Amplification primer and detection method for pkd2 gene mutation detection
A target gene and genome technology, applied in biochemical equipment and methods, recombinant DNA technology, and microbial assay/inspection, etc., can solve the problems of heavy Sanger sequencing workload, many types of primers, lack of mutation hotspots, etc., and achieve sequencing depth The effect of good uniformity, improved range and throughput, and shortened detection cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Genomic DNA Extraction
[0032] Collect 2ml of peripheral blood from the subject and place it in an EDTA anticoagulant tube. Take 0.2 ml of EDTA anticoagulated peripheral blood sample, and extract DNA according to the instructions of the whole blood DNA extraction kit. The DNA concentration and purity were analyzed with a Nanodrop 2000 photometer, and the extracted genomic DNA was ready to be used as a template for the next step of LR-PCR.
Embodiment 2
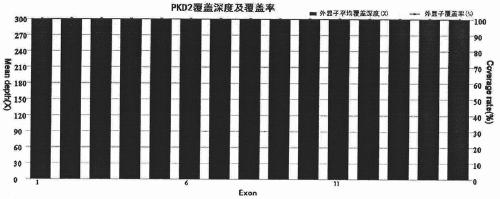
[0034] LR-PCR amplification
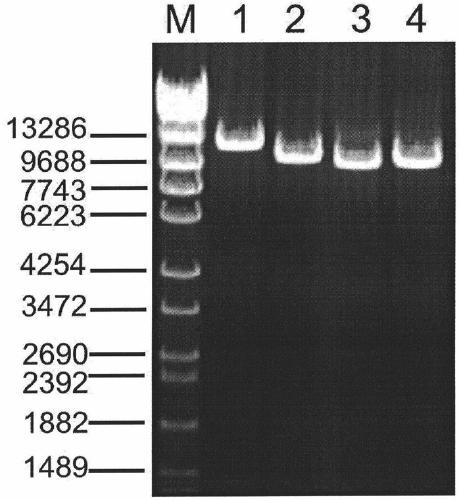
[0035] According to the sequence information of the PKD2 gene, 4 pairs of PCR primers were designed, and GeneAmp High Fidelity PCR System was used for LR-PCR to amplify 4 large fragment sequences, namely fragment 1, fragment 2, fragment 3 and fragment 4. The primer sequences and amplification The product size is shown in Table 1. The total volume of the PCR reaction is 50 μl, including 5 μl of 10×GeneAmp High Fidelity PCR buffer, 2 μl of 10mmol / LdNTP, 1 μl of 10 μmol / L upstream and downstream primers, 10 μl of 5mol / L betaine, 1 μl of 5U / μl polymerase mixture, 20ng / μL DNA template 5μl, make up to 50μl with ultrapure water. PCR cycle parameters: denaturation at 95°C for 2min; 20sec at 94°C, 14min at 68°C, a total of 35 cycles; last extension at 72°C for 10min. The PCR reaction was completed on a Veriti 96-well Thermal Cycler PCR instrument (ABI Company, USA).
[0036] Table 1, LR-PCR primers for amplifying PKD2 gene
[0037]
[0038] Note: F...
Embodiment 3
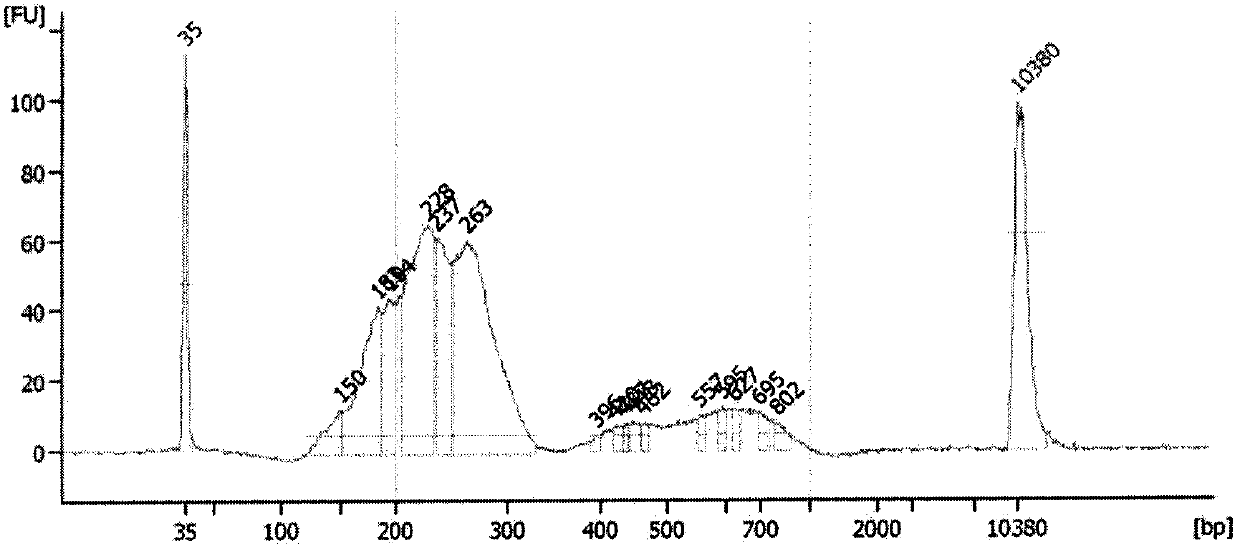
[0041] PCR product purification and pooling
[0042] The PCR product was purified using a common DNA product purification kit (TIANGEN biology), and the purified product was quantified using a Qubit 2.0 fluorescence photometer. Quantify.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com