A Fetal Chromosome Detection System Based on DNA Variation Counting
A chromosome and variant number technology, applied in the field of medical testing, can solve problems such as low coverage depth, missed detection of chromosome 3, and insufficient accuracy, and achieve the effect of improving sequencing depth, reducing adverse effects, and increasing the range of genetic diseases
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Example 1 A method for detecting fetal chromosomes based on DNA variation counting
[0065] (1) Use the Cell-Free DNA BCT test tube of STRECK Company to collect 5ml of peripheral blood of the 5-week pregnant woman to be tested according to the instruction manual of the corresponding equipment, and use the QIAamp Circulating Nucleic Acid Kit kit of QIAGEN Company to extract according to the instruction manual of the corresponding kit Plasma cell-free DNA in the sample to be tested;
[0066] (2) Use the Nextseq500 high-throughput sequencing instrument of Illumina to sequence the DNA obtained in step (1) to obtain the short sequence data of the plasma cell-free DNA of the pregnant woman;
[0067] (3) positioning the short sequence obtained in step (2) to the position with the highest similarity on the long sequence of 24 chromosomes in the human genome;
[0068] (4) Use bwa and GATK software to calculate the characters and character strings with differences in the corresp...
Embodiment 2
[0078] Example 2 A fetal chromosome detection method based on DNA variation counting
[0079] (1) Use the Cell-Free DNA BCT test tube of STRECK Company to collect 5ml of peripheral blood of the 10-week pregnant woman to be tested according to the instruction manual of the corresponding equipment, and use the QIAamp Circulating Nucleic Acid Kit kit of QIAGEN Company to extract according to the instruction manual of the corresponding kit For the plasma free DNA in the sample to be tested, use the KAPA Hyper Prep Kit kit from KAPA Biosystems to build a library for the extracted plasma free DNA according to the corresponding kit instruction manual. During the library construction process, use probe primers from DNA sequences of the same DNA template have the same probe;
[0080] (2) Use the Nextseq500 high-throughput sequencing instrument of Illumina to sequence the DNA sample library obtained in step (1), and obtain the short sequence data of the plasma cell-free DNA of the pregn...
Embodiment 3
[0091] Example 3 A Fetal Chromosome Detection System Based on DNA Variation Counting
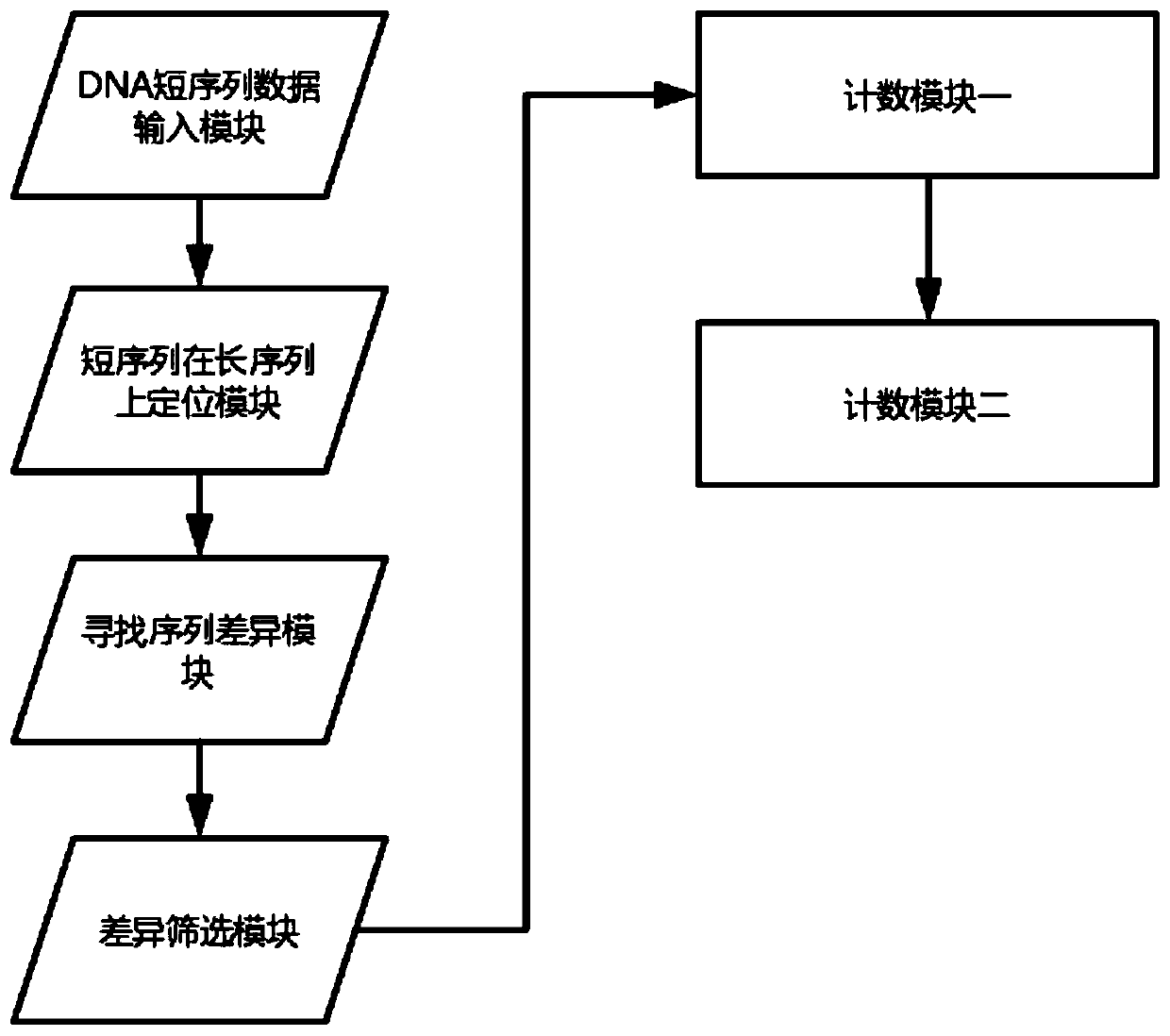
[0092] Including the following six modules: DNA short sequence data input module, short sequence positioning module on long sequence, sequence difference search module, difference screening module, counting module 1, counting module 2, the modules are electrically connected in sequence; The DNA short sequence data input module inputs the short sequence data from the free plasma DNA of pregnant women; the short sequence positioning module on the long sequence locates the short sequence of the free plasma DNA of pregnant women input by the DNA short sequence data input module to the human genome The position with the highest similarity on the long sequence of the 24 chromosomes; the module for finding sequence differences, based on the output of the short sequence positioning module on the long sequence, compares the short sequence of the plasma cell-free DNA of pregnant women with the correspo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com