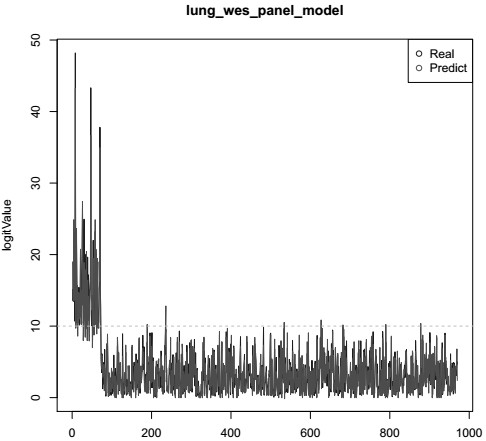
The invention relates to the technical field of biomedicine, and in particular relates to a method and prediction model for detecting the tumour mutation burden. The method comprises the following steps of: S1, performing DNA extraction on tumour tissue and blood samples to be detected; S2, for tumour-related genes, constructing a target region targeted capture sequencing library; S3, obtaining original offline data by utilizing a high-throughput sequencing platform; and S4, obtaining the tumour mutation burden of the sample through a check-out process. The invention aims to provide the methodand prediction model for detecting the tumour mutation burden. Gene mutation is detected and calculated through the method and the prediction method; the method can achieve the same result as TMB detection by whole exon sequencing; and thus, the evaluation accuracy of high tumour mutation burden can be improved.
Owner:求臻医学科技(浙江)有限公司
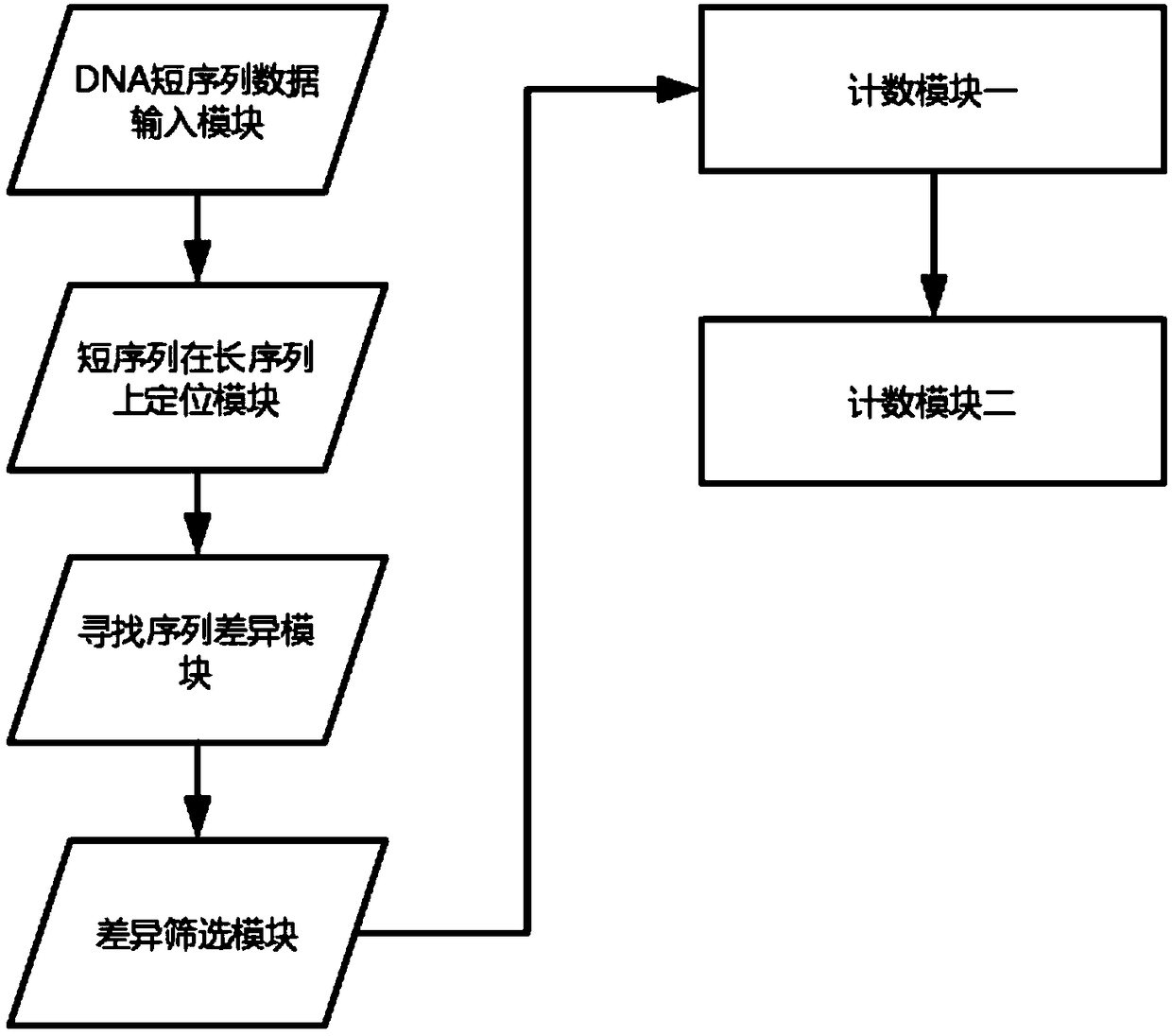
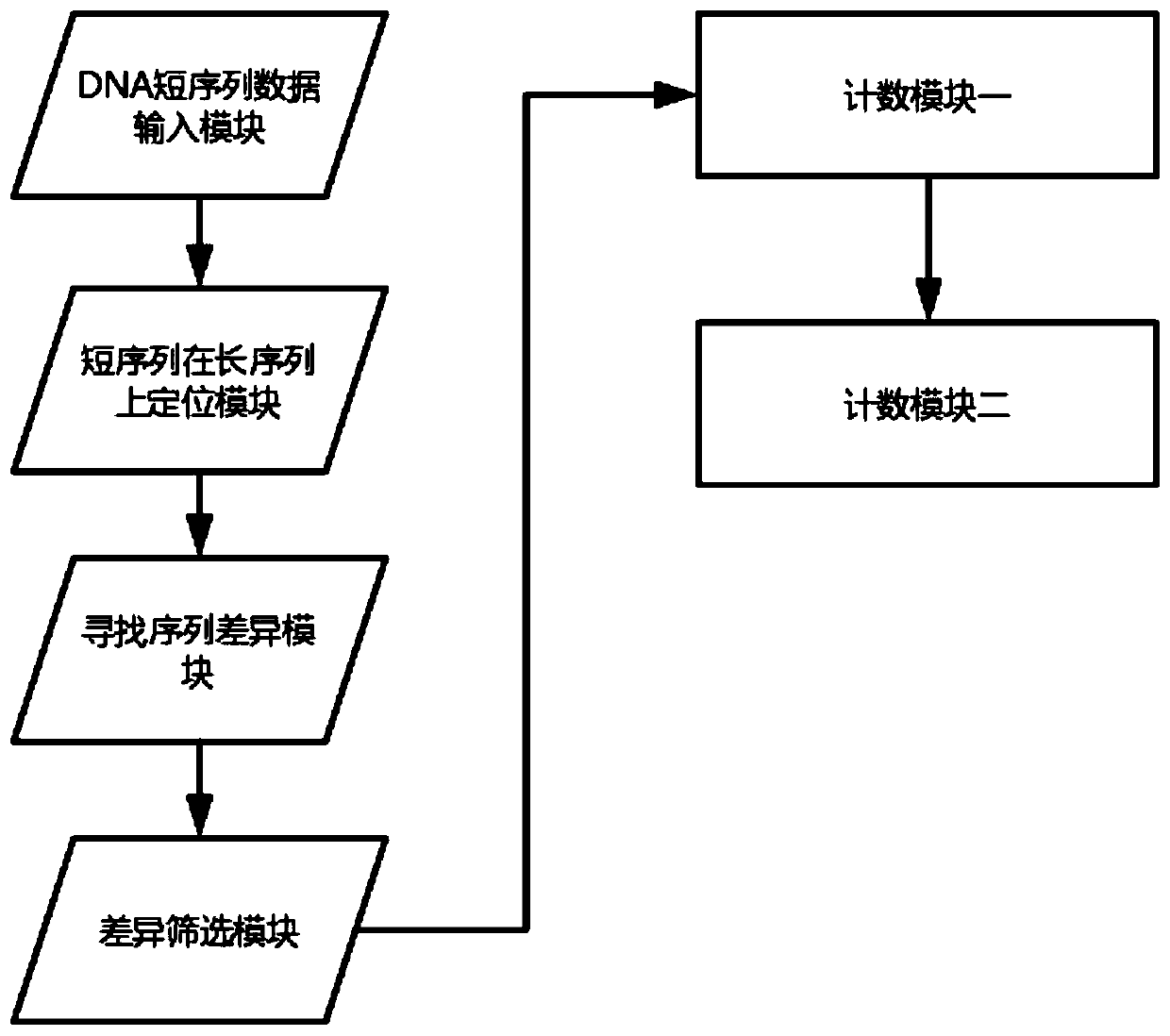
Fetus chromosome detecting system based on DNA variation counting
ActiveCN109402247AReduce adverse effectsPredict chromosomal abnormalitiesMicrobiological testing/measurementPlasma glucoseDNA
The invention provides a fetus chromosome detecting method based on DNA variation counting. The method comprises the steps of obtaining and sequencing free DNA of plasma in a peripheral blood sample of a pregnant woman, comparing the free DNA of the plasma with human reference sequences, namely DNA long sequences on 24 chromosomes, counting the number of variation in the free DNA of the plasma andconducting comparison. The invention further discloses a system for achieving the detecting method. The system comprises a DNA short sequence data input module, a module for positioning short sequences on the long sequences, a module for founding sequence difference, a difference screening module, a first counting module and a second counting module, wherein the modules are sequentially and electrically connected. According to the fetus chromosome detecting method based on DNA variation counting and the system for achieving the method, by detecting chromosomes in peripheral blood of the pregnant woman, whether there is chromosome abnormality or not in a fetus is judged, thus harmful effects on the pregnant woman and the fetus in a detecting process are greatly reduced, and by calculatingthe variation number to judge the chromosome abnormality of the fetus, compared with a method that a sequence number is calculated simply, the fetus chromosome detecting method based on DNA variationcounting is more accurate.
Owner:苏州首度基因科技有限责任公司
Probe set for detecting tuberous sclerosis gene mutation and kit thereof
ActiveCN113388676AIncrease coverageImprove capture efficiencyMicrobiological testing/measurementLibrary creationMedicineTSC1
The invention relates to a probe set for detecting tuberous sclerosis gene mutation and a kit thereof. The probe set can specifically capture at least one of a TSC1 gene segment and a TSC2 gene segment. The probe set has the advantages of high coverage, high capture efficiency, high sequencing depth, high detection sensitivity and low detection cost, and can detect possible pathogenic new point mutation, copy number variation and chimeric mutation in a certain proportion.
Owner:AEGICARE (SHENZHEN) TECH CO LTD
Semi-specific amplification primer group, method and kit for quickly detecting chromosome number abnormality
ActiveCN103397089AIncreased sequencing depthReduce background noiseMicrobiological testing/measurementDNA/RNA fragmentationMedicineGenetics
The invention provides a semi-specific amplification primer group, a method and a kit for quickly detecting chromosome number abnormality. The invention relates to two groups of amplification primers. The kit provided by the invention consists of the following components: a 10*PCR buffer solution, ddH2O, a dNTP solution, PlatinumPf*DNA polymerase (2.5U / mu L), primer pairs of amplification primer 1 with concentration of 25 mu m / mu l, an amplification primer 2 and purified buffer (QIAGENMinElutePCRPurificationKit). Another object of the invention is to provide a method for quickly detecting chromosome number abnormality through the semi-specific amplification primer group. By adopting the invention, the chromosome number abnormality can be detected quickly.
Owner:ANNOROAD GENE TECH BEIJING +2
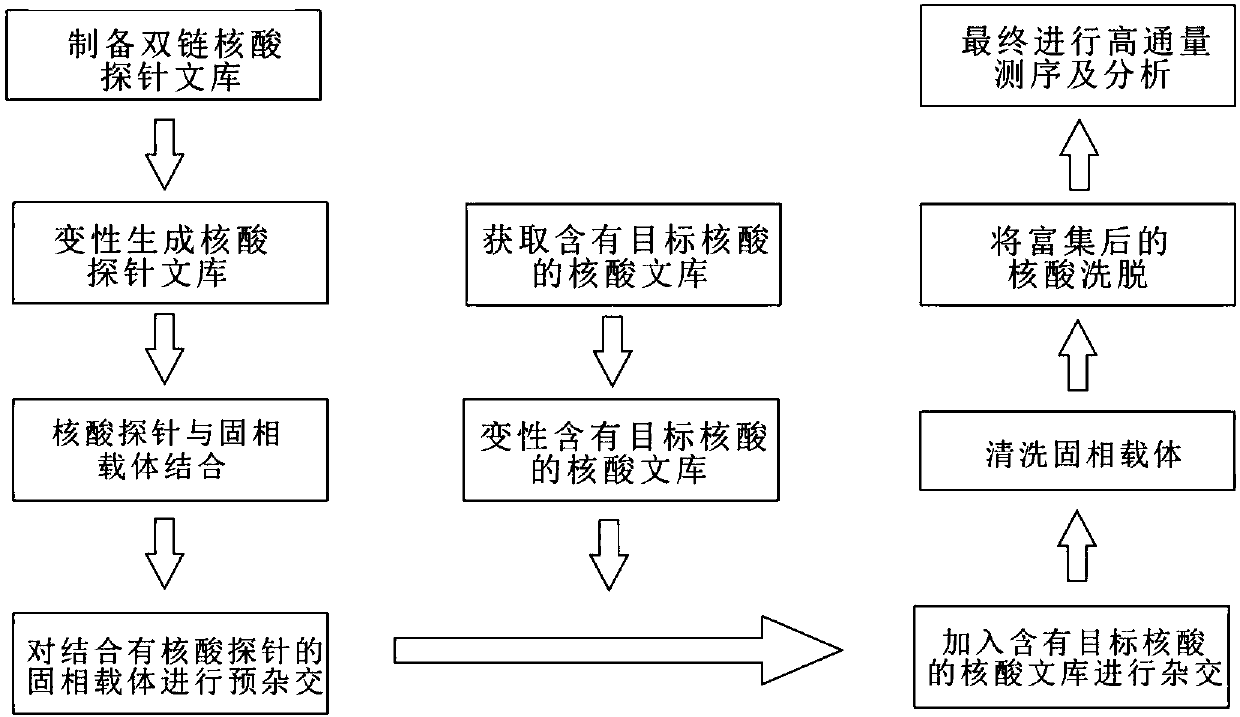
Solid-phase-carrier-membrane-based target nucleic acid enrichment method for high-throughput sequencing
InactiveCN107893067ALow costSuitable for independent developmentMicrobiological testing/measurementLibrary creationEnrichment methodsNucleic Acid Probes
The invention relates to the field of biotechnology, and specifically provides a solid-phase-carrier-membrane-based target nucleic acid enrichment method for high-throughput sequencing. The solid-phase-carrier-membrane-based target nucleic acid enrichment method comprises: 1) combining a nucleic acid probe library and a solid phase carrier membrane to form a probe-solid phase carrier membrane complex; 2) adding target nucleic acid to a hybridization solution containing the probe-solid phase carrier membrane complex, and enriching the target nucleic acid; 3) washing the obtained probe-solid phase carrier membrane complex with the target nucleic acid by using a washing solution, eluting from the solid phase carrier membrane by using an eluent, and purifying the enriched target nucleic acid library; and 4) applying the target nucleic acid library enriched in the step 3) in high-throughput sequencing. According to the present invention, with the method, the high-multiple coverage and single-base displacement nucleic acid probe for enriching the target nucleic acid can be rapidly produced, and is immovably fixed on the solid phase carrier membrane; and the method has advantages of simple operation, flexible probe acquisition and low cost.
Owner:朱静
Virus integrated DNA enrichment method, sequencing data analyzing method and device
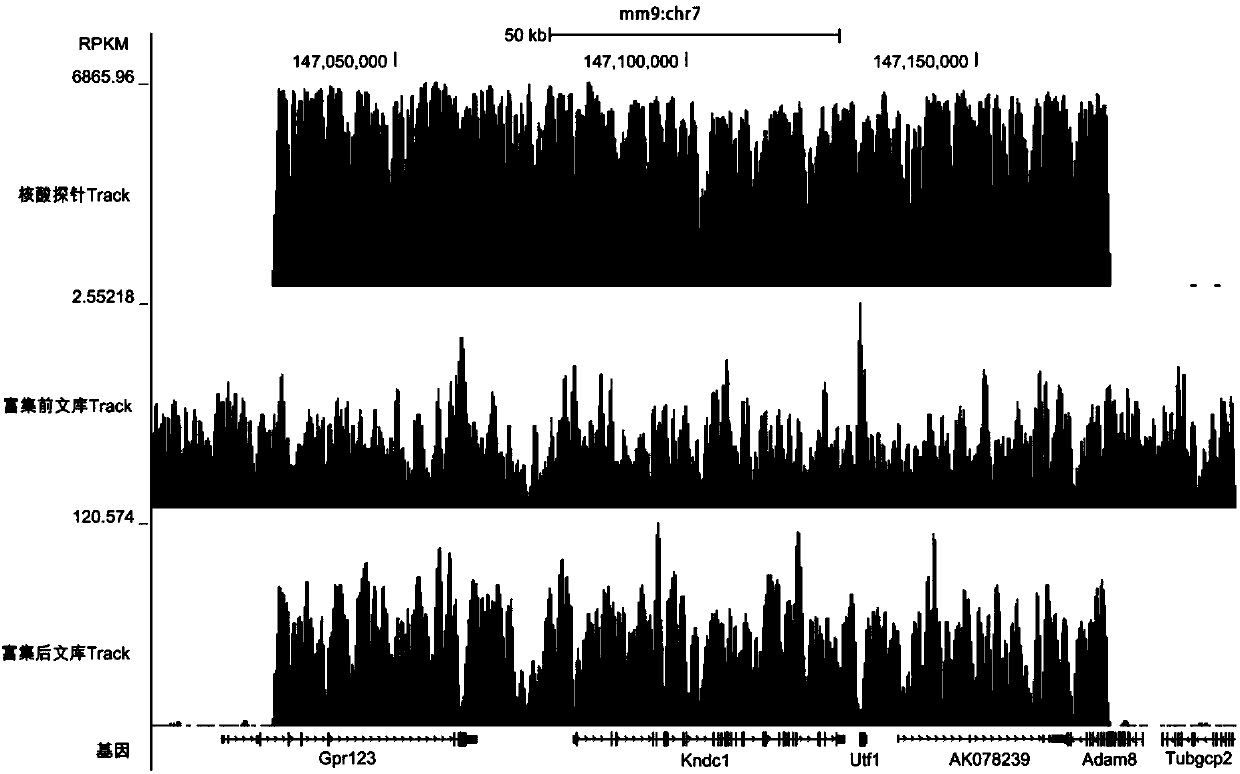
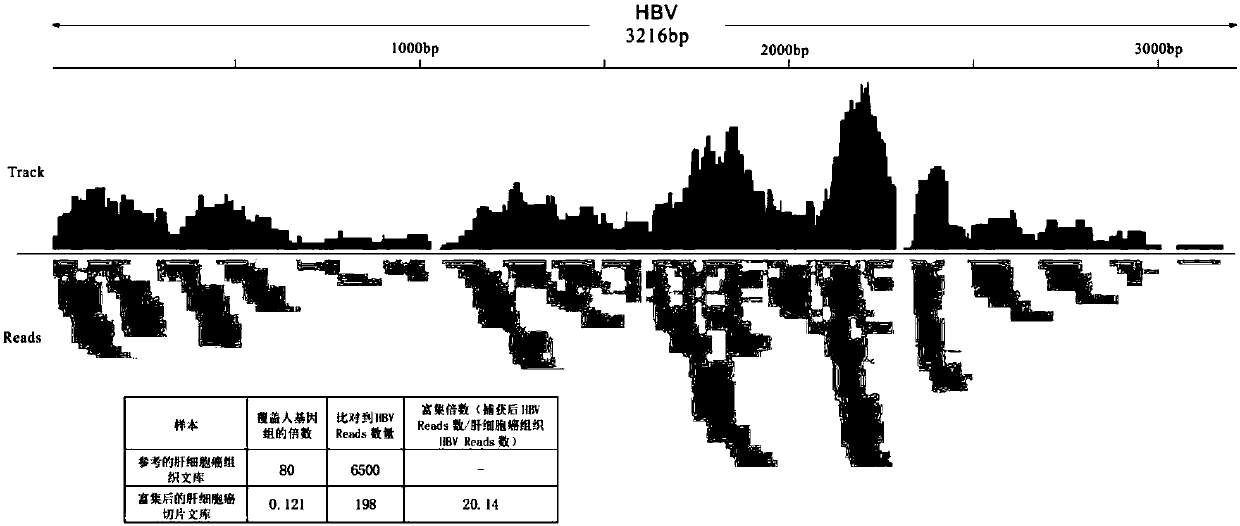
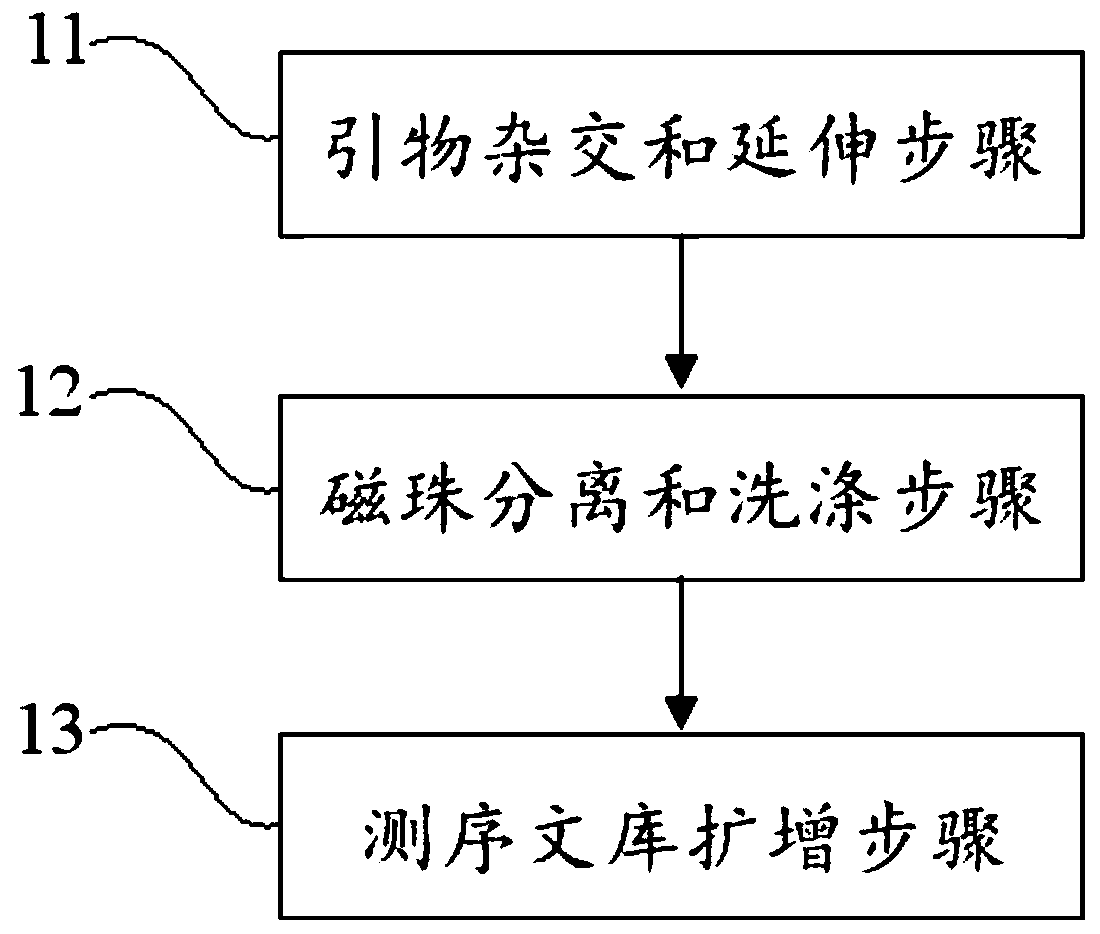
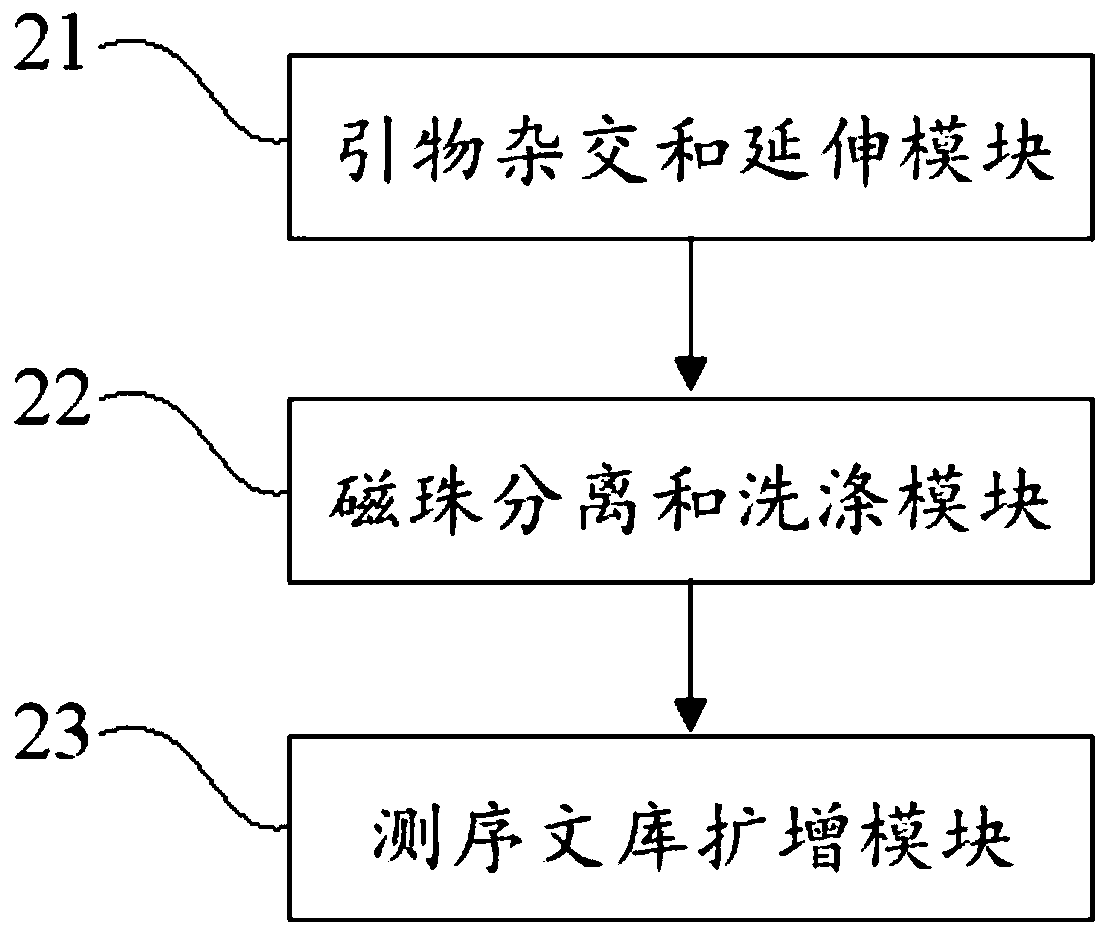
PendingCN110273028AReduce data volumeIncreased sequencing depthMicrobiological testing/measurementMicroorganism based processesMagnetic beadEnrichment methods
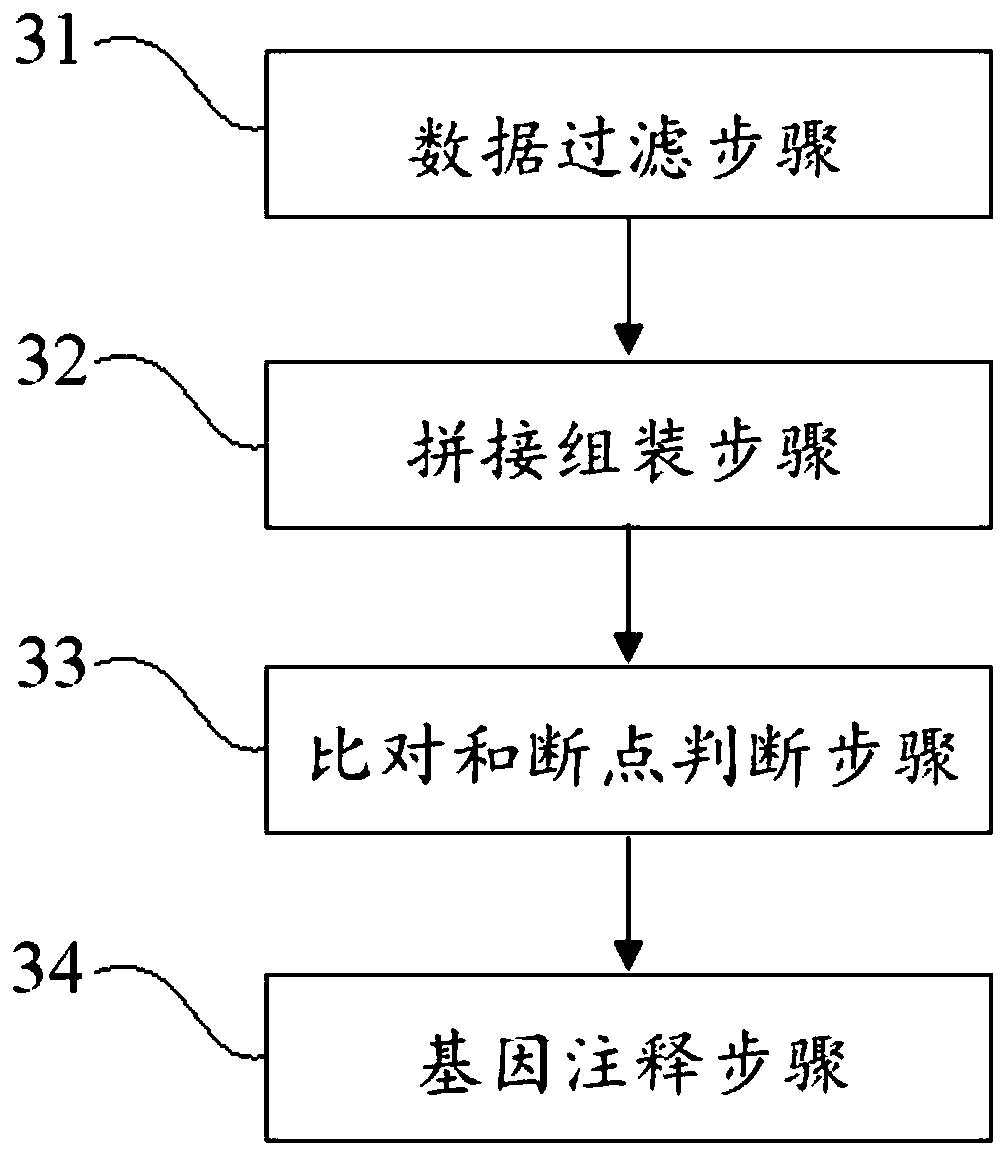
The application discloses a virus integrated DNA enrichment method, a sequencing data analyzing method and a device. The enrichment method of the application comprises the steps of primer hybridizing and extending, magnetic bead separating and washing. The sequencing data analyzing method disclosed by the application comprises the steps of data filtering, splicing and assembling, comparing and breakpoint judging and gene noting. According to the enrichment method disclosed by the application, specific primers are adopted for extending virus genome target genes so as to replace a long probe, so that the cost is low, the efficiency is high, the specificity is high, and the enrichment time is shortened. Compared with a complete genome, the sequencing data analyzing method disclosed by the application can save data amount, can obtain higher sequencing depth, can obtain more virus DNA breakpoint information and integration position information on the host genome, and can obtain clonal integration events supported by more reads, and besides, can detect non-clonal integration supported by single reads, so as to construct a comprehensive prospect integrated on the host genome.
Owner:SHENZHEN HAPLOX BIOTECH
Kit and use thereof
InactiveCN106916881AQuality improvementMeet on-machine sequencingNucleotide librariesMicrobiological testing/measurementStomach cancerGene
The invention discloses a kit. The kit comprises a probe, the probe is fixed on a solid matrix or is free in a solution, and the probe can specifically identify at least one part of regions in every gene of seventeen genes in table 1. The invention also provides a use of the kit, a method for constructing a target region sequencing library, a sequencing method, a method for detecting the variation of the target region, and a device for detecting the variation of the target region. The kit and / or the method or the device are / is used to once, simply, conveniently and highly-specifically obtain relevant gene sequences of stomach cancer and accurately detect and analyze the relevant gene sequences in order to make the detection and analysis result used for medication guidance and / or risk predication of the stomach cancer in an auxiliary manner.
Owner:BGI GUANGZHOU MEDICAL LAB CO LTD +1
Probes and uses thereof
ActiveCN105603052AEfficient detectionRealize detectionBioreactor/fermenter combinationsBiological substance pretreatmentsCoding regionGC-content
The present invention discloses probes and uses thereof. The group of the probes specifically recognize at least one part of a FLG gene coding region, and meet at least one condition selected from the following conditions that (1) the length of the probe is 75 bp; (2) the probe specifically recognizes the sequence between the upstream 10 bp and the downstream 10 bp of the FLG gene coding region; (3) the multiplier of the probe specifically recognizing the region having the GC content of higher than 0.6 and lower than 0.3 is more than 2; (4) the melting temperatures of the probe and the target sequence is 60-10 DEG C, preferably 80 DEG C; (5) the probe does not contain a hairpin structure; (6) the probe is matched with two sites on a reference genome at the most; and (7) the window sliding size is 10 bp during the probe selection. According to the present invention, the group of the probes provides good capture specificity, high sensitivity and high coverage for at least one part of the specifically-recognized FLG gene coding region, and can be effectively used for the capture detection of the FLG gene coding region.
Owner:WUHAN BGI CLINICAL LAB CO LTD +1
Probe composition for detecting three lumen organ tumors
PendingCN112662759AProlong lifeReduce testing costsMicrobiological testing/measurementDNA/RNA fragmentationParanasal Sinus CarcinomaOncology
The invention discloses a probe composition. According to the composition, through meta analysis of methylation data of a TCGA database and a GEO database, a 4.29 Kbp capture region is screened out, the 4.29 Kbp capture region comprises up to 37 methylation change genes highly related to cancer and non-coding DNA regions, and the method can detect methylation level changes of three kinds of lumen organ cancers such as esophageal cancer, gastric cancer and colorectal cancer at a time. The probe composition can be used for early screening of asymptomatic people and prognosis detection of cancer patients, and the breadth of detection genes of the probe compositino is superior to that of detection genes in the prior art and products.
Owner:BIOCHAIN BEIJING SCI & TECH
Gene chip for high-depth sequencing of gene mutation, preparation method and applications thereof
PendingCN111118610AIncreased sequencing depthNucleotide librariesMicrobiological testing/measurementNucleotideDeletion mutation
The invention discloses a gene chip for high-depth sequencing of gene mutation, a preparation method and applications thereof. The preparation method of the gene chip comprises the following steps: constructing capture interval libraries by combining a gene mutation online database and a local database according to different mutation types; and designing a gene chip by the capture interval library, so that the capture quality and the capture efficiency are improved, and high-depth sequencing is realized, wherein the mutation types comprise at least one selected from mononucleotide variation, insertion and deletion mutation, copy number mutation and structural variation, and the capture interval libraries comprise a mononucleotide variation and insertion and deletion mutation capture interval library, a copy number mutation capture interval library and a structural variation capture interval library. According to the preparation method of the gene chip, mutant people can be covered to the maximum extent, cancers and related genes thereof can be covered to the maximum extent, and the gene chip suitable for Chinese people is designed. The gene chip disclosed by the invention is high in sequencing depth, and is particularly suitable for detecting extremely trace circulating DNA in blood.
Owner:BGI GENOMICS CO LTD +2
Probe composition for detecting five tumors in digestive tracts
PendingCN112662762AProlong lifeExcellent in breadthMicrobiological testing/measurementDNA/RNA fragmentationPancreas CancersOncology
Disclosed herein is a probe composition. According to the composition, methylation data of a TCGA database and a GEO database are meta-analyzed, a 4.37 Kbp capture region is screened out and comprises as many as 38 methylation change genes highly related to cancer and a non-coding DNA region, and the method can detect methylation level changes of five common cancers such as esophageal cancer, gastric cancer, colorectal cancer, liver cancer and pancreatic cancer at a time. The probe composition can be used for early screening of asymptomatic people and prognosis detection of cancer patients, and the breadth of detection genes of the probe composition is superior to that of detection genes in the prior art and products.
Owner:BIOCHAIN BEIJING SCI & TECH
Kit for detecting single gene mutations of 50 hereditary diseases, and probe group used by kit
ActiveCN112301123AEnsure screening accuracyAccurate discoveryNucleotide librariesMicrobiological testing/measurementMonogenic inheritanceMolecular biology
The invention discloses a kit for detecting single gene mutations of 50 hereditary diseases, and a probe group used by the kit. The probe group consists of 1059 specific probes, and nucleotide sequences of the 1059 specific probes are sequentially shown as SEQ ID NO:1 to SEQ ID NO:1059. The inventors of the invention develop the kit based on the liquid phase capture technology through a large number of experiments, and on the premise of ensuring the gene screening accuracy, the sequencing cost is greatly reduced, the sequencing depth is improved, and genetic variation information of specific regions of 50 single-gene hereditary diseases are more accurately found. The kit has important application value.
Owner:北京迈基诺基因科技股份有限公司
Probe composition for detecting 11 cancers
PendingCN112662764AProlong lifeExcellent in breadthMicrobiological testing/measurementDNA/RNA fragmentationPancreas CancersOncology
The invention discloses a probe composition. According to the composition, through meta analysis of methylation data of a TCGA database and a GEO database, a 6.5 Kbp capture region is screened out, and the 6.5 Kbp capture region comprises up to 66 methylation change genes highly related to cancer and a non-coding DNA region. The method can be used for detecting methylation level changes of 11 common cancers such as esophageal cancer, gastric cancer, colorectal cancer, lung cancer, liver cancer, pancreatic cancer, prostate cancer, breast cancer, ovarian cancer, cervical cancer and endometrial cancer at one time. The probe composition can be used for early screening of asymptomatic people and prognosis detection of cancer patients, and the breadth of detection genes of the probe composition is superior to that of detection genes in the prior art and products.
Owner:BIOCHAIN BEIJING SCI & TECH
Probe composition for detecting three substantive organ tumors
PendingCN112662761AProlong lifeReduce testing costsMicrobiological testing/measurementDNA/RNA fragmentationPancreas CancersCoding region
Disclosed herein is a probe composition. According to the composition, through meta analysis of methylation data of a TCGA database and a GEO database, a 3.75 Kbp capture area is screened out, the 3.75 Kbp capture area comprises as many as 36 methylation change genes highly related to cancer and a non-coding DNA area, and the method can detect methylation level changes of three substantive organ tumors such as lung cancer, liver cancer and pancreatic cancer at a time. The probe composition can be used for early screening of asymptomatic people and prognosis detection of cancer patients, and the breadth of detection genes of the probe composition is superior to that of detection genes in the prior art and products.
Owner:BIOCHAIN BEIJING SCI & TECH
Probes and their uses
ActiveCN105603052BHigh sensitivityIncrease coverageBioreactor/fermenter combinationsBiological substance pretreatmentsCoding regionGC-content
The present invention discloses probes and uses thereof. The group of the probes specifically recognize at least one part of a FLG gene coding region, and meet at least one condition selected from the following conditions that (1) the length of the probe is 75 bp; (2) the probe specifically recognizes the sequence between the upstream 10 bp and the downstream 10 bp of the FLG gene coding region; (3) the multiplier of the probe specifically recognizing the region having the GC content of higher than 0.6 and lower than 0.3 is more than 2; (4) the melting temperatures of the probe and the target sequence is 60-10 DEG C, preferably 80 DEG C; (5) the probe does not contain a hairpin structure; (6) the probe is matched with two sites on a reference genome at the most; and (7) the window sliding size is 10 bp during the probe selection. According to the present invention, the group of the probes provides good capture specificity, high sensitivity and high coverage for at least one part of the specifically-recognized FLG gene coding region, and can be effectively used for the capture detection of the FLG gene coding region.
Owner:WUHAN BGI CLINICAL LAB CO LTD +1
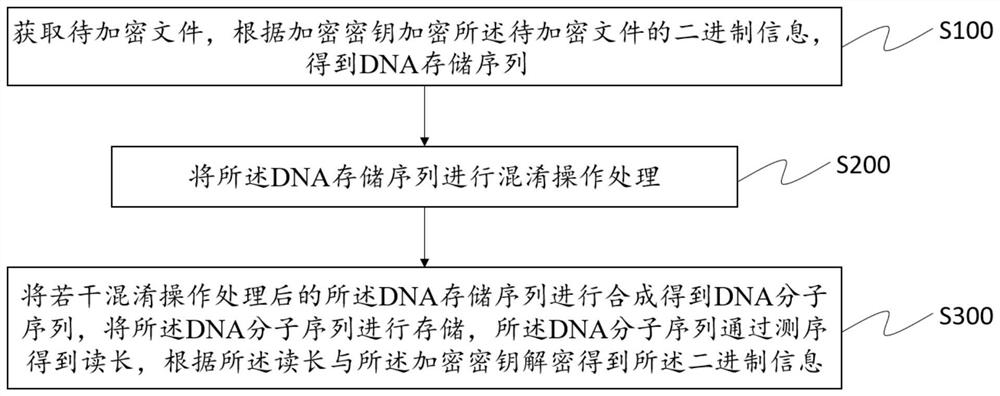
Symmetrical encryption method for DNA storage
ActiveCN113315623AReduce redundancyIncreased sequencing depthKey distribution for secure communicationEncryption apparatus with shift registers/memoriesAlgorithmConfidentiality
The invention provides a symmetric encryption method for DNA storage, and the method comprises the following steps: obtaining a to-be-encrypted file, encrypting the binary information of the to-be-encrypted file according to an encryption key, and obtaining a DNA storage sequence; carrying out confusion operation processing on the DNA storage sequence; synthesizing the plurality of DNA storage sequences processed by the confusion operation to obtain a DNA molecular sequence, storing the DNA molecular sequence, sequencing the DNA molecular sequence to obtain a read length, and decrypting according to the read length and the encryption key to obtain binary information. According to the method, under different error rates, the sequencing depth is increased, the data recovery rate is in a rising trend, the information redundancy in the encryption and decryption process can be effectively reduced, the robustness is higher, the malicious cracking difficulty is large, the confidentiality effect is better, and the method can be widely applied to the technical field of system biology research.
Owner:GUANGZHOU UNIVERSITY
Methylated primer, kit, model and construction method for cervical cancer related genes
ActiveCN113930516AIncreased sequencing depthShorten the timeMicrobiological testing/measurementSequence analysisCervical caLogistische regression
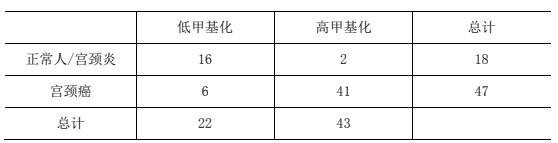
The invention relates to the field of molecular biological detection, in particular to a methylated primer, a kit, a model and a construction method for cervical cancer related genes. The construction method comprises the following steps: designing a methylated primer; methylating a cervical cell DNA by extracting the cervical cell DNA; performing first PCR amplification and purification on the methylated cervical cell DNA by using a PCR amplification kit; performing second PCR amplification and purification on the treated product to obtain a library; performing high-pass sequencing on the library through a next-generation sequencing platform; performing bioinformatic analysis; calculating each PCR amplification area to obtain a methylation score as an independent variable based on a cervical cancer sample and a normal sample; and constructing a cervical cancer related gene methylation evaluation model by adopting a logistic regression mode. According to the method, the methylation state of the sample is obtained through multiple PCR methylation library establishment and high-throughput sequencing, the sequencing depth is high, the time is short, the cost is low, treatment or observation is determined according to the assessed risk, and excessive treatment can be effectively reduced.
Owner:北京迈基诺基因科技股份有限公司
Method for capturing genetic colorectal cancer genome target sequence
InactiveCN113234822AEfficient captureCapture completeMicrobiological testing/measurementLibrary creationHuman DNA sequencingExon
The invention belongs to the technical field of gene detection, and particularly discloses a method for capturing a genetic colorectal cancer genome target sequence, wherein the sequence of a capture probe adopted by the method is as shown in SEQ ID NO: 1-2641, and the capture probe covers a region of 80-300Kb, preferably 80-100Kb, of the genome, and the Tm value of the capture probe is 62-76 DEG C. The probe designed and synthesized by the invention can efficiently and completely capture a coding exon region and an exon-intron junction region of a human genome target region, reduces non-specific capture, is uniform in hybridization temperature, greatly reduces the detection cost and the sequencing data volume, shortens the single sample analysis time, and meets the personalized user requirements; the method is especially suitable for screening of high-risk population of hereditary colorectal cancer and public physical examination markets.
Owner:XIEHE HOSPITAL ATTACHED TO TONGJI MEDICAL COLLEGE HUAZHONG SCI & TECH UNIV
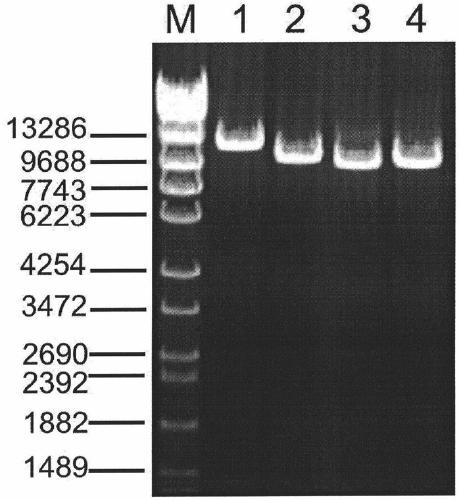
Amplification primer and detection method for pkd2 gene mutation detection
ActiveCN105886605BIncreased sequencing depthGood amplification effectMicrobiological testing/measurementDNA/RNA fragmentationInteinExon
The invention belongs to a technology of medical in vitro diagnosis, and in particular relates to an amplification primer for detecting mutation of autosomal dominant polycystic kidney disease-causing gene PKD2 as well as a detection method. The amplification primer, in accordance with the sequence information of the PKD2 gene, designs four pairs of PCR primers; the PKD2 is subjected to targeted amplification by virtue of long-chain PCR; and an amplification region includes all exon sequences and most intron sequences, as well as partial 5' untranslated region sequences and 3' untranslated region sequences. According to the method, by combining long-chain PCR targeted amplification to the newest developed next generation of sequencing technology, a targeted sequencing process is simplified, the scope and flux of detecting the PKD2 gene mutation are improved, a detection cycle is shortened and cost is reduced.
Owner:NANJING MATERNITY & CHILD HEALTH CARE HOSPITAL
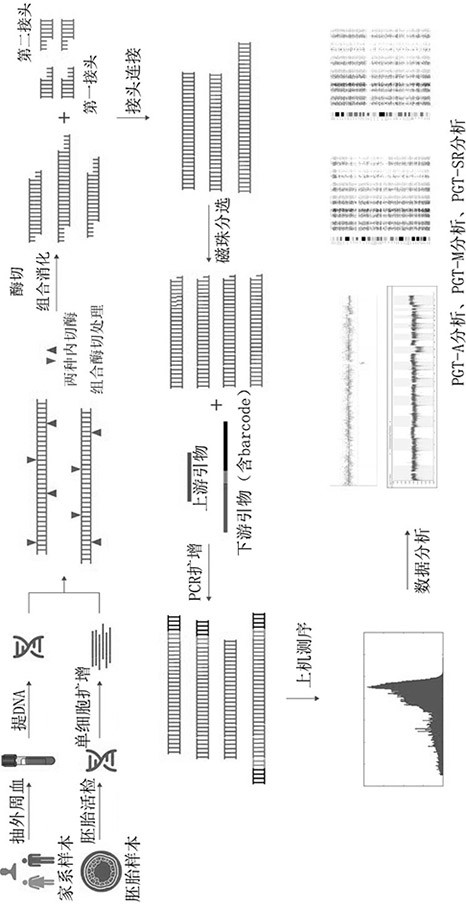
A nucleic acid library construction method and its application in the analysis of abnormal chromosomal structure of preimplantation embryos
ActiveCN111961707BSimplify the build processIncreased sequencing depthMicrobiological testing/measurementLibrary creationSequencing dataChromosomal Abnormality
The invention relates to a method for constructing a nucleic acid library and its application in the analysis of abnormal chromosome structure of pre-implantation embryos. The combination of the first endonuclease and the second endonuclease performs enzyme cutting and breaking, and captures a DNA sequence within a fixed fragment range. , and then sequence the captured specific sequence. When the average sequencing depth of the genome is more than 3×, SNP analysis can be performed on the whole genome, and embryo balance translocation can be detected through linkage analysis of family samples. This method reduces the amount of data required for whole genome sequencing, while ensuring effective SNP sites and their depth, increasing the number of SNP sites that can be constructed from haplotypes, and obtaining sufficient coverage of the entire genome with a very low amount of sequencing data. Genomes can be used to analyze haplotype SNPs and indels, and there is no need to design multiple PCR primers for SNPs, which greatly reduces the amount of data required and detection costs.
Owner:SUZHOU BASECARE MEDICAL DEVICE CO LTD
A mitochondrial genome library based on high-throughput sequencing and its construction method
ActiveCN105907748BGuaranteed Accurate AmplificationSensitive and accurate mutation detectionNucleotide librariesMicrobiological testing/measurementHuman DNA sequencingA-DNA
A mitochondrial genomic library based on high-throughput sequencing and a method for constructing the library. The method comprises the following steps: S1. performing a one-step PCR amplification of total mitochondrial DNA from total DNA; S2. fragmenting the total mitochondrial DNA to obtain a DNA fragment; S3. performing end repair on the DNA fragment to obtain a blunt-end DNA fragment; S4. A-tailing a 3' end of the blunt-end DNA fragment to obtain an A-tailed DNA fragment; S5. adding a linker on the 3' end of the A-tailed DNA fragment to obtain a DNA fragment with linkers; S6. performing LM-PCR on the DNA fragment with linkers; and S7. performing PCR on an LM-PCR product. The method for constructing the mitochondrial genomic library performs PCR amplification on total DNA to obtain total mitochondrial DNA, thereby ensuring accurate amplification of a human mitochondrial genome and improving the accuracy and precision of mutation detection by preventing contamination or mixing of a homologous sequence from a human genome.
Owner:GUANGZHOU JIAJIAN MEDICAL TESTING CO LTD
Single cell capturing system based on image analysis
PendingCN114755229AUnlimited live rateNo preferenceBioreactor/fermenter combinationsBiological substance pretreatmentsComputer hardwareInformation control
The invention discloses a single cell capture system based on image analysis. The single cell capture system comprises a liquid storage unit, an optical imaging unit, an image analysis unit, a sampling unit and a control unit which is respectively connected with the sampling unit and the image analysis unit, the optical imaging unit is used for acquiring image information in the liquid storage unit and comprises a light source and an image acquisition element; the sampling unit comprises a sampling suction head and a sampling pipeline which are communicated; the sampling suction head sucks a target object from the liquid storage unit and transfers the target object through the sampling pipeline communicated with the sampling suction head; the image analysis unit is connected with the image acquisition element to receive and output image information; the control unit comprises a first displacement control module for controlling the sampling suction head; the first displacement control module is connected with the image analysis unit; and the first displacement control module controls the displacement of the sampling suction head according to information fed back by the image analysis unit. The single cell capture system provided by the invention can realize capture of single cells and single cell analogues with high efficiency and high specificity.
Owner:TONGJI UNIV
A method and prediction model for detecting tumor mutational burden
ActiveCN111826447BStrong specificitySensitive highMedical simulationMicrobiological testing/measurementTarget captureMedicine
The present invention relates to the field of biomedical technology, in particular to a method for detecting tumor mutation load and a prediction model; it includes the following steps: S1: extracting DNA from tumor tissue and blood samples to be tested; S2: constructing target regions for tumor-related genes Targeted capture sequencing library; S3: Using a high-throughput sequencing platform to obtain the original off-machine data; S4: Obtain the tumor mutation load of the sample through the detection process. The purpose of the present invention is to provide a method for detecting tumor mutation load and a prediction model, through which the detection and calculation of gene mutations can achieve the same results as the TMB detection of whole exome sequencing, so as to improve Accuracy of Tumor Mutational Burden Estimation.
Owner:求臻医学科技(浙江)有限公司
A Fetal Chromosome Detection System Based on DNA Variation Counting
ActiveCN109402247BReduce adverse effectsPredict chromosomal abnormalitiesMicrobiological testing/measurementMedicineA-DNA
The invention provides a fetus chromosome detecting method based on DNA variation counting. The method comprises the steps of obtaining and sequencing free DNA of plasma in a peripheral blood sample of a pregnant woman, comparing the free DNA of the plasma with human reference sequences, namely DNA long sequences on 24 chromosomes, counting the number of variation in the free DNA of the plasma andconducting comparison. The invention further discloses a system for achieving the detecting method. The system comprises a DNA short sequence data input module, a module for positioning short sequences on the long sequences, a module for founding sequence difference, a difference screening module, a first counting module and a second counting module, wherein the modules are sequentially and electrically connected. According to the fetus chromosome detecting method based on DNA variation counting and the system for achieving the method, by detecting chromosomes in peripheral blood of the pregnant woman, whether there is chromosome abnormality or not in a fetus is judged, thus harmful effects on the pregnant woman and the fetus in a detecting process are greatly reduced, and by calculatingthe variation number to judge the chromosome abnormality of the fetus, compared with a method that a sequence number is calculated simply, the fetus chromosome detecting method based on DNA variationcounting is more accurate.
Owner:苏州首度基因科技有限责任公司
Primers, kits, models and construction methods for methylation of cervical cancer-related genes
ActiveCN113930516BIncreased sequencing depthShorten the timeMicrobiological testing/measurementSequence analysisCervical cellsIndependent predictor
The invention relates to the field of molecular biology detection, in particular to a primer, a kit, a model and a construction method for the methylation of cervical cancer-related genes. The methylation primer is designed, and cervical cell DNA is extracted, and the cervical cell DNA is methylated. Methylation treatment, using a PCR amplification kit to perform the first PCR amplification and purification on the cervical cell DNA after methylation treatment, and performing the second PCR amplification and purification on the treated product to obtain a library. The library was subjected to high-pass sequencing on the next-generation sequencing platform for bioinformatics analysis. Based on cervical cancer samples and normal samples, a methylation score was calculated for each PCR amplification region as an independent variable, and the correlation between cervical cancer and cervical cancer was constructed by logistic regression. Gene methylation evaluation model; the present invention builds a database through multiple PCR methylation, and obtains the methylation status of the sample through high-throughput sequencing. The sequencing depth is high, the time is short, and the cost is low. According to the assessed risk, it is decided whether to treat or observe , can effectively reduce overtreatment.
Owner:北京迈基诺基因科技股份有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com