Probe composition for detecting three lumen organ tumors
A composition and probe technology, applied in the field of DNA sequence probe composition, can solve problems such as the influence of cancer occurrence and development, change the tumor phenotype of cell development tendency, and achieve the goal of overcoming tumor heterogeneity and saving probe synthesis cost. , to achieve the effect of real-time monitoring
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Embodiment 1
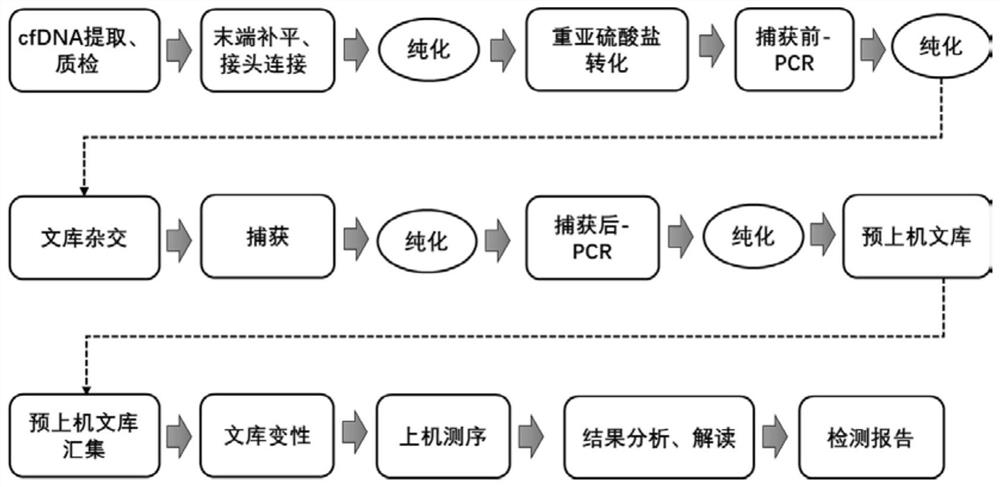
[0081] Such as figure 1 As shown, the implementation process of this application is as follows:
[0082] 1.1.cfDNA extraction and purification
[0083] 1.1.1. Plasma sample preparation:
[0084] Centrifuge the blood sample at 2000g for 10min at 4°C and transfer the plasma to a new centrifuge tube. Centrifuge the plasma sample at 16,000g at 4°C for 10 minutes, and proceed to the next step according to the type of collection tube used. The type of collection tube used in this experiment is other.
[0085] Table 2
[0086]
[0087]
[0088] 1.1.2. Lysis and conjugation
[0089] 1.1.2.1. Prepare the binding solution / bead mixture according to the table below and mix thoroughly.
[0090] table 3
[0091]
[0092] Add an appropriate volume of plasma sample.
[0093] 1.1.2.2. Thoroughly mix the plasma sample and binding solution / bead mixture.
[0094] 1.1.2.3. Combine cfDNA fully on the rotary mixer for 10 minutes to bind the cfDNA to the magnetic beads.
[0095] 1.1...
Embodiment 2
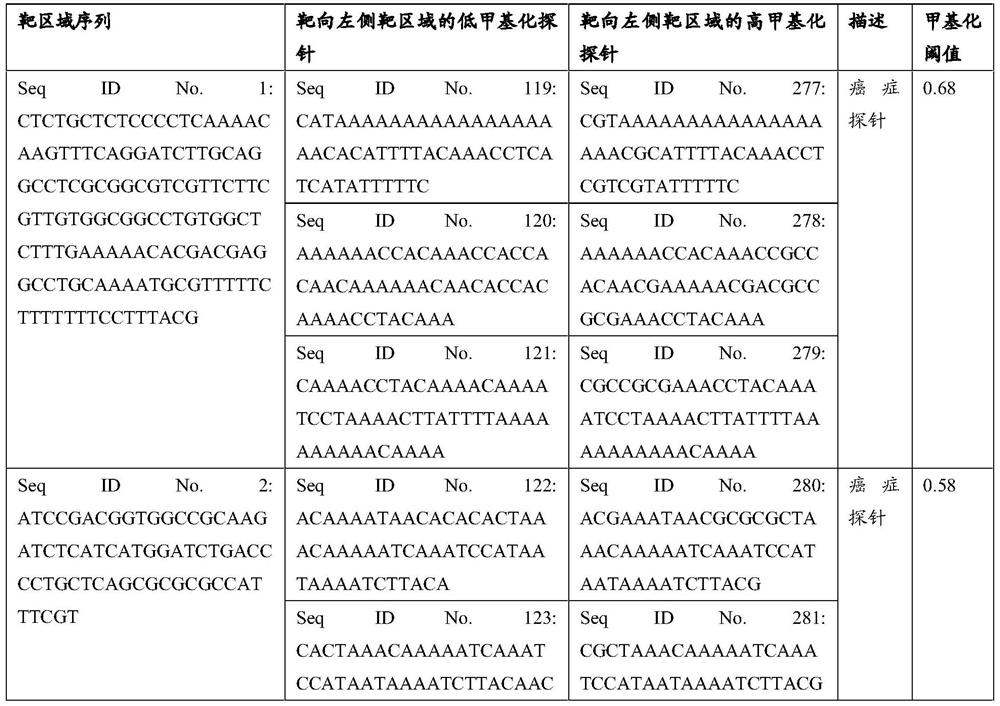
[0244] A case of gastric cancer sample that has been pathologically identified was detected by the Panel of the present application, and peripheral blood was collected according to the method in Example 1; the database was built and sequenced through the Illumina platform; the sequencing data was analyzed through the above-mentioned biological information to obtain the methylation level. The results are shown in Table 19 below (Table 19 shows the detected target regions greater than or equal to the methylation threshold).
[0245] Table 19
[0246]
[0247]
[0248] Carry out pattern recognition classification and identification on the test samples, firstly judge the methylation level of the pan-cancer specific markers TBX15 and CRYGD genes greater than or equal to 55% and 60%, then preliminarily judge that the sample is a sample with cancer; secondly, judge the methylation level of the gene If the methylation levels of cancer-specific markers OTX1, SFRP2, CDO1, TRIM15, ...
Embodiment 3
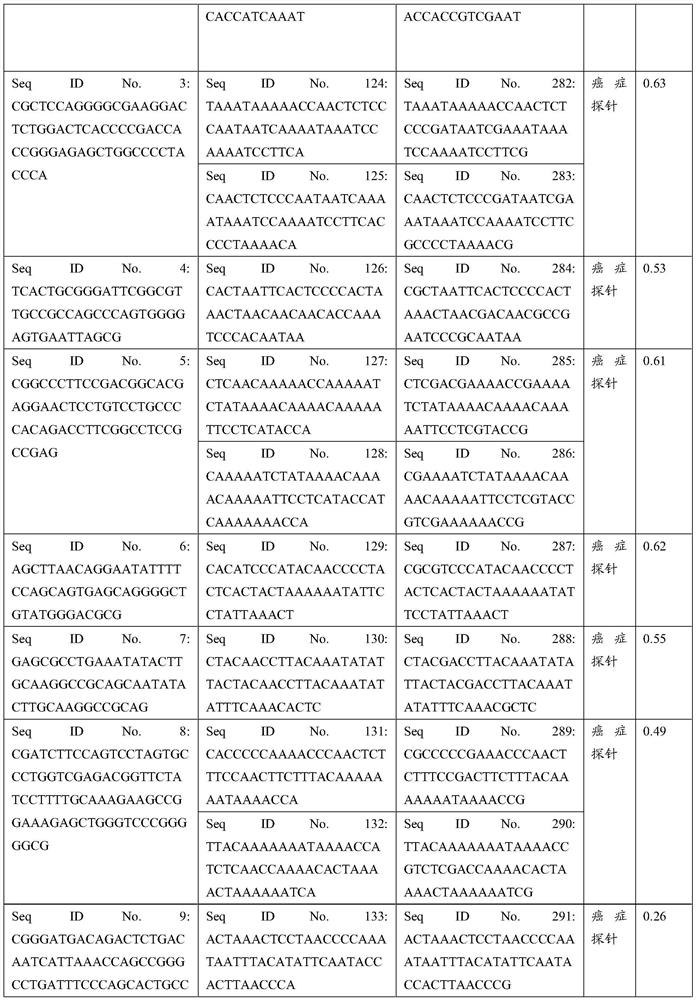
[0251] A sample of colorectal cancer was detected by the Panel of the present application, and peripheral blood was collected according to the method in Example 1; the library was constructed and sequenced through the Illumina platform; the sequencing data was analyzed through the above-mentioned biological information analysis process to obtain the methylation level, and the results are as follows Shown in Table 20 (Table 20 shows the detected target regions greater than or equal to the methylation threshold).
[0252] Table 20
[0253] Gene CHR start termination methylation ratio target region serial number TBX15 1 119527108 119527157 0.55 Seq ID No.63 CRYGD 2 208989200 208989249 0.60 Seq ID No.64 C6orf155 6 72130359 72130408 0.56 Seq ID No.87 C6orf155 6 72130553 72130602 0.71 Seq ID No.88 C6orf155 6 72130641 72130690 0.65 Seq ID No.89 C6orf155 6 72130755 72130804 0.69 Seq ID No.90 SHIS...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com