A nucleic acid library construction method and its application in the analysis of abnormal chromosomal structure of preimplantation embryos
A construction method and nucleic acid library technology, which is applied in the construction of nucleic acid library and the analysis of abnormal chromosome structure of pre-implantation embryos, and can solve the problems of long detection cycle, unfavorable clinical application promotion, and analysis errors, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] Example 1 Balanced Translocation Genetic Family Embryo Detection
[0072] A family of chromosomal balanced translocation carriers who underwent assisted reproduction was recruited (the chip test results were available), and the family information is shown in Table 3. 5 mL of peripheral blood samples (a total of three blood samples) were collected from the balanced translocation carrier, wife, and carrier inherited from one parent, and stored in EDTA anticoagulated blood collection tubes. augmented product. Genomic DNA of the pedigree was extracted using a whole blood extraction kit. Preimplantation chromosomal balanced translocation analysis was performed using the method of the present invention.
[0073] Table 3 Chromosomal karyotype table of balanced translocation family (chip results)
[0074]
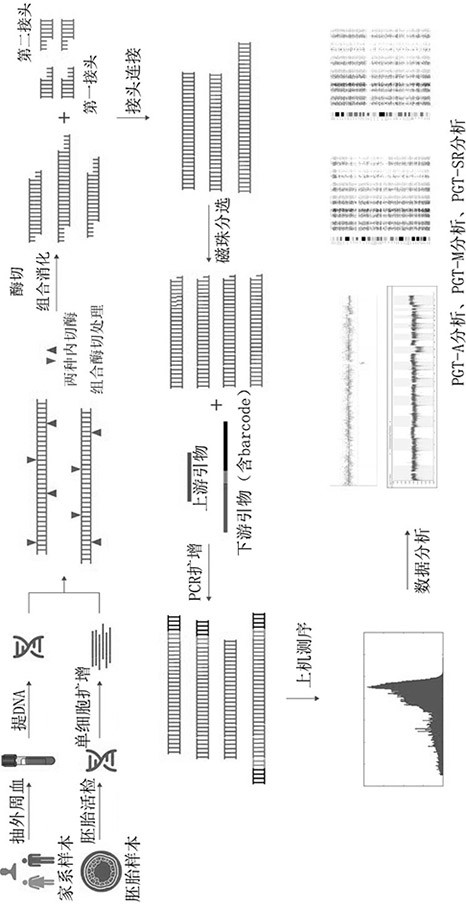
[0075] principle such as figure 1 As shown, firstly, a group of endonucleases recognize specific enzyme cutting sites on the genome, and the genomic DNA is cut into t...
Embodiment 2
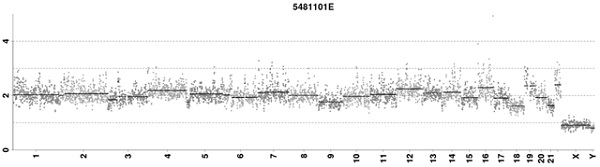
[0126] Chromosomal aneuploidy cell line samples (5 cells) are used for details, see Table 12 below, and whole genome amplification is performed using the MDA method.
[0127] Table 12 Cell line sample details
[0128]
[0129] Detect according to the method of embodiment 1, cell line PGT-A data analysis result is as follows Figure 10 As shown, the detection results show that the method can detect more than 5M deletions.
Embodiment 3
[0136] The method of this embodiment is basically the same as that of Example 1, the only difference is that the endonuclease combination adopts BfaI and TaqI, or MboI and MspI respectively, and can also obtain sufficient SNP sites and indel sites under low data volume. Construct a genetic map to identify whether there is an abnormality in the structure of the embryo's chromosomes, and then distinguish between balanced translocation carrier embryos and normal embryos.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com