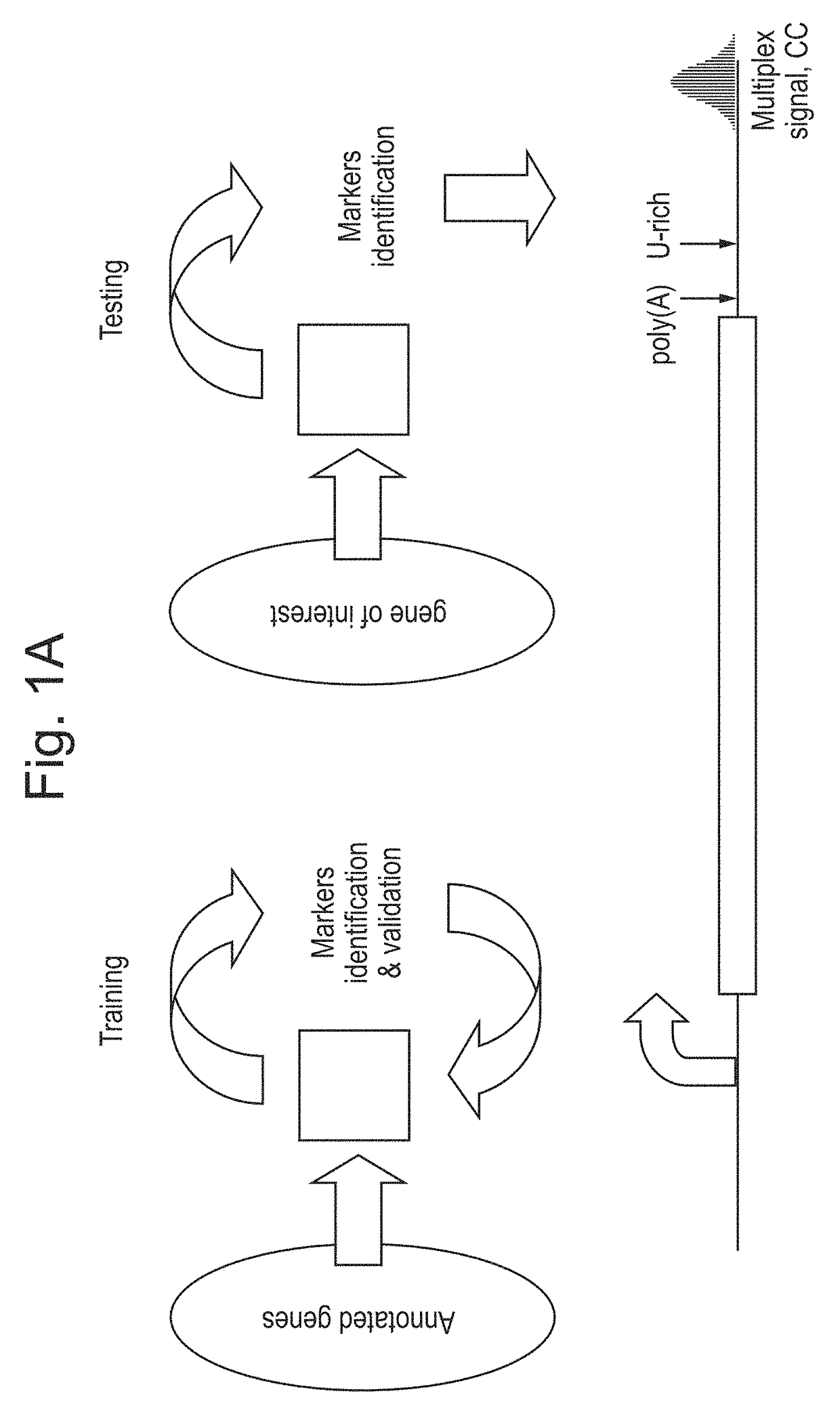
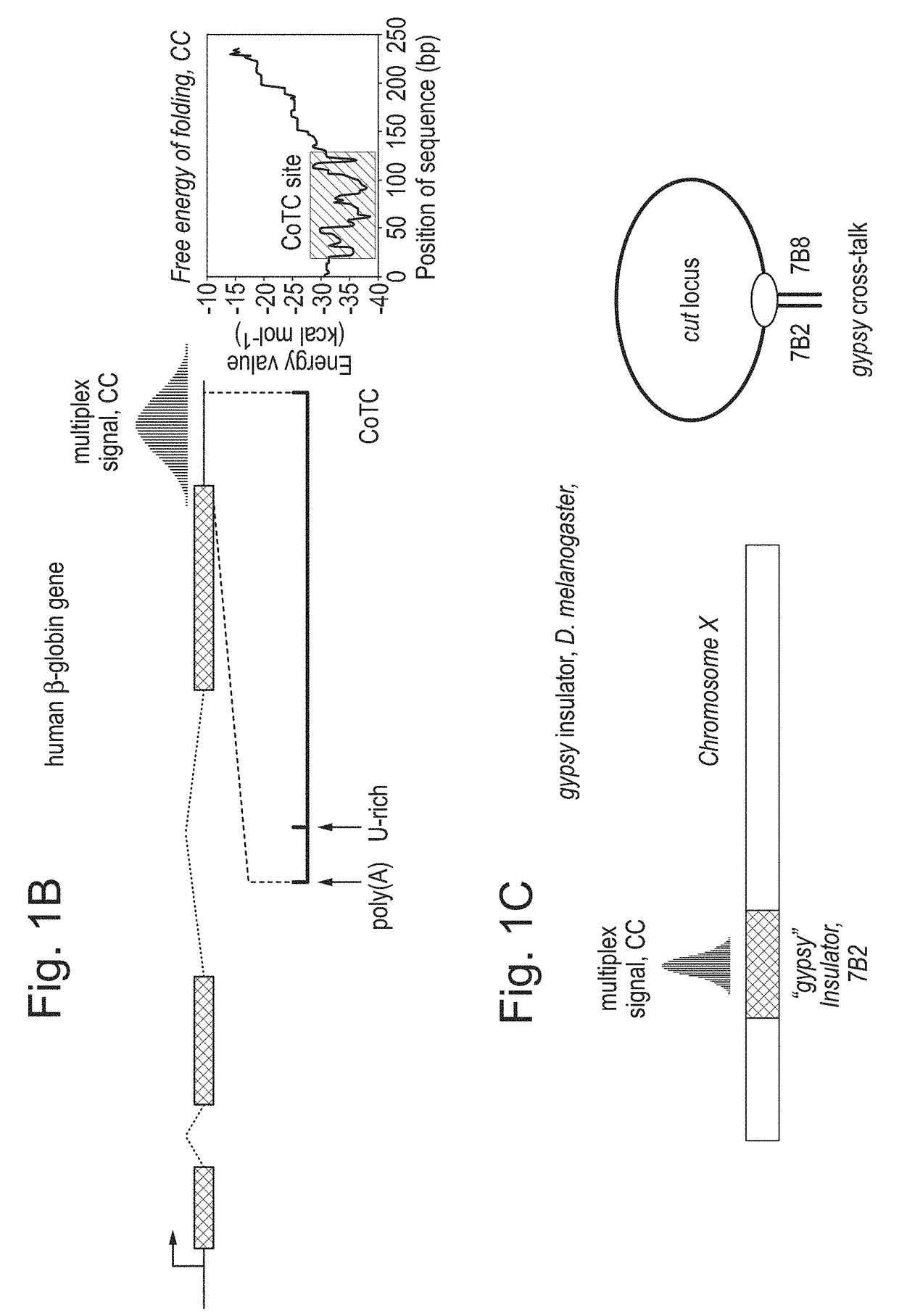
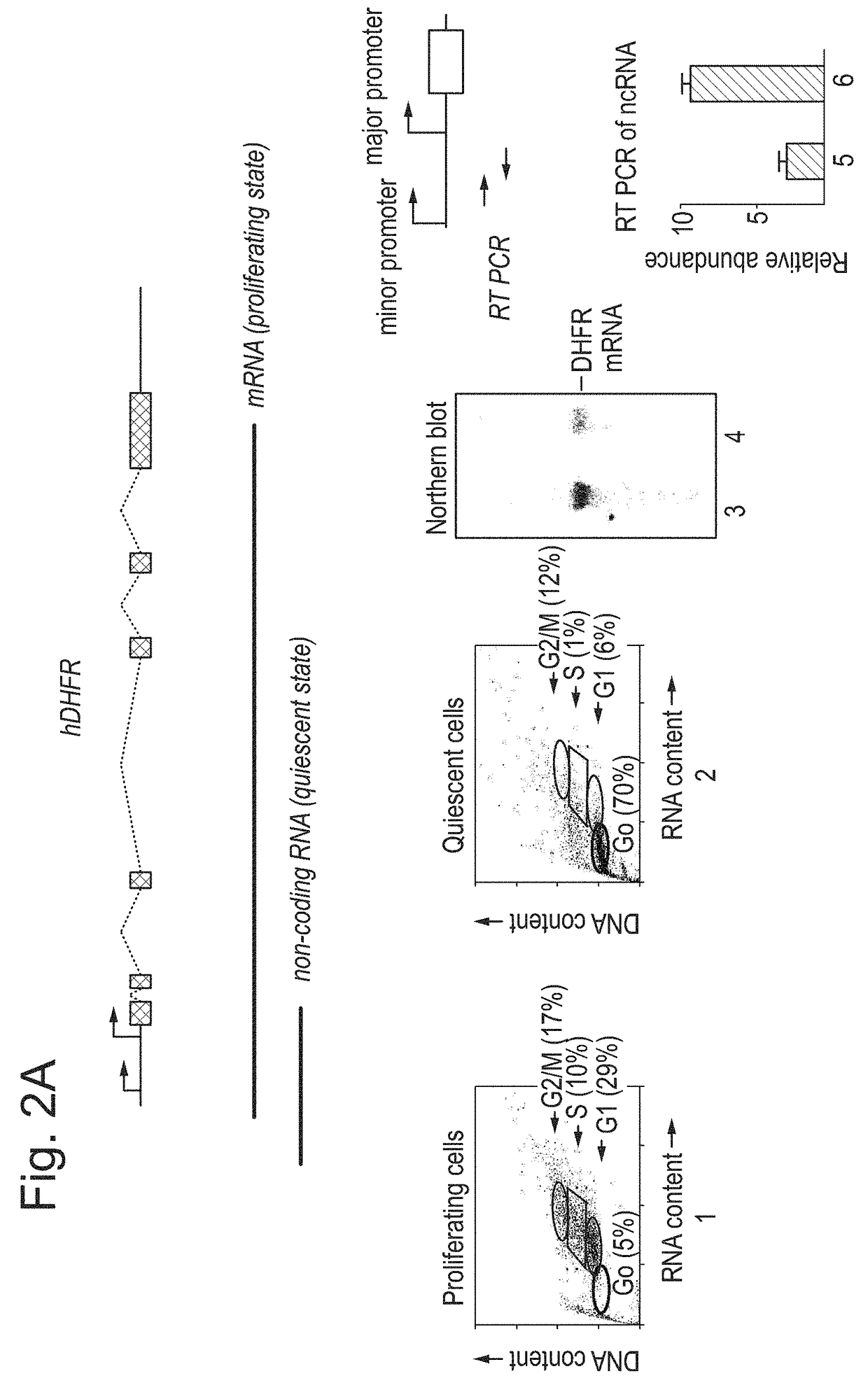
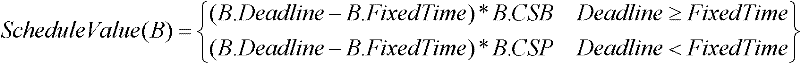Patents
Literature
43 results about "Chromosome Structures" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
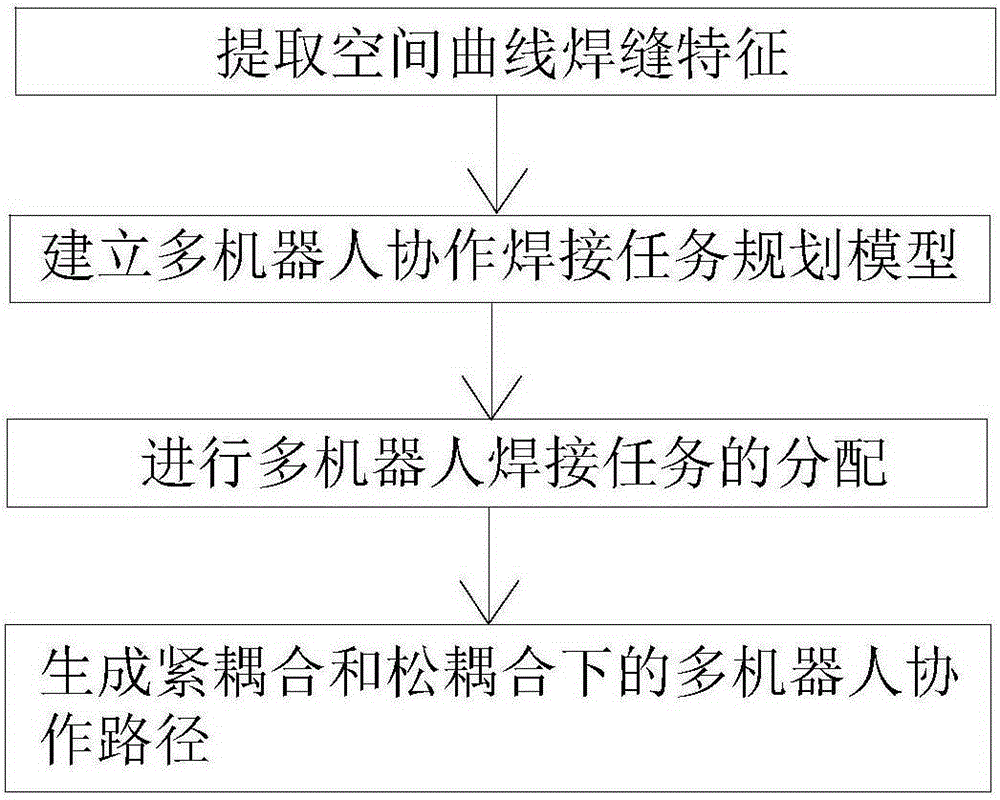
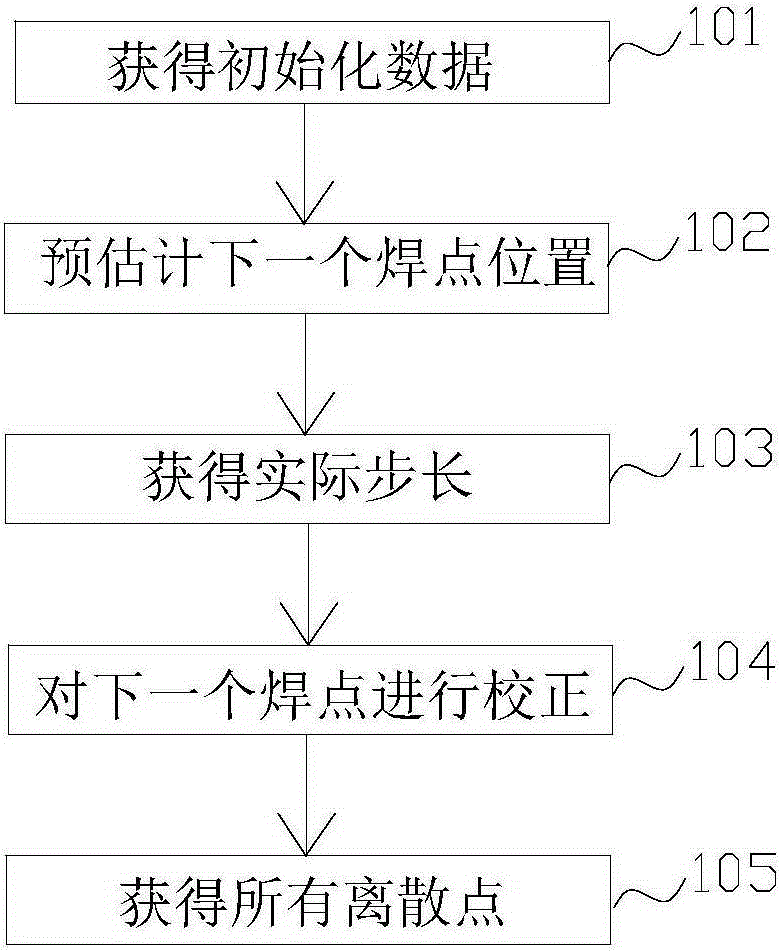
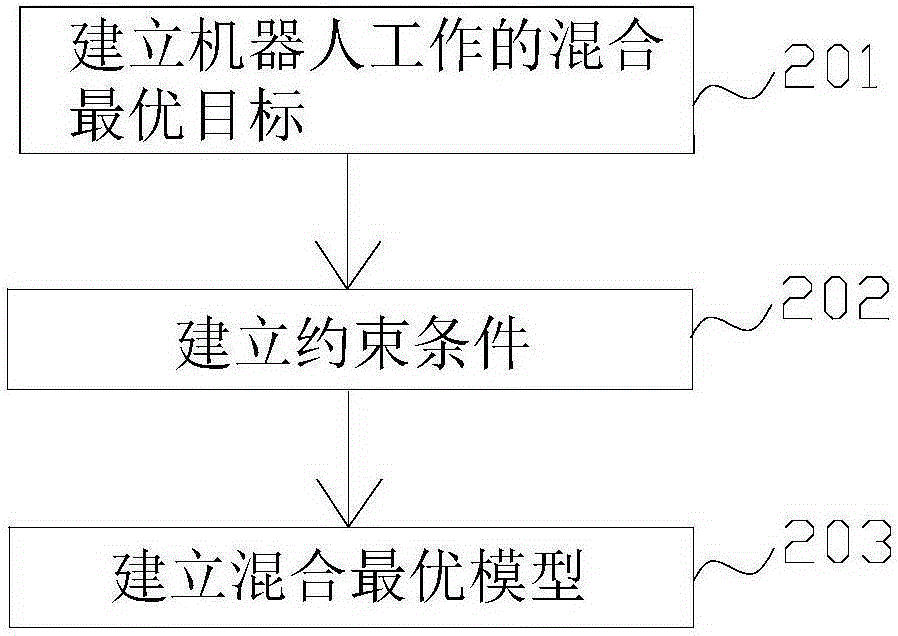
Task decoupling-based multi-robot collaboration welding path generation method
The invention discloses a task decoupling-based multi-robot collaboration welding path generation method. The method includes the following steps that: first step, based on a to-be-welded workpiece model, a chord tolerance and minimum step-combined method is adopted to perform discretization processing on welding seams through non-equidistant point sampling, so that space curve welding seam features can be extracted; second step, a multi-robot cooperative welding task planning model is established; third step, a multi-operator multi-chromosome structured improved genetic algorithm is adopted to distribute multi-robot welding tasks; and fourth step, a motion closed chain-based multi-robot master-slave type collaboration motion model is established, so that multi-robot cooperation paths under tight coupling and loose coupling are generated. According to the method of the invention, the unified multi-robot cooperation welding model is established according to the coupling relations of multiple master-slave robots, and therefore, task planning efficiency of multi-robot cooperation welding can be effectively improved.
Owner:SOUTHEAST UNIV
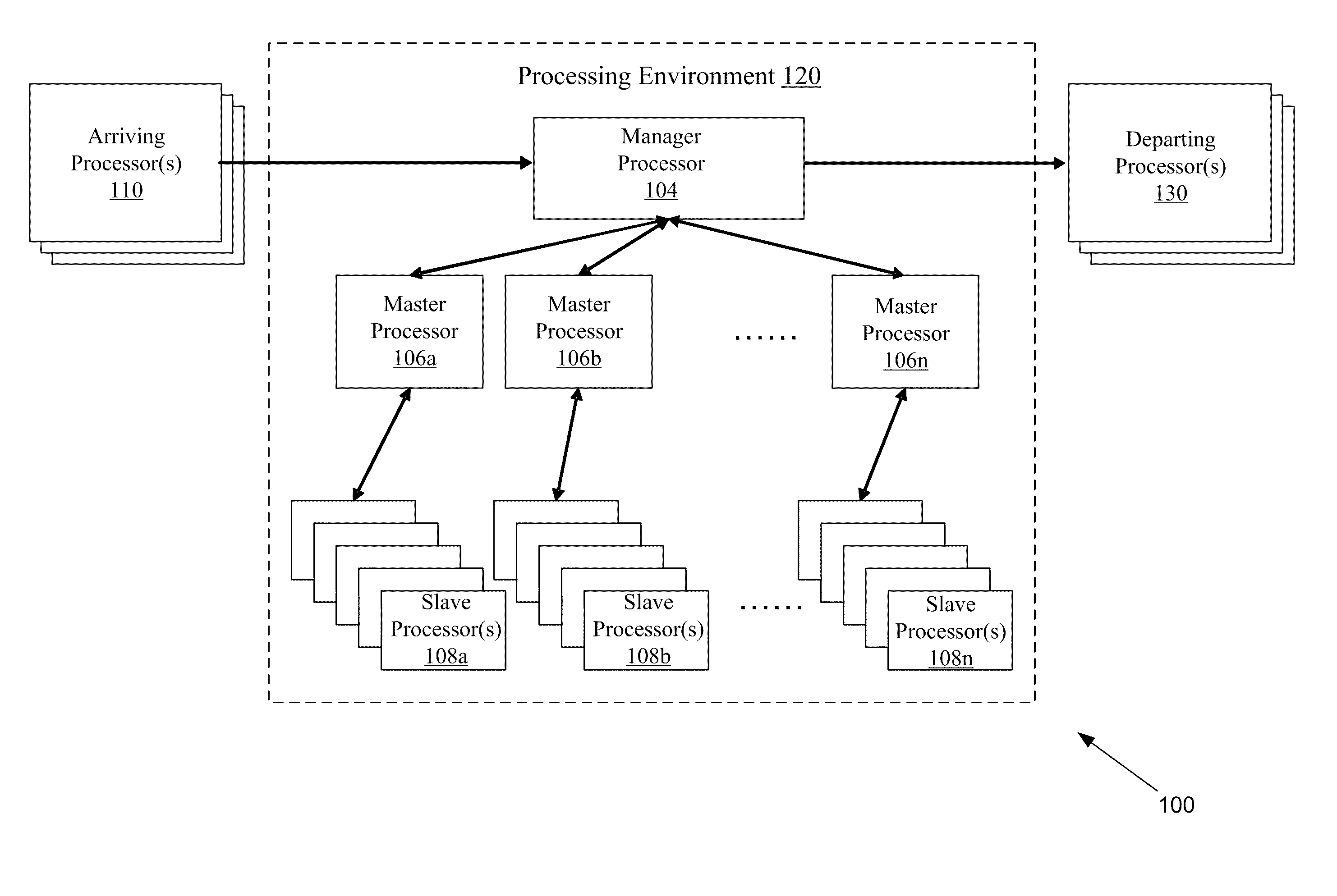
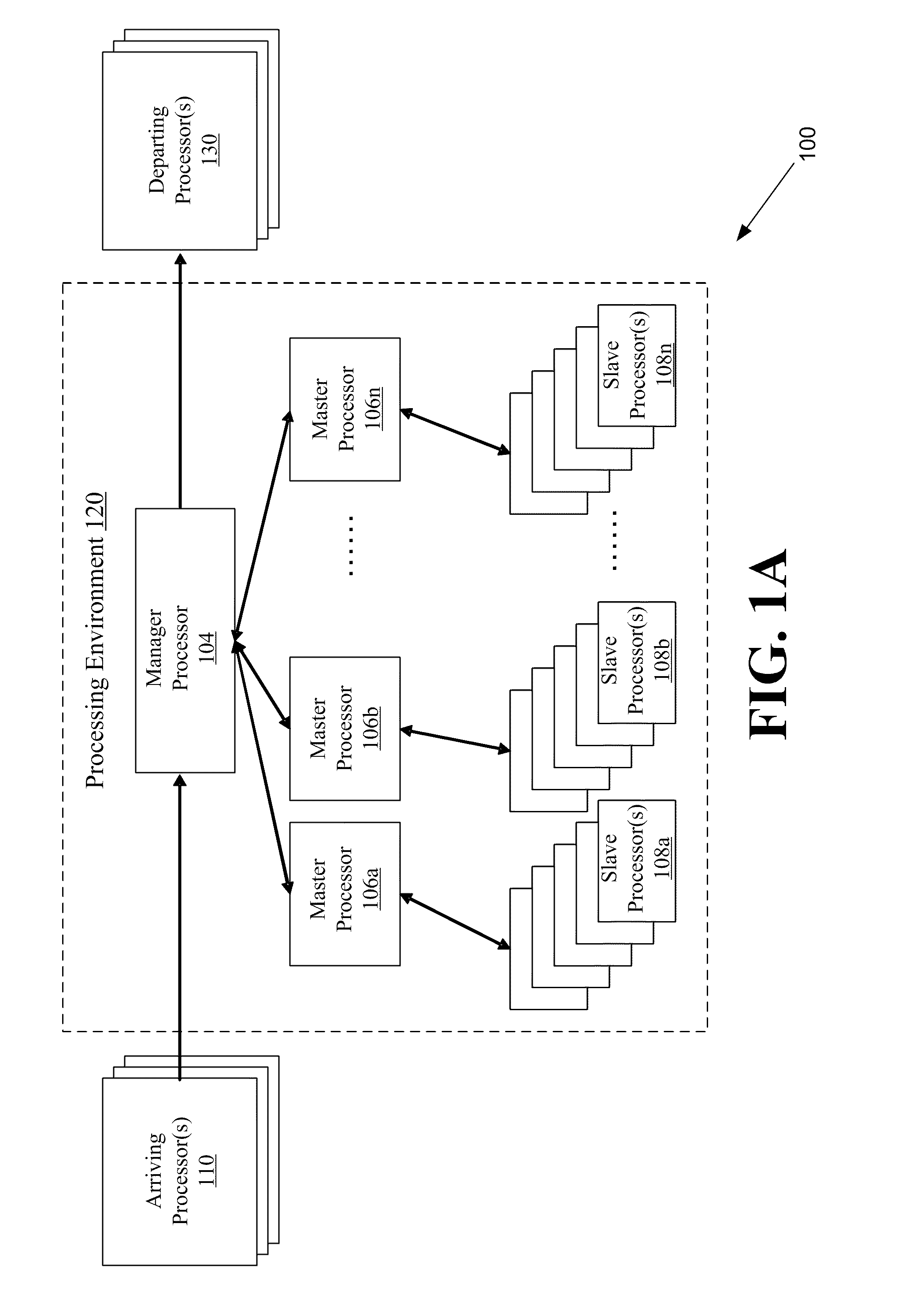
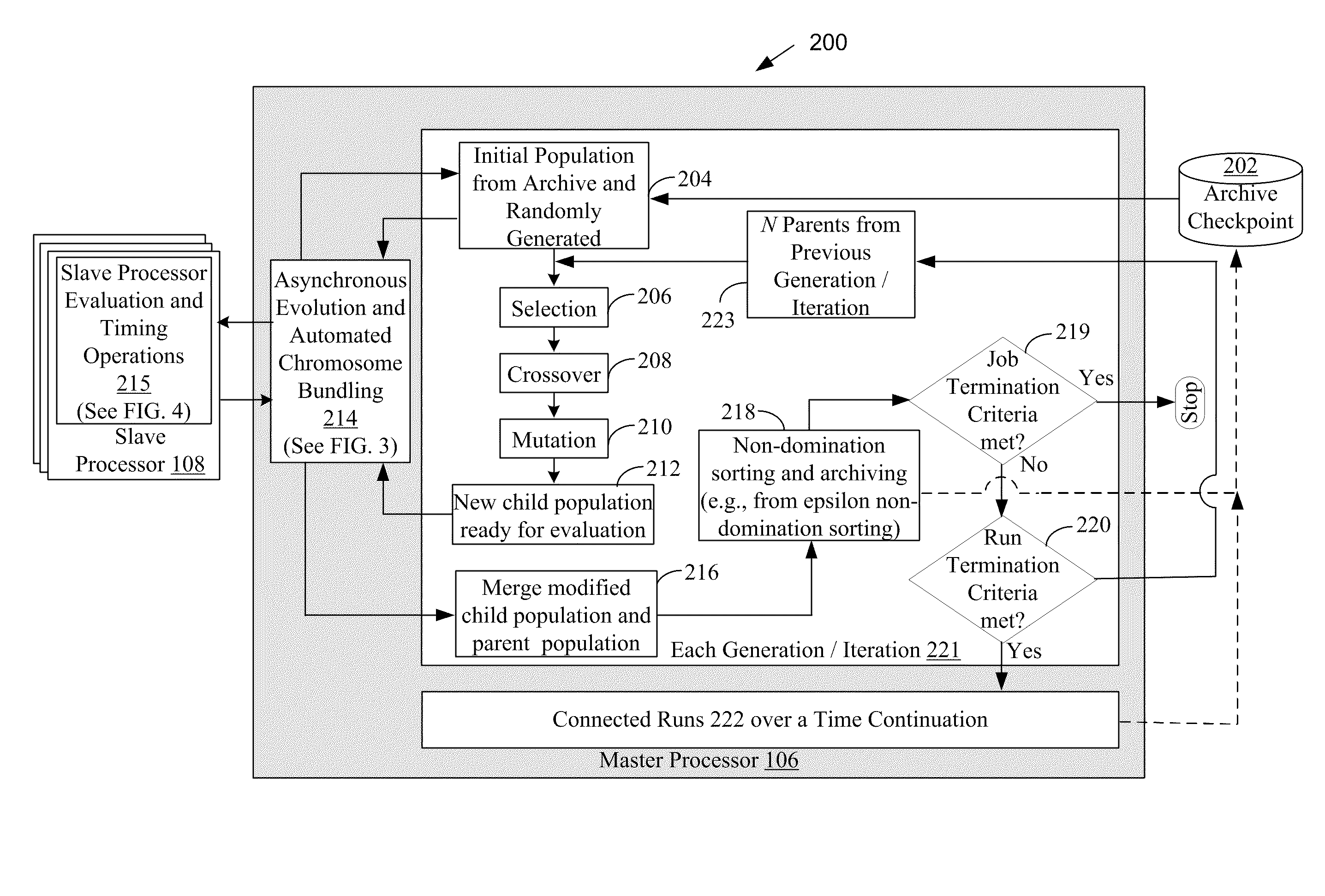
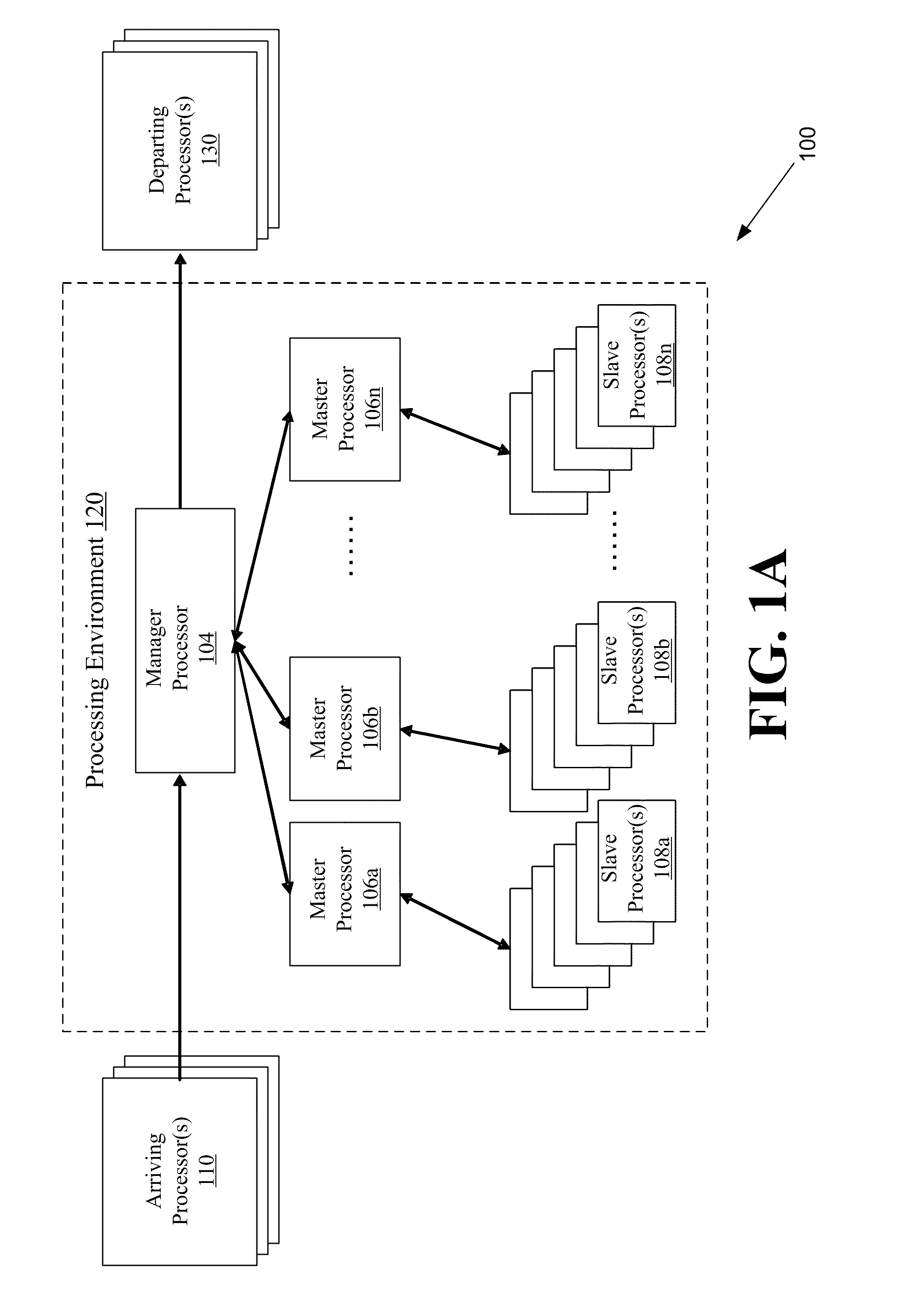
Systems and methods for parallel processing optimization for an evolutionary algorithm
ActiveUS20100293119A1Program controlNeural learning methodsParallel processingEvolutionary operators
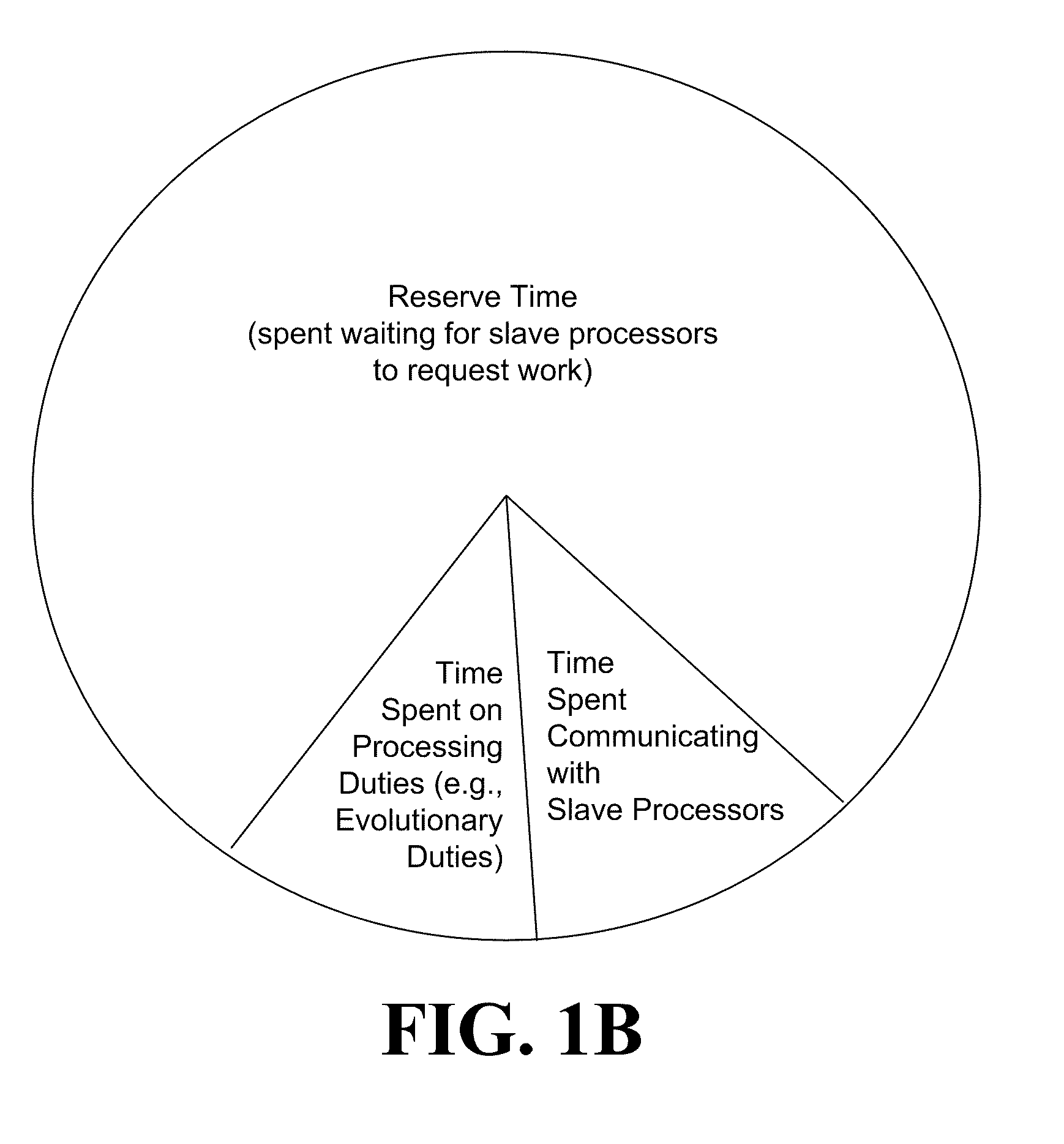
The systems and methods may include receiving an initial population of parent chromosome data structures, where each parent chromosome data structure provides a plurality of genes; selecting pairs of parent chromosome data structures; applying at least one evolutionary operator to the genes of the selected pairs to generate a plurality of child chromosome data structures; allocating, the generated plurality of child chromosome structures to a plurality slave processors, where each slave processor evaluates one or more of the plurality of child chromosome data structures and generates respective objective function values; receiving objective function values for a portion of the plurality of allocated child chromosome data structures; merging the parent chromosome data structures with the received portion of the child chromosome data structures for which objective function values have been received; and identifying a portion of the merged set of chromosome data structures as an elite set of chromosome data structures.
Owner:THE AEROSPACE CORPORATION
Systems and methods for parallel processing optimization for an evolutionary algorithm
ActiveUS8255344B2General purpose stored program computerNeural learning methodsAlgorithmGene selection
Owner:THE AEROSPACE CORPORATION
Chromosome sequencing method based on stripe recognition
ActiveCN110533672AAccurate identificationEasy to observeImage enhancementImage analysisImaging processingMonosomal karyotype
The invention discloses a chromosome sorting method based on stripe recognition, and belongs to the technical field of chromosome image processing. According to an existing chromosome sequencing method, due to the fact that manual recognition is prone to interference, selection and arrangement accuracy is low, and structural comparison of doctors is affected. The method comprises the following steps: step 1, obtaining a chromosome karyotype diagram; step 2, carrying out chromosome cutting on the karyotype diagram to obtain a plurality of chromosome images, and identifying chromosome types; 3,extracting a plurality of images of chromosomes of the same number, and identifying chromosome stripe types based on an intelligent algorithm model; and step 4, sorting the plurality of divided chromosome images according to the chromosome stripe type changes of the chromosome images. The strip types of the chromosome images can be predicted, the chromosome strip number can be accurately recognized, reasonable sorting is performed according to the chromosome strip number, a doctor can conveniently observe the chromosome structure, the burden of the doctor can be effectively reduced, and time and labor are saved.
Owner:HANGZHOU DIAGENS BIOTECH CO LTD
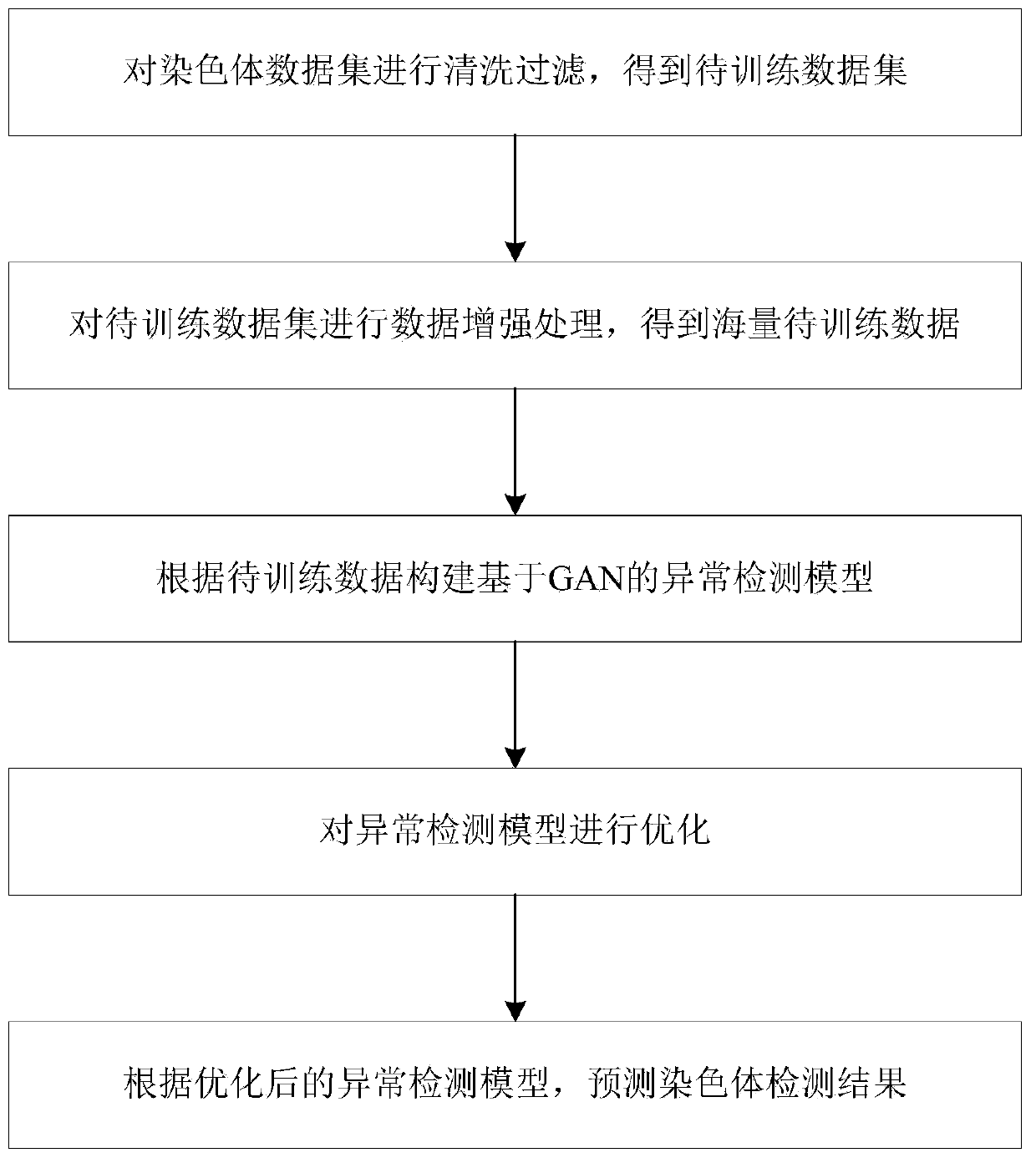
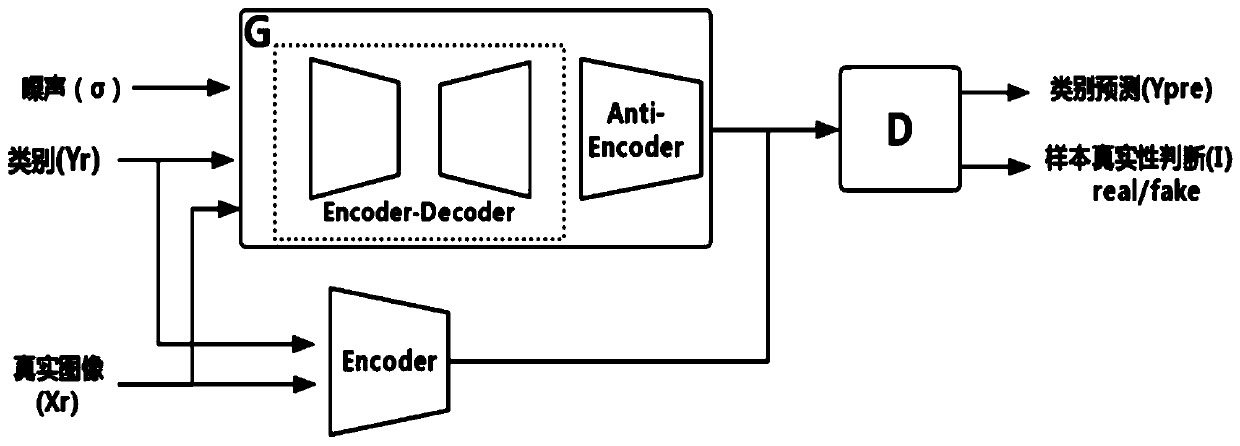
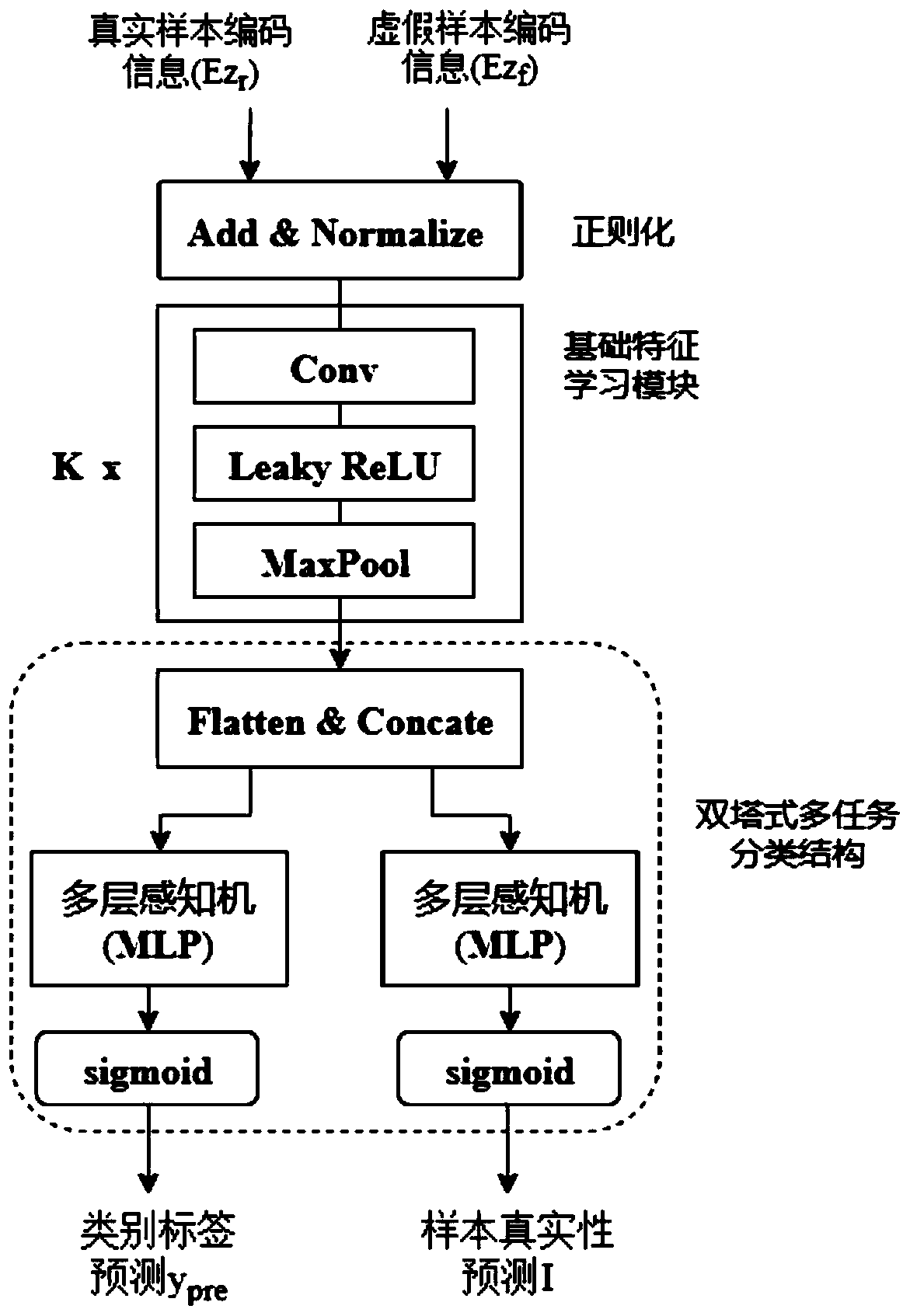
GAN-based chromosome structure anomaly detection method and system and storage medium
ActiveCN111105032AAvoid missingImprove practicalityMachine learningNeural learning methodsData setAnomaly detection
The invention discloses a GAN-based chromosome structure anomaly detection method and system and a storage medium, and the method comprises the steps: carrying out cleaning and filtering of a chromosome data set, and obtaining a to-be-trained data set; performing data enhancement processing on the to-be-trained data set to obtain massive to-be-trained data; constructing a GAN-based anomaly detection model according to the to-be-trained data; optimizing the anomaly detection model; and predicting a chromosome detection result according to the optimized anomaly detection model. The GAN-based anomaly detection model is constructed, only normal chromosomes need to be trained and feature information of the normal chromosomes needs to be learned, and chromosome structure anomaly detection is performed by fully utilizing the difference between the feature information of the normal chromosomes and the feature information of abnormal chromosomes; besides, the invention can be used for carryingout chromosome classification and anomaly detection on chromosomes, is high in practicability, and can be widely applied to the technical field of machine learning.
Owner:SOUTH CHINA NORMAL UNIVERSITY +1
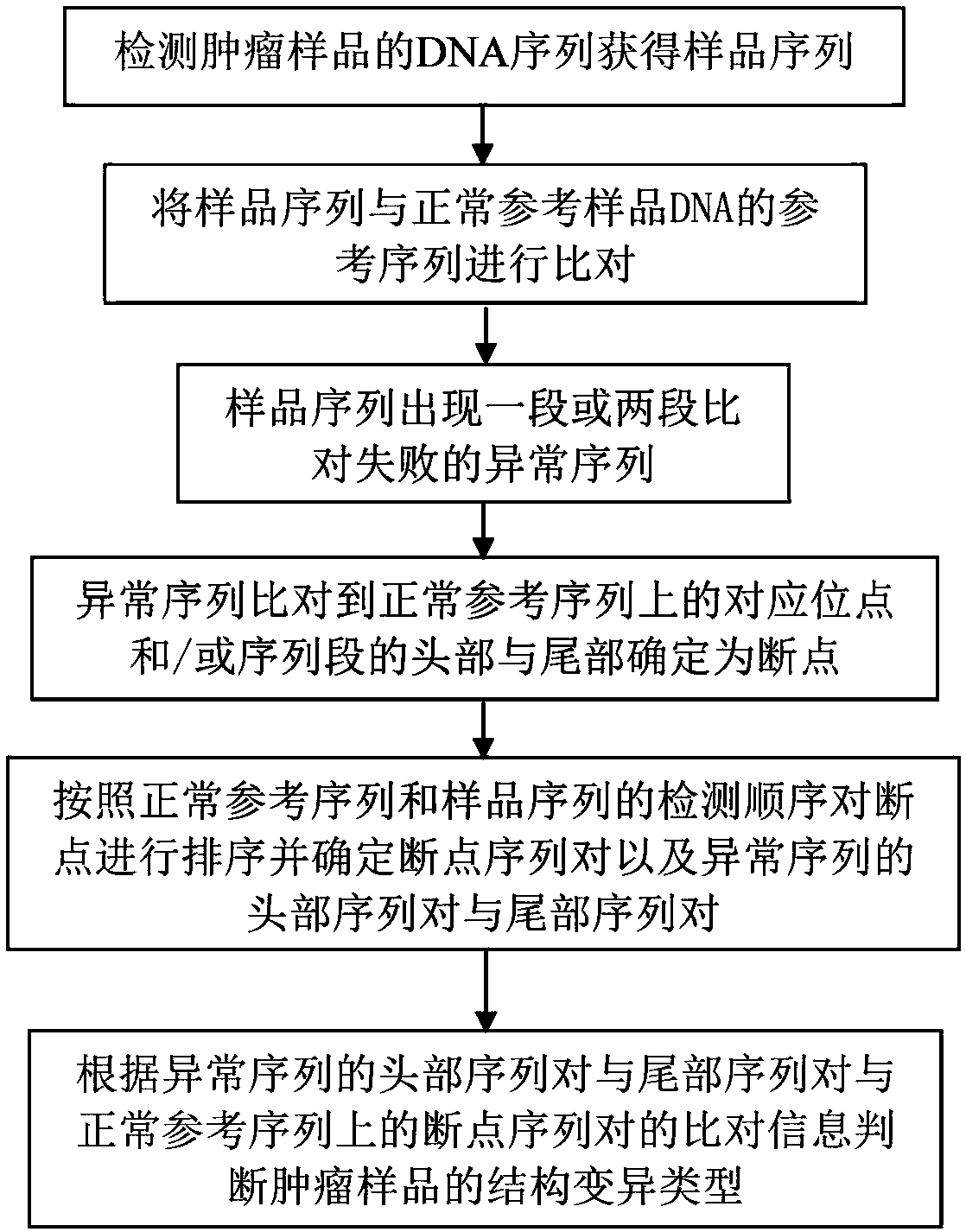
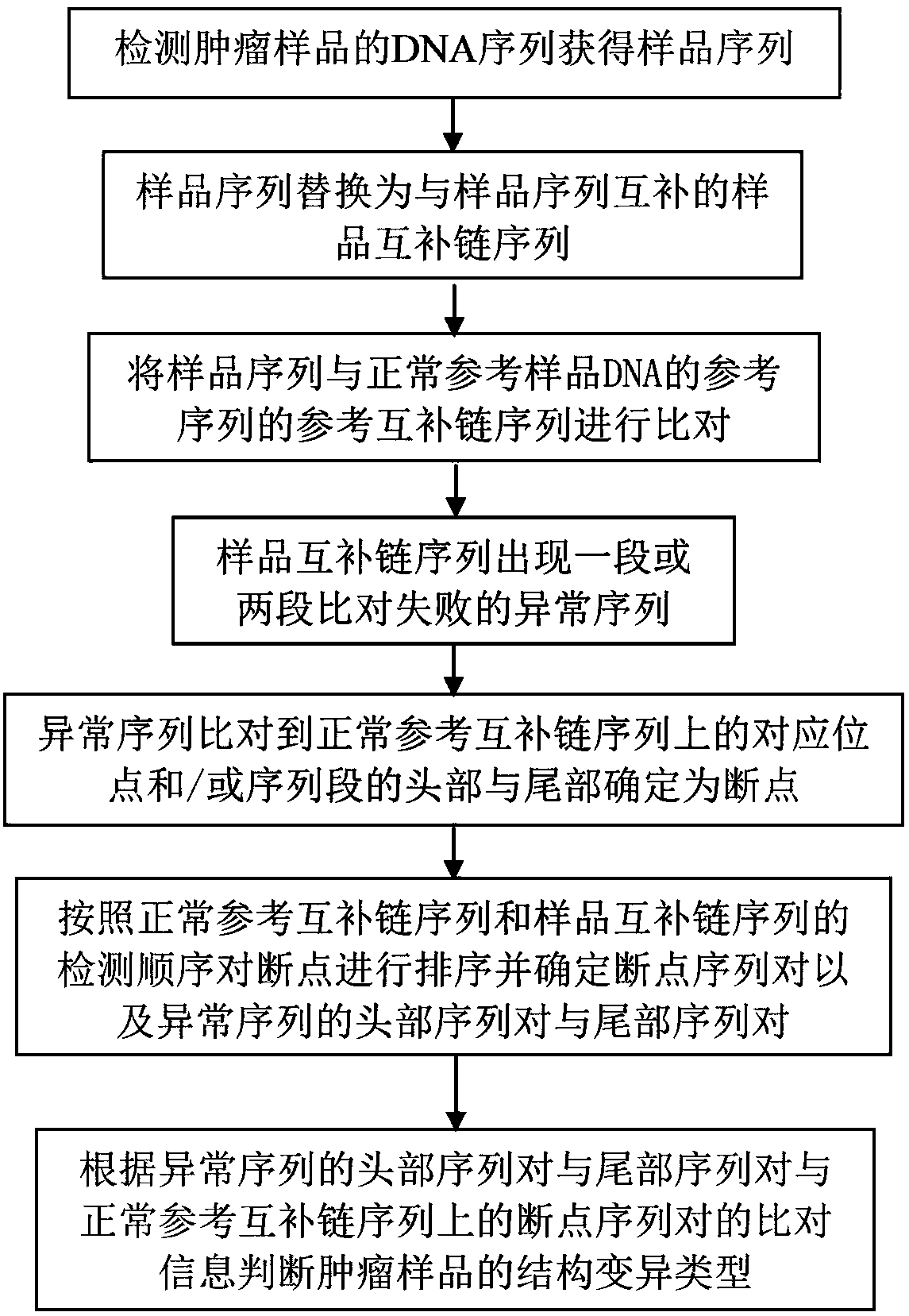
Method for determining chromosome structure variation signal intensity and insert fragment length distribution characteristics of sample, and application thereof
The invention provides a method for determining chromosome structure variation signal intensity and insert fragment length distribution characteristics of a sample, and application thereof. Specifically, the invention relates to a method for determining a sample source. The method comprises the following steps: (1) comparing sequencing read data of chromosomes in a sample with a reference genome,and determining a low-quality comparison rate and a high-quality comparison rate of the sequencing read data of the chromosomes of the sample; (2) determining the structural variation proportion of the chromosomes of the sample, the content of mitochondrial DNA and the proportion of an insert fragment with a predetermined length based on reads corresponding to the high-quality comparison rate; (3)determining the probability of sample sources based on a predetermined tumor prediction model, the structural variation proportion of the chromosomes obtained in the step (2), the content of the mitochondrial DNA and the proportion of the insert fragment with the predetermined length; and (4) determining the source of the sample based on the probability of the sample source.
Owner:深圳思勤医疗科技有限公司
Method and system for determining individual chromosome structure abnormity
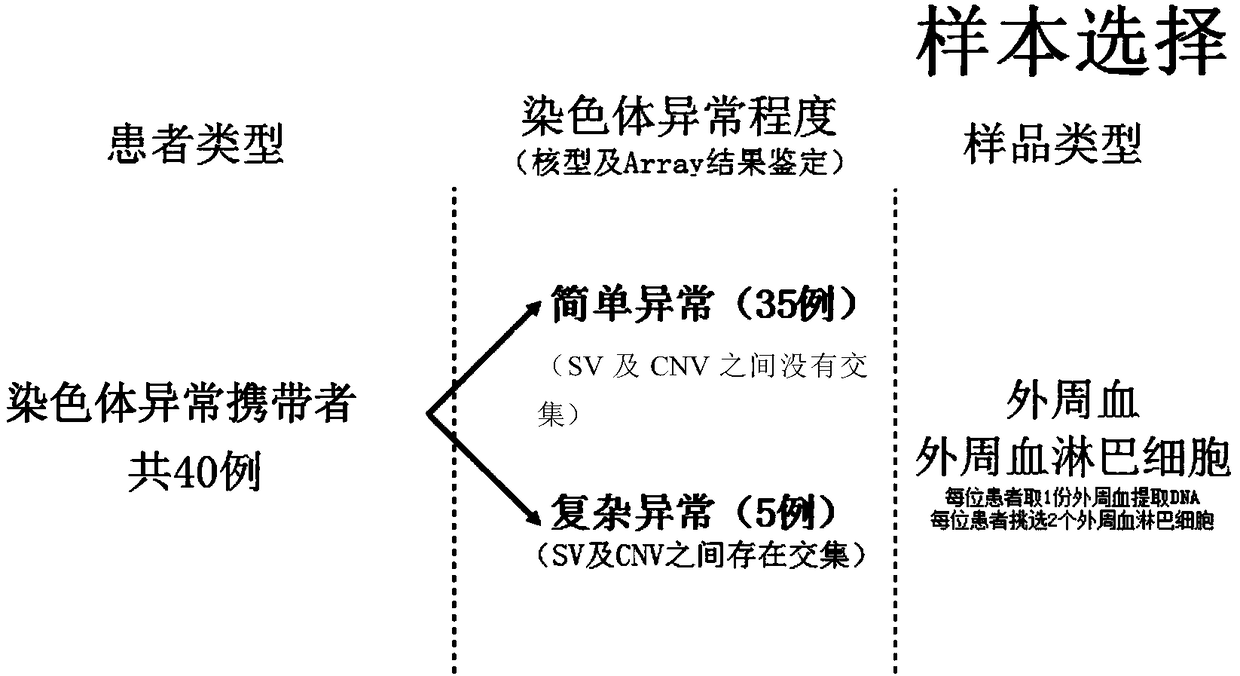
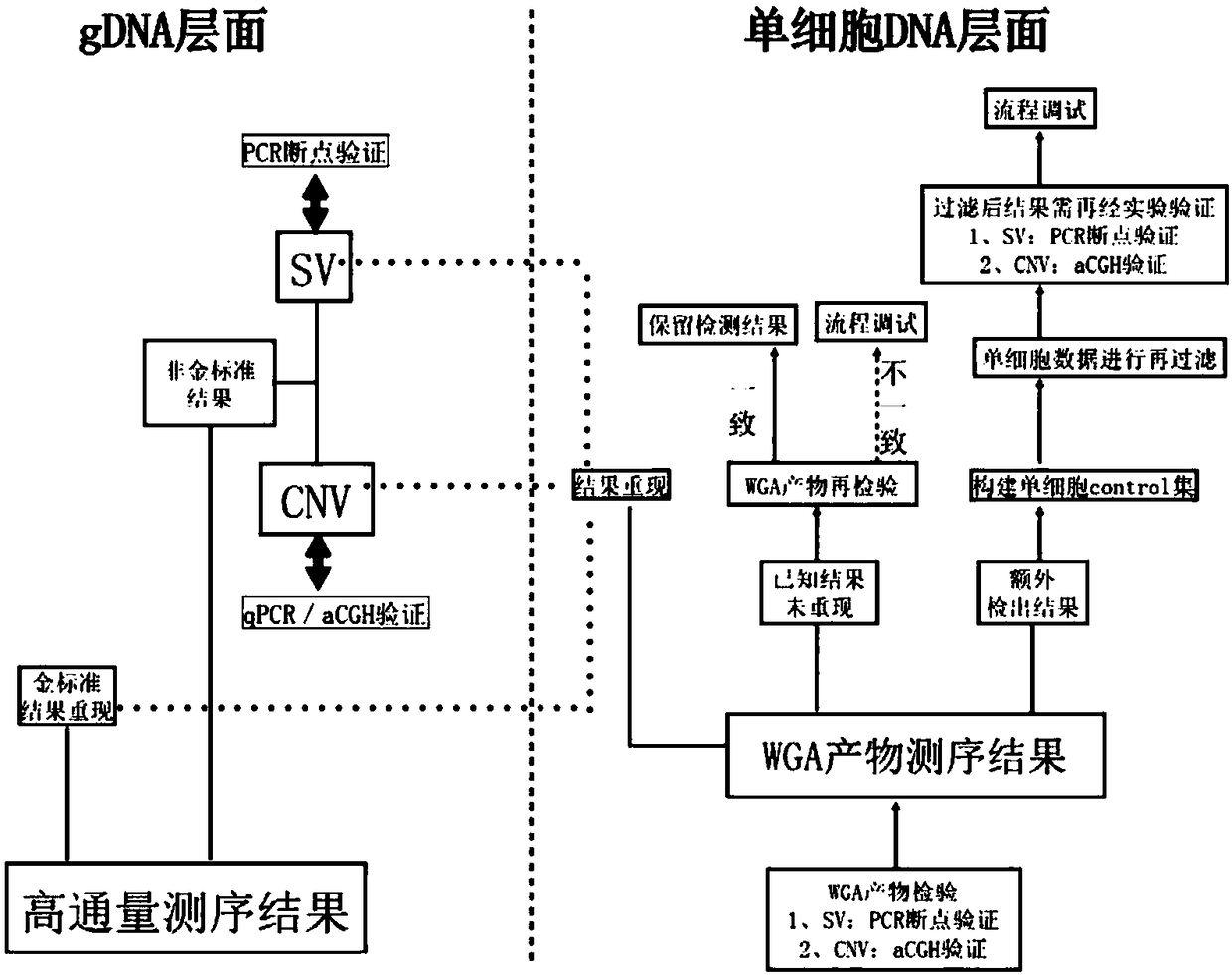
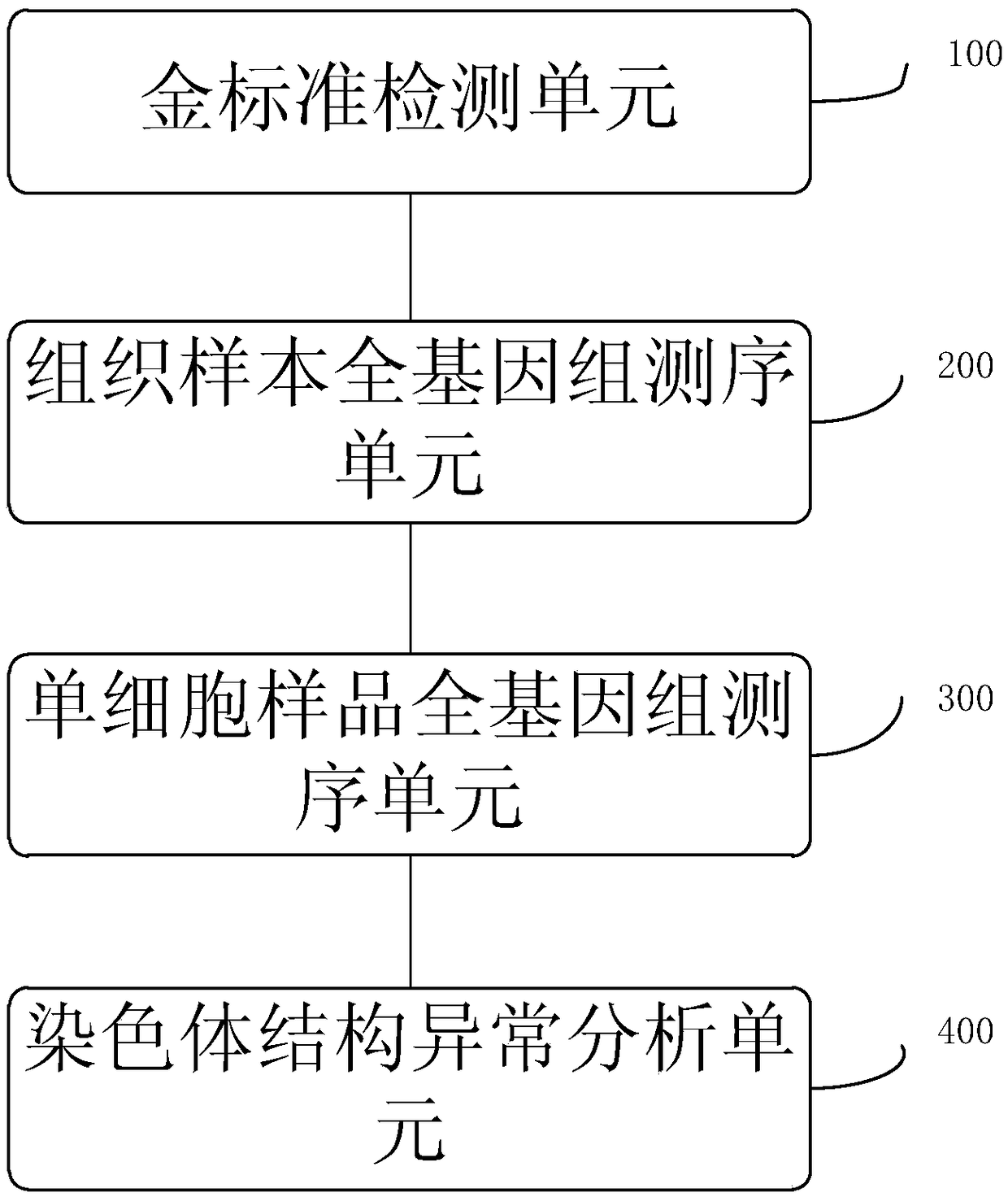
PendingCN109280702AImprove the detection rateMicrobiological testing/measurementWhole genome sequencingTissue sample
The invention provides a method for determining an individual chromosome structure abnormity. The method includes the following steps: (1) determining first candidate chromosome structure abnormity types of an individual by using at least one of karyotyping and microarray chip analysis to obtain a first candidate chromosome abnormality type set; (2) performing whole genome sequencing on tissue samples of the individual and preforming first data analysis on obtained sequencing results to obtain a second candidate chromosome abnormality type set; (3) performing whole genome sequencing on singlecell samples of the individual and preforming second data analysis on obtained sequencing results to obtain a third candidate chromosome abnormality type set; and (4) determining a final chromosome structure abnormity type of the individual based on the first candidate chromosome abnormality type set, the second candidate chromosome abnormality type set and the third candidate chromosome abnormality type set.
Owner:SHENZHEN HUADA GENE INST
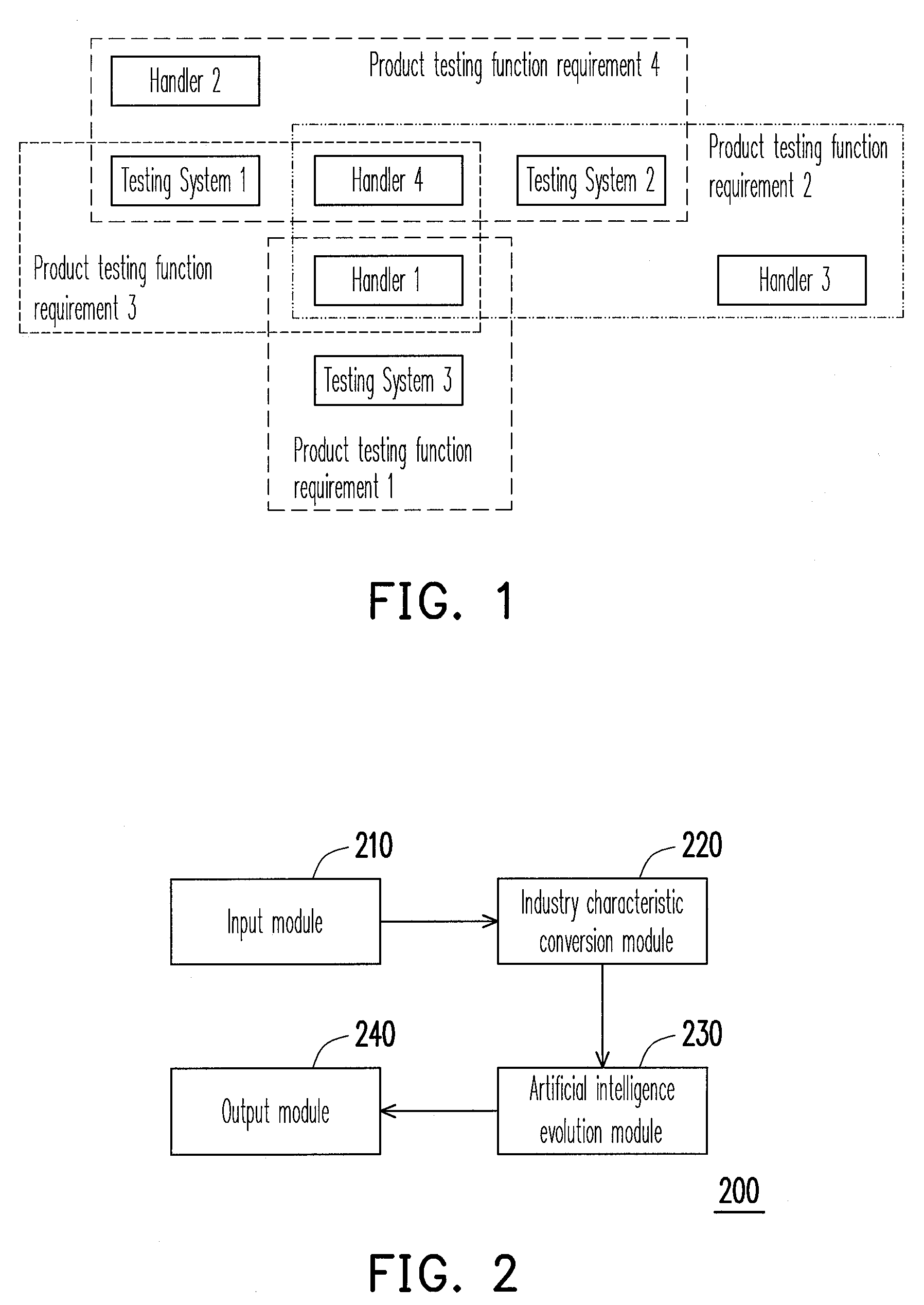
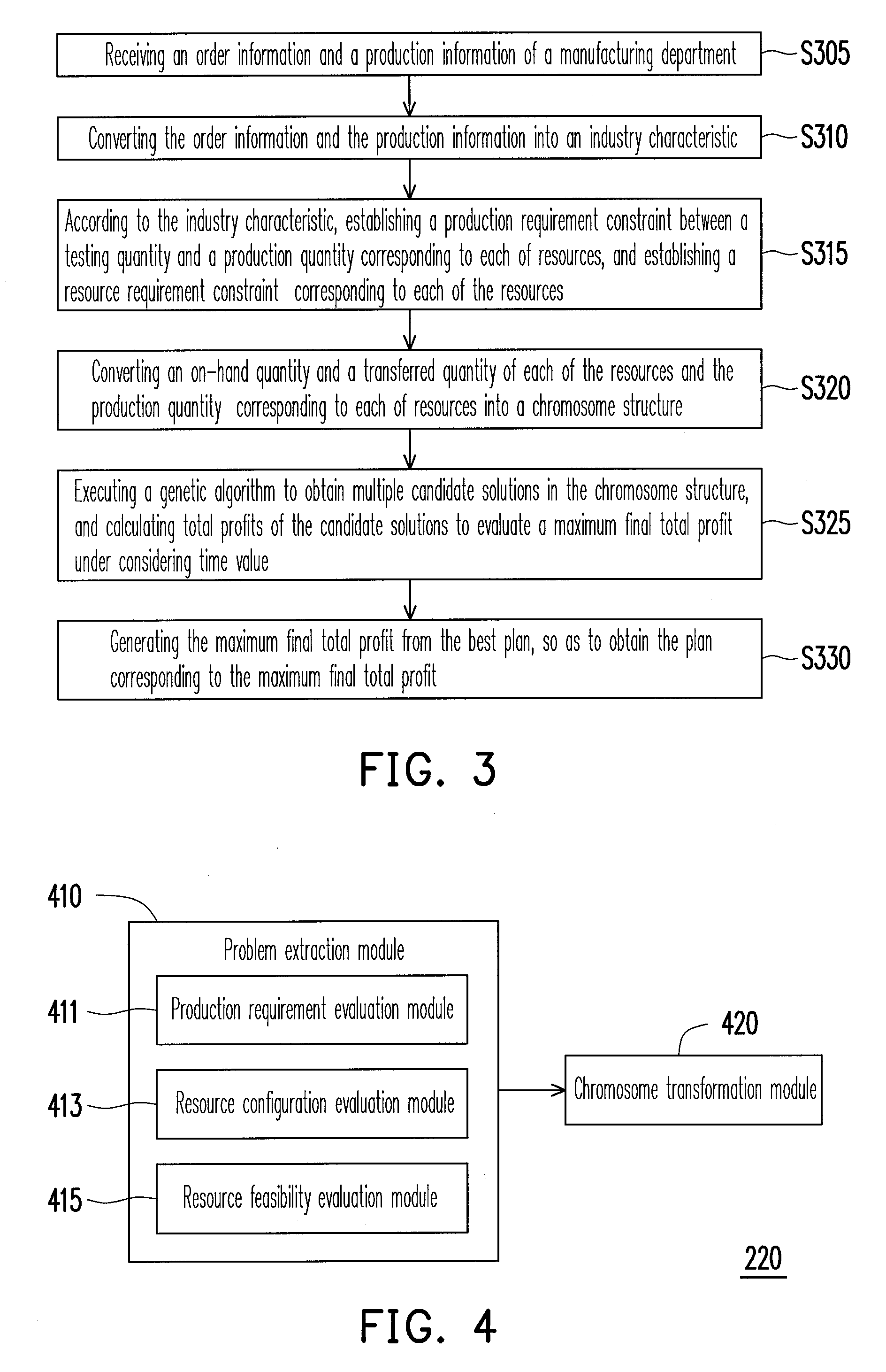
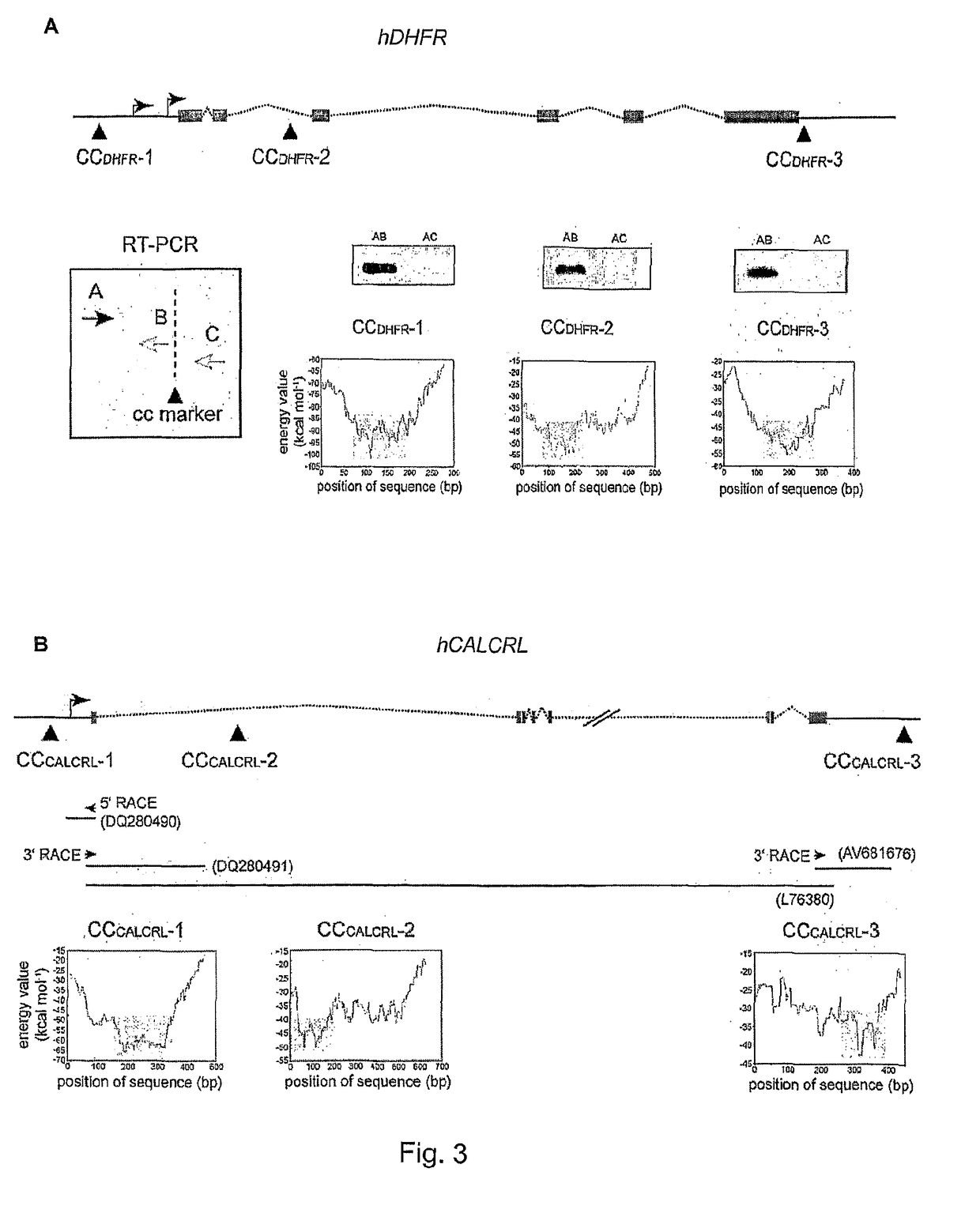
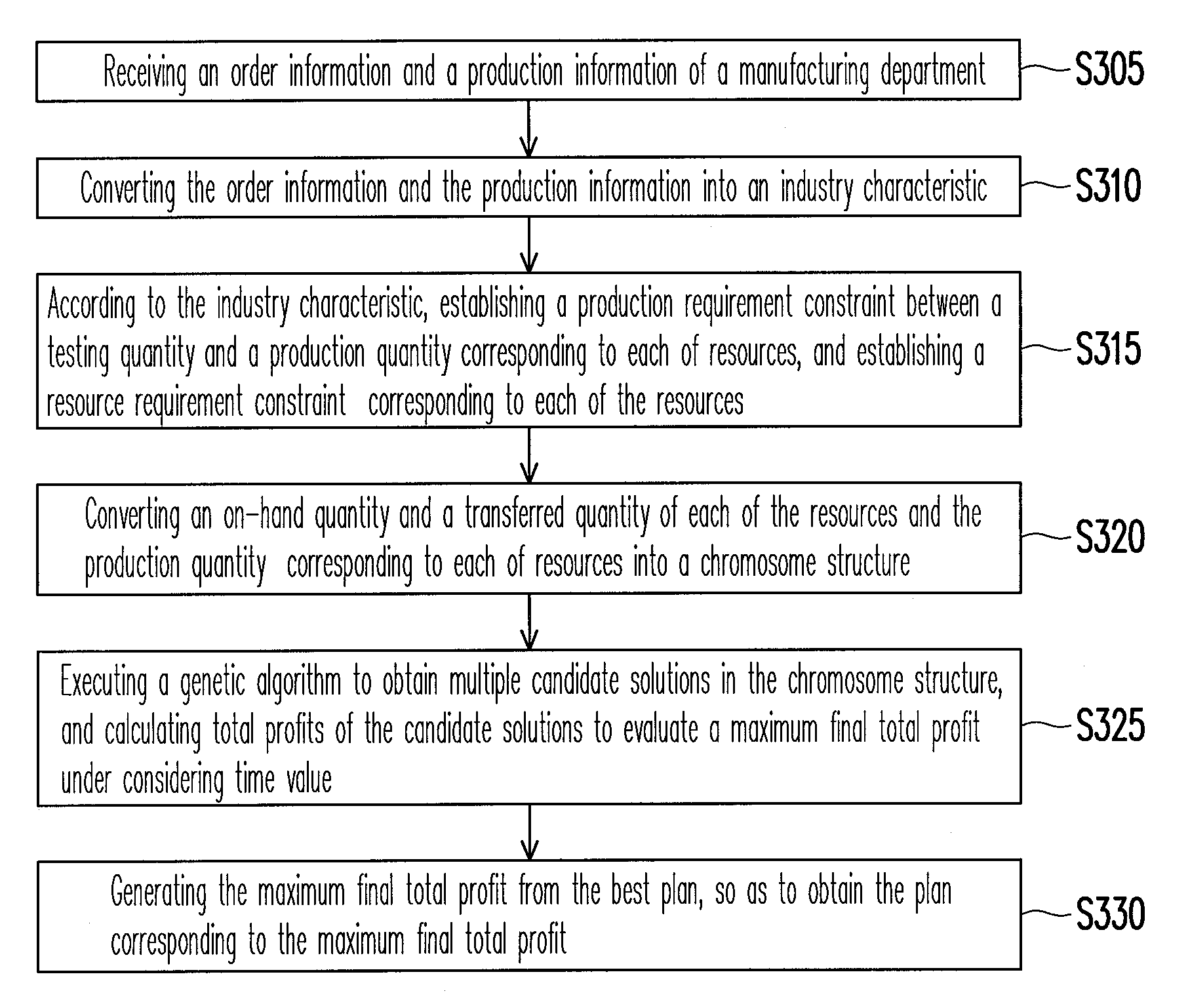
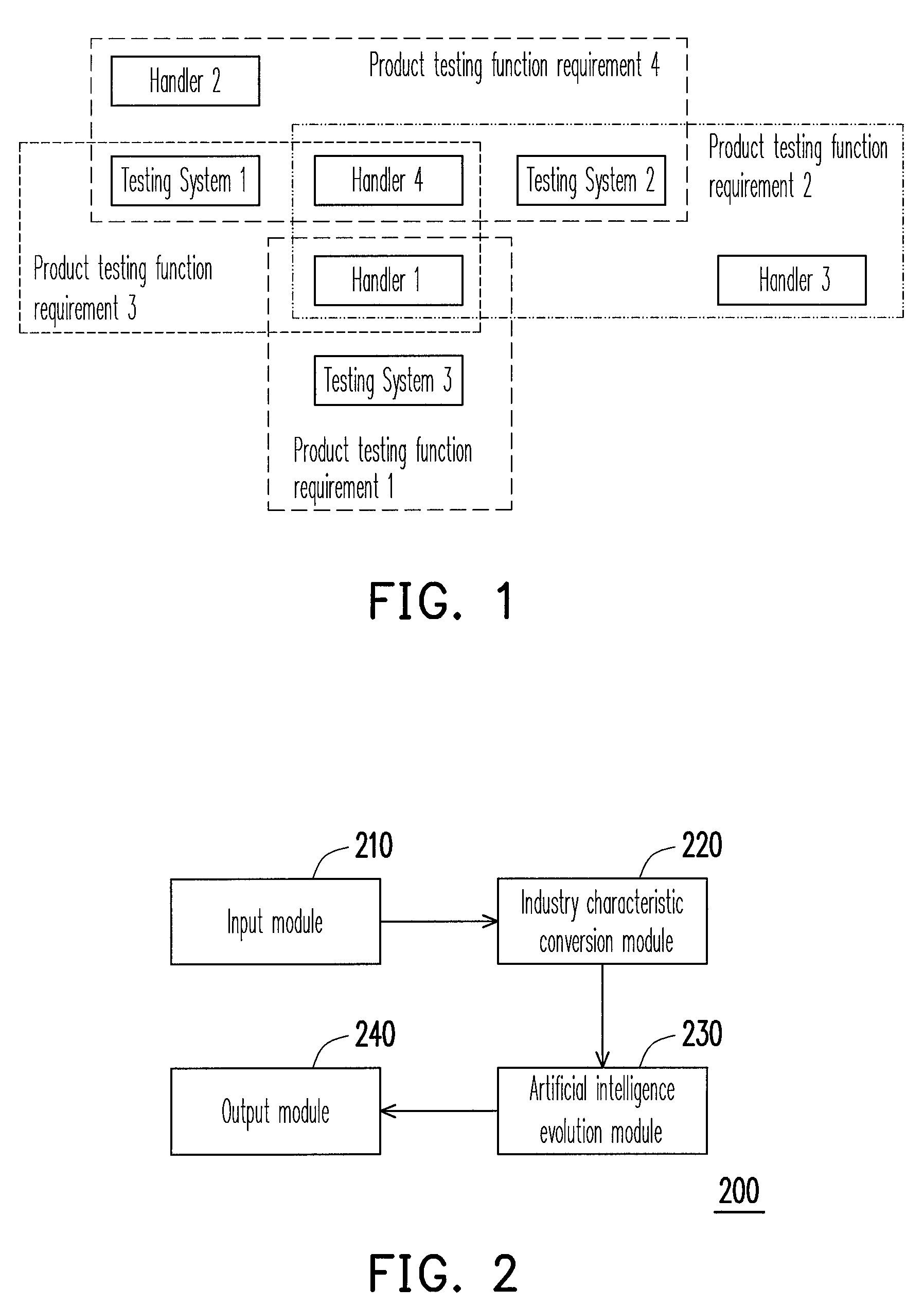
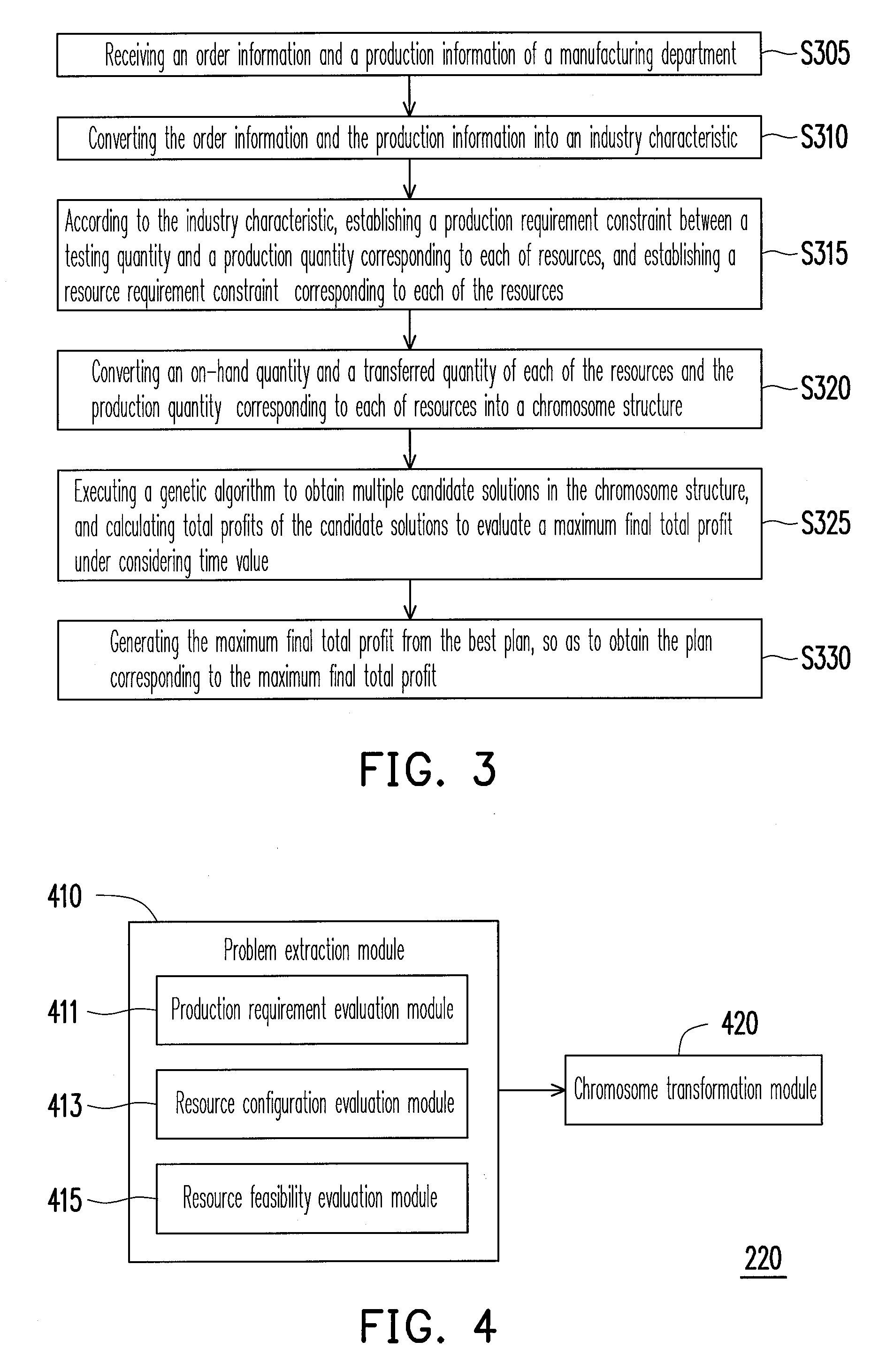
System and method for resource allocation of semiconductor testing industry
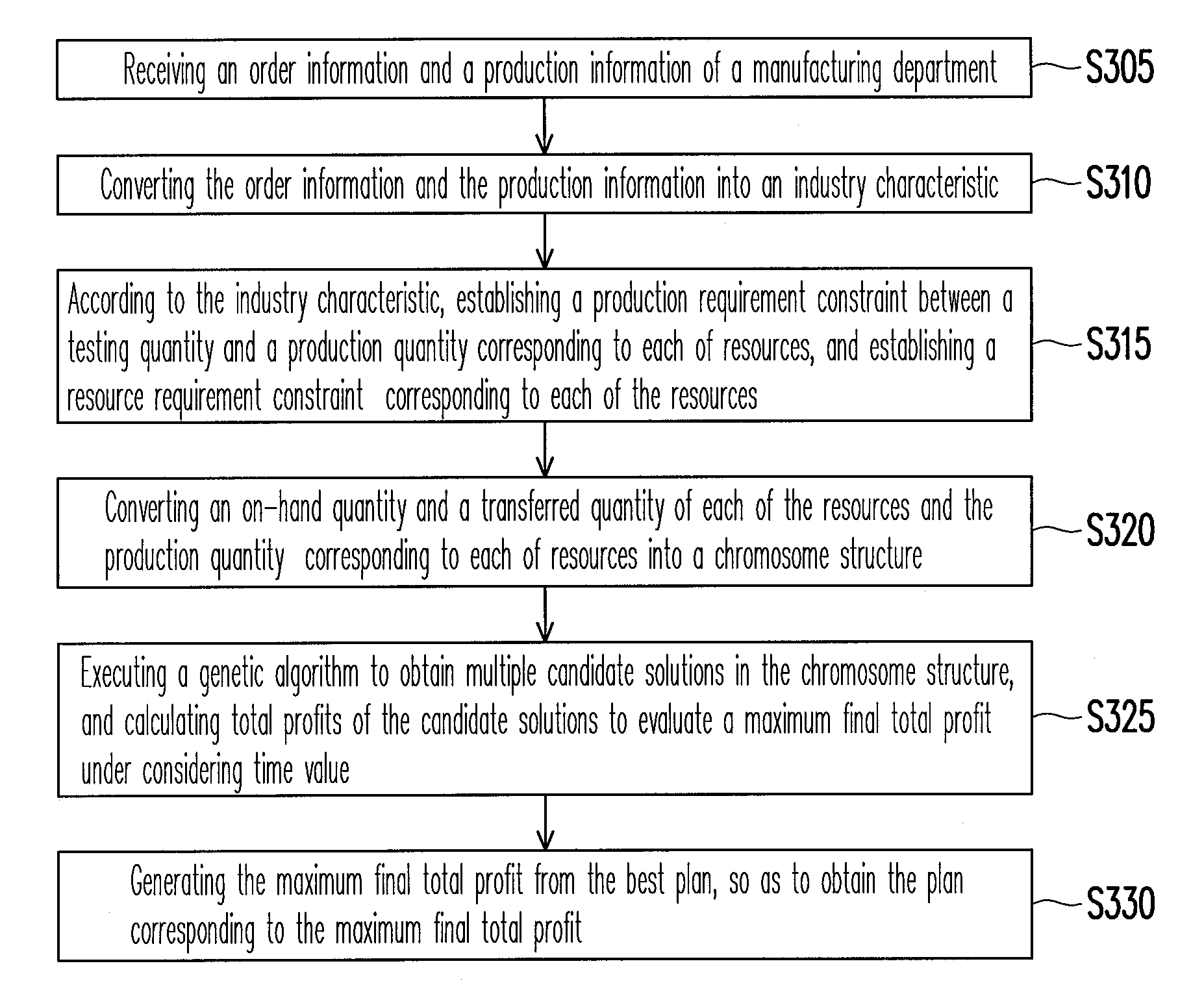
InactiveUS20100185480A1Efficient developmentFacilitate decision-makingGenetic modelsDigital computer detailsChromosome StructuresResource allocation
A system and a method for resource allocation in the semiconductor testing industry are provided. In the system, an industry characteristic conversion module is used to transform the industry characteristic obtained from an input module into a chromosome structure. Next, an artificial intelligence evolution module proceeds to find an optimal solution by handling the chromosome structure until a candidate solution having a maximum final total profit converges to a value.
Owner:NAT TAIWAN UNIV OF SCI & TECH
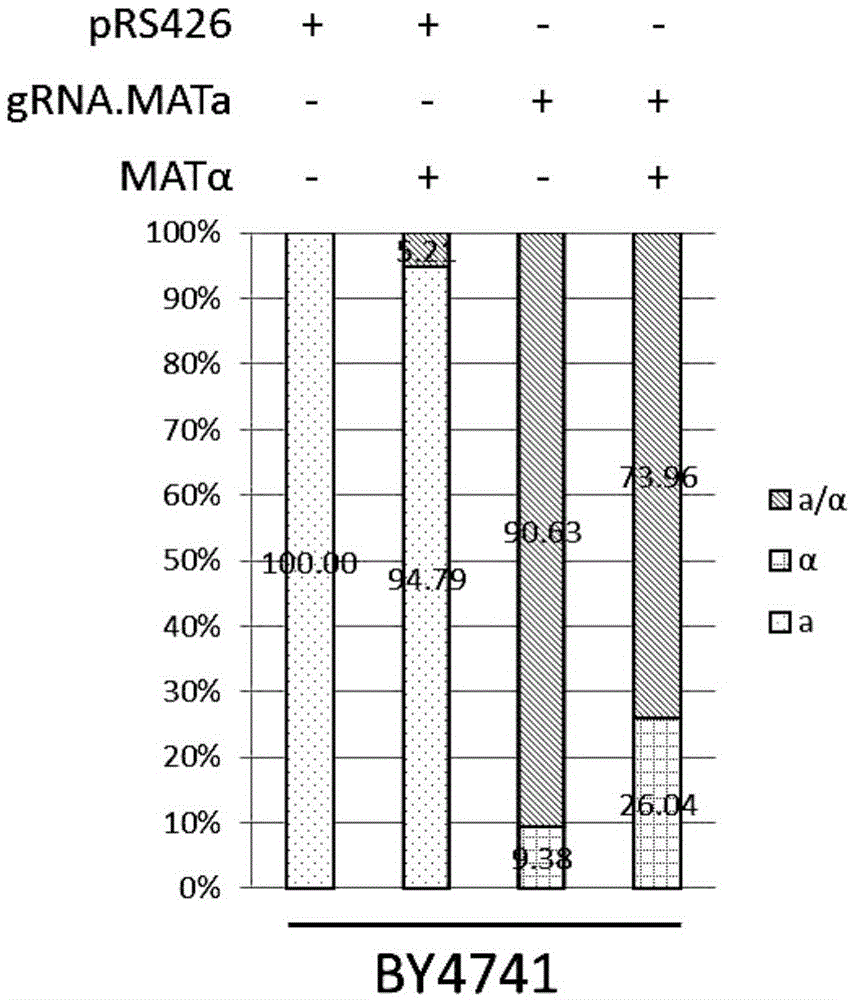
Saccharomyces cerevisiae mating type conversion method
The invention relates to the field of molecular biology, in particular to a saccharomyces cerevisiae mating type conversion method. According to the saccharomyces cerevisiae mating type conversion method, a Cas9 protein is utilized to produce a double-strand break at an MAT locus, the double-strand break is repaired by an exogenous introduced MAT fragment for conversion of saccharomyces cerevisiae mating types. The saccharomyces cerevisiae mating type conversion method can efficiently and quickly convert saccharomyces cerevisiae haploid mating types and does not rely on saccharomyces cerevisiae chromosome structures HML alpha and HMRa itself, a diploid saccharomyces cerevisiae stage is omitted, induction and other treatment conducted on saccharomyces cerevisiae strains to be converted are not needed, anti-mating type saccharomyces cerevisiae strains are directly obtained, and an experiment period is short.
Owner:TIANJIN UNIV
Marker-free chromosome screening
InactiveUS20110058177A1Radiation pyrometryInterferometric spectrometryChromosome StructuresMicroscope
The present invention relates to a method for analyzing chromosomes by preparing a chromosome preparation, measuring at least one interference property of the chromosome preparation and characterizing at least one chromosome structure by way of the interference property. Also, the invention relates to the use of a near field microscope for analyzing unstained chromosomes.
Owner:HOCHSCHULE REUTLINGEN
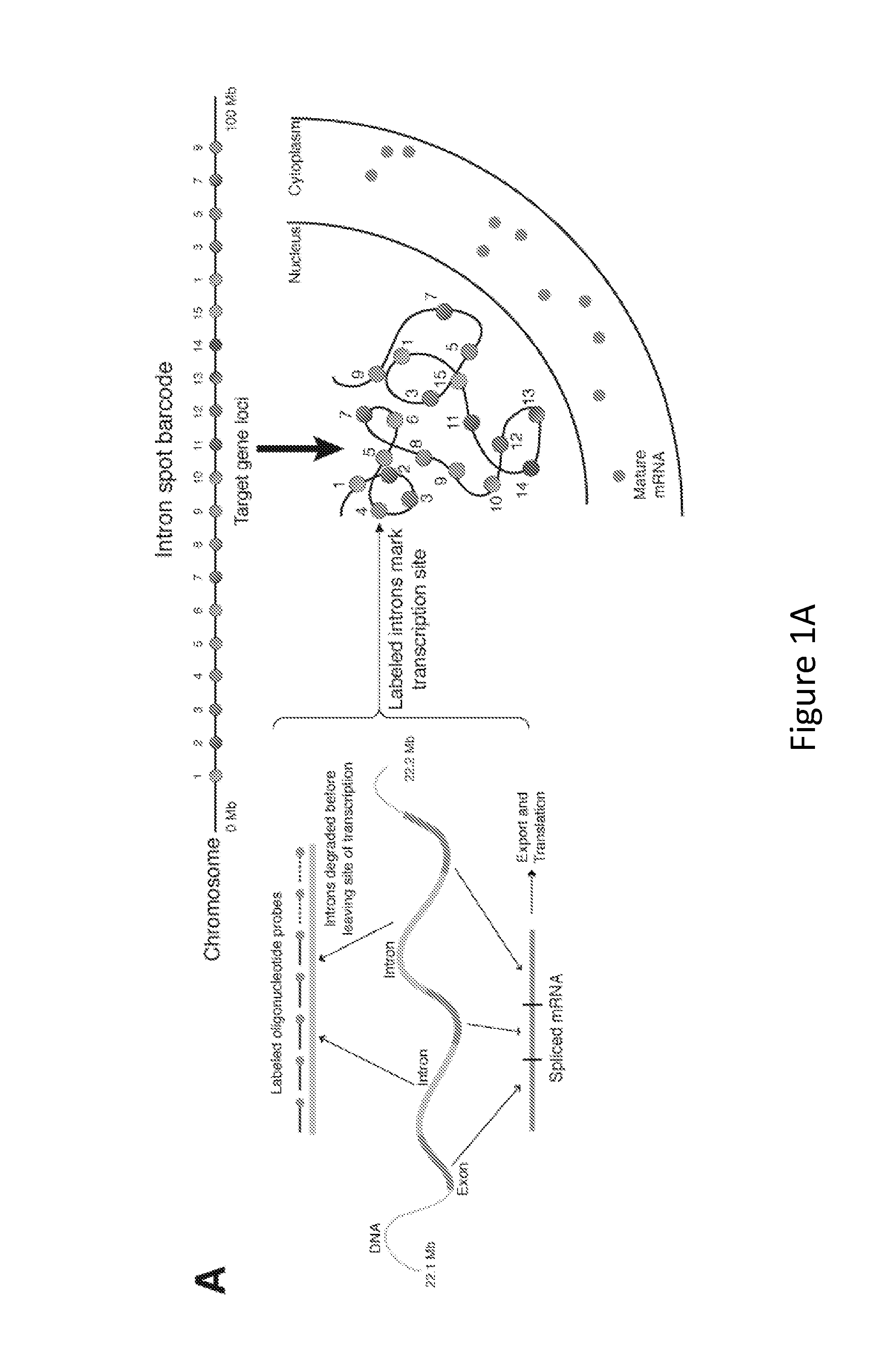
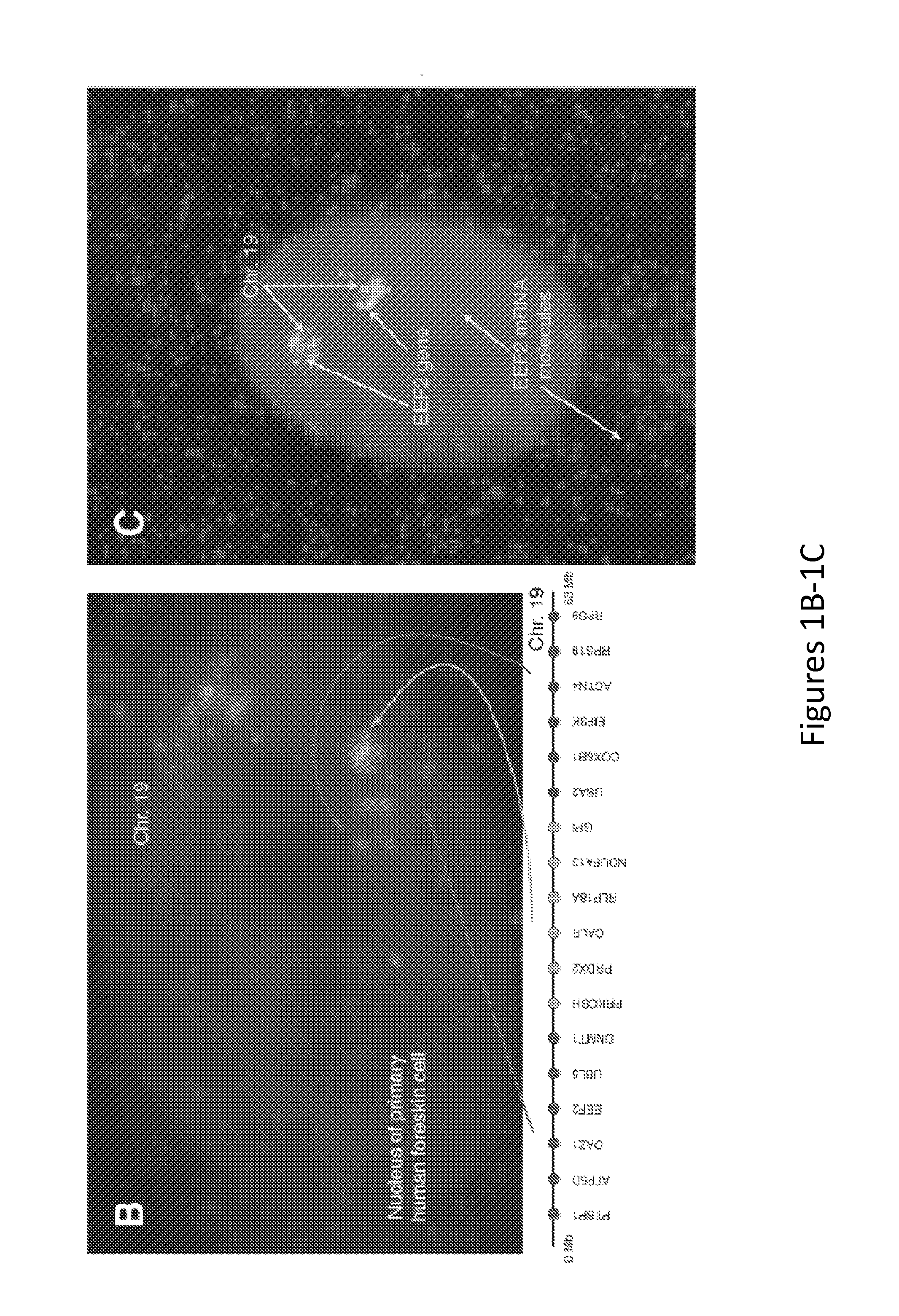
Method for Detecting Chromosome Structure and Gene Expression Simultaneously in Single Cells
The present invention relates to systems and methods for measuring chromosome structure and gene expression. In one embodiment, the present invention includes an assay for determining the spatial organization of gene expression in the nucleus. The present invention also includes a method based on fluorescence in situ hybridization (FISH) that simultaneously yields information on the physical position and expression of individual genes. By lighting up a large number of targets on a particular chromosome using a bar-coding scheme, the large scale structure of an entire chromosome can be determined.
Owner:THE TRUSTEES OF THE UNIV OF PENNSYLVANIA
Chromosome structure variation molecular marker related to tomato fruit color, specific primers and application thereof
ActiveCN110257550AReduce investmentSpeed up the breeding processMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceMutant
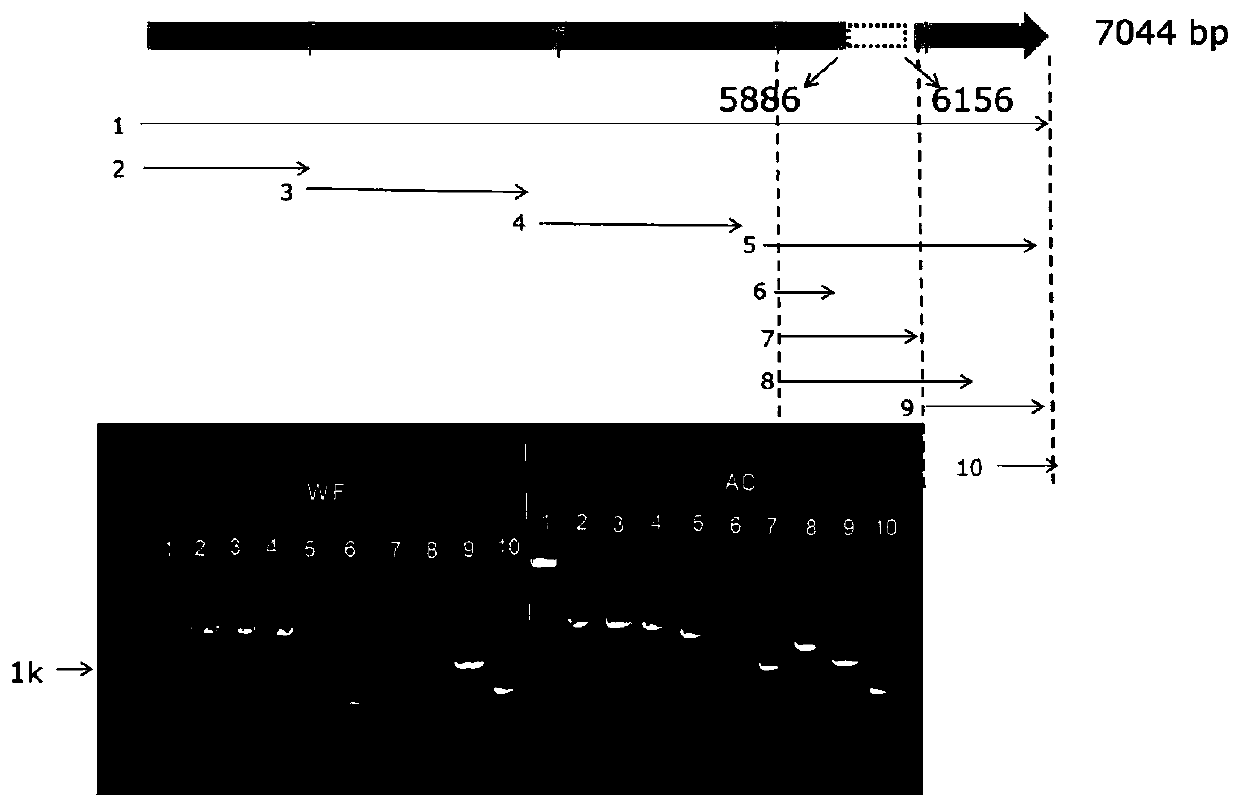
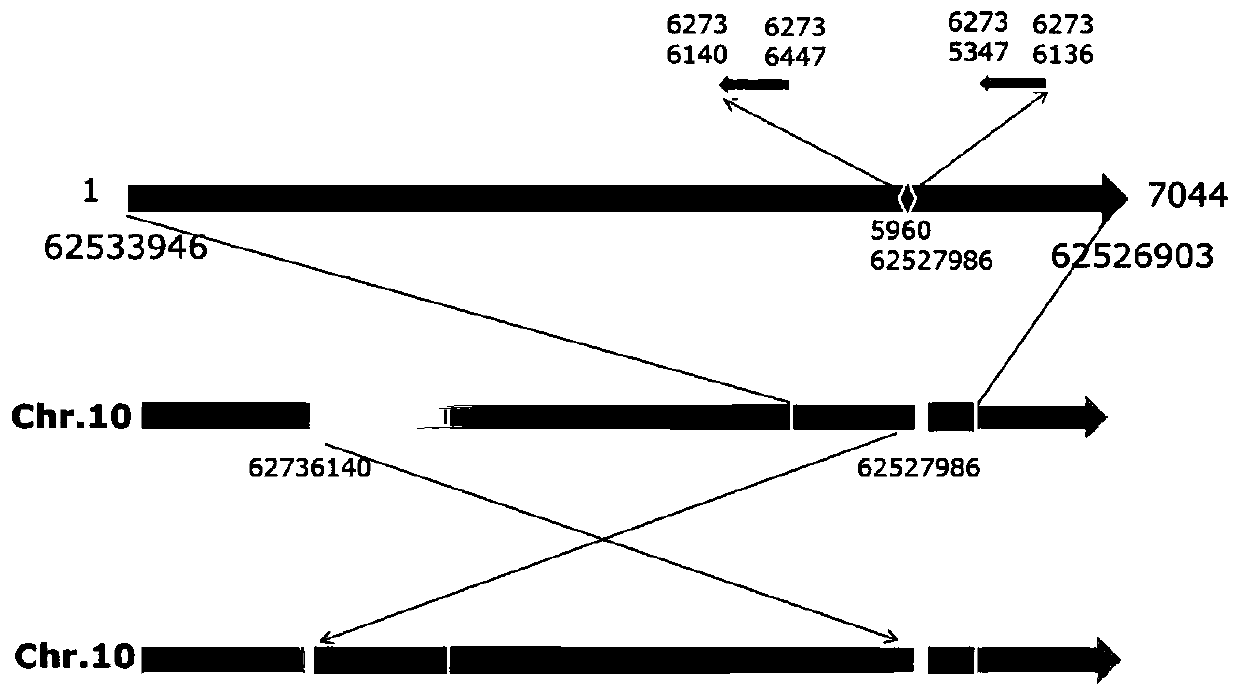
The invention belongs to the field of plant genetic engineering and molecular biology, particularly relates to an identification method for identifying whether the filial generation has white tomato fruits or not, specific primers used by the method and application of the specific primers and the method, and provides a chromosome structure variation (SV) molecular marker related to tomato fruit color. The molecular marker utilizes the 208kb inversion, at chromosome 10, of a white-fruit mutant, and the physical position of the inversion is located from the 62736140th base of chromosome 10 to the 62527986th base of chromosome 10. The two pairs specific primers can be used to identify whether the tomato filial generation has white fruits or not and whether the tomato filial generation is in a heterozygosis state or not. The method uses the molecular marker and the specific primers to identify whether the tomato filial generation has white fruits. By the molecular marker, the specific primers and the method, the white-fruit tomato materials can be fast selected, breeding cycle can be shortened, and breeding efficiency can be increased.
Owner:HUAZHONG AGRI UNIV
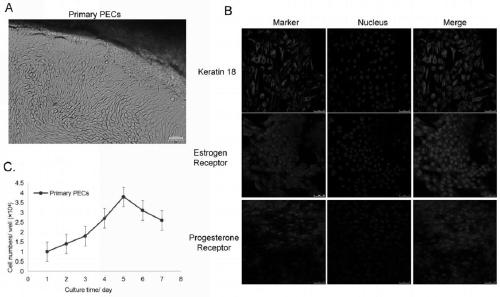
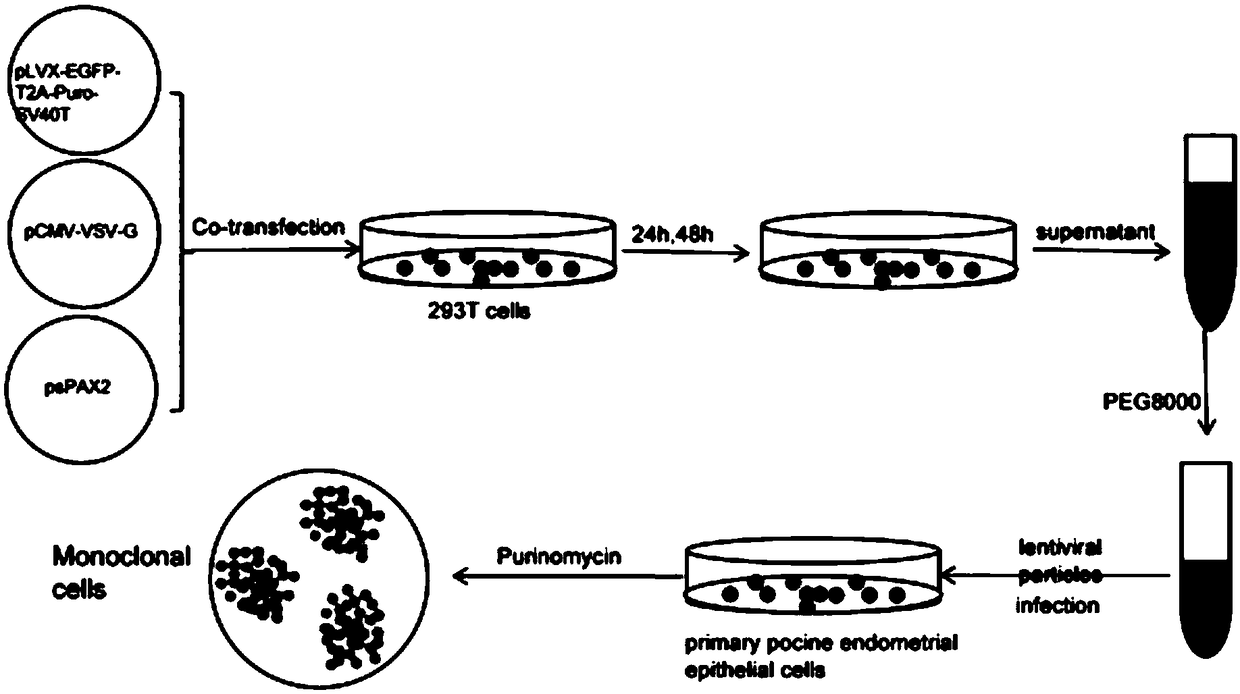
Porcine endometrium epithelial cell line susceptible to PRRSV and construction method of cell line
InactiveCN109486769AFluorescence experiments are not disturbedLong cycleSsRNA viruses positive-senseGenetically modified cellsKaryotypeSV40 T-antigen
The invention discloses a porcine endometrium epithelial cell line susceptible to PRRSV and a construction method of the cell line. The porcine endometrium epithelial cell line is obtained by taking porcine endometrium epithelial cells as host cells, transfecting lentivirus expression plasmids with SV40T antigen genes, and performing Puromycin drug screening and cell expansion culture. The porcineendometrial epithelial cell line has the function of infinite passage, can maintain the characteristics of the porcine endometrial epithelial cells, can maintain normal karyotype in the passage process, and does not have changes in chromosome structure and number; and moreover, the porcine endometrial epithelial cell line is sensitive to PRRSV virus, and can be used for the passage of PRRSV and related research.
Owner:SHANDONG AGRICULTURAL UNIVERSITY +1
Method for automatically generating Bug repair plan and Bug repair method
InactiveCN102226913AImprove accuracyReduce workloadGenetic modelsComputer Software EngineeringGenetic algorithm
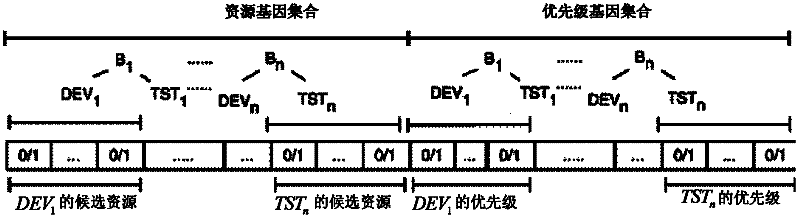
The invention discloses a method for automatically generating a Bug repair plan and a Bug repair method, belonging to the field of computer software engineering. The method for automatically generating the Bug repair plan comprises the following steps: 1) using a resource modeling module to extract the attribute information of an input Bug report set; 2) using the resource modeling module to extract the attribute information of stored human resources; 3) using a task scheduling module to acquire a candidate human resource set required by each Bug repair activity in accordance with the extracted attribute information; 4) configuring a binary-system chromosome structure which can be identified by a genetic algorithm, wherein the binary-system chromosome structure comprises candidate resources of the repair activities and executed priority of the repair activities; and 5) using the task scheduling module to optimize the binary-system chromosome structure by the genetic algorithm to obtain the optimal repair plan of the input Bug report set. Through the invention, the accuracy of configuring the human resources is improved, the dependence on subjective consciousness of the human in the planning process is reduced, and the utilization rate of the human resources is improved.
Owner:INST OF SOFTWARE - CHINESE ACAD OF SCI
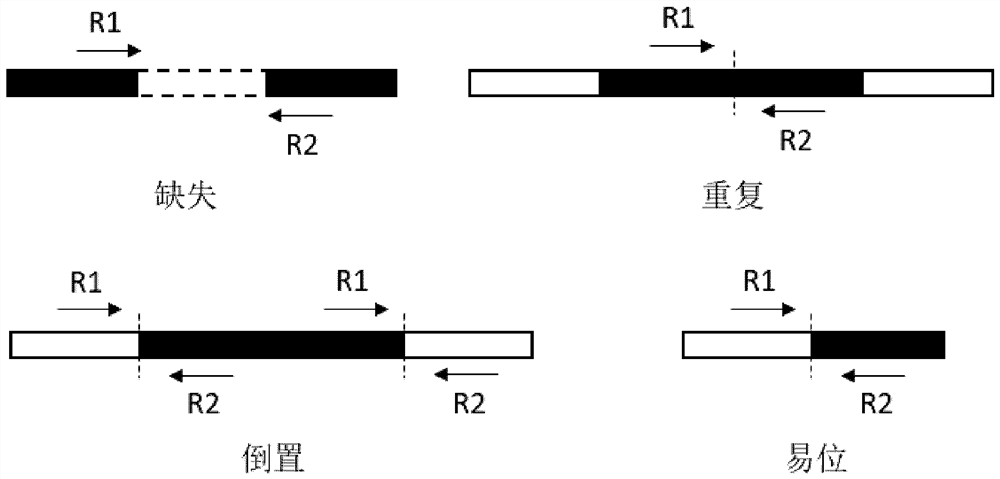
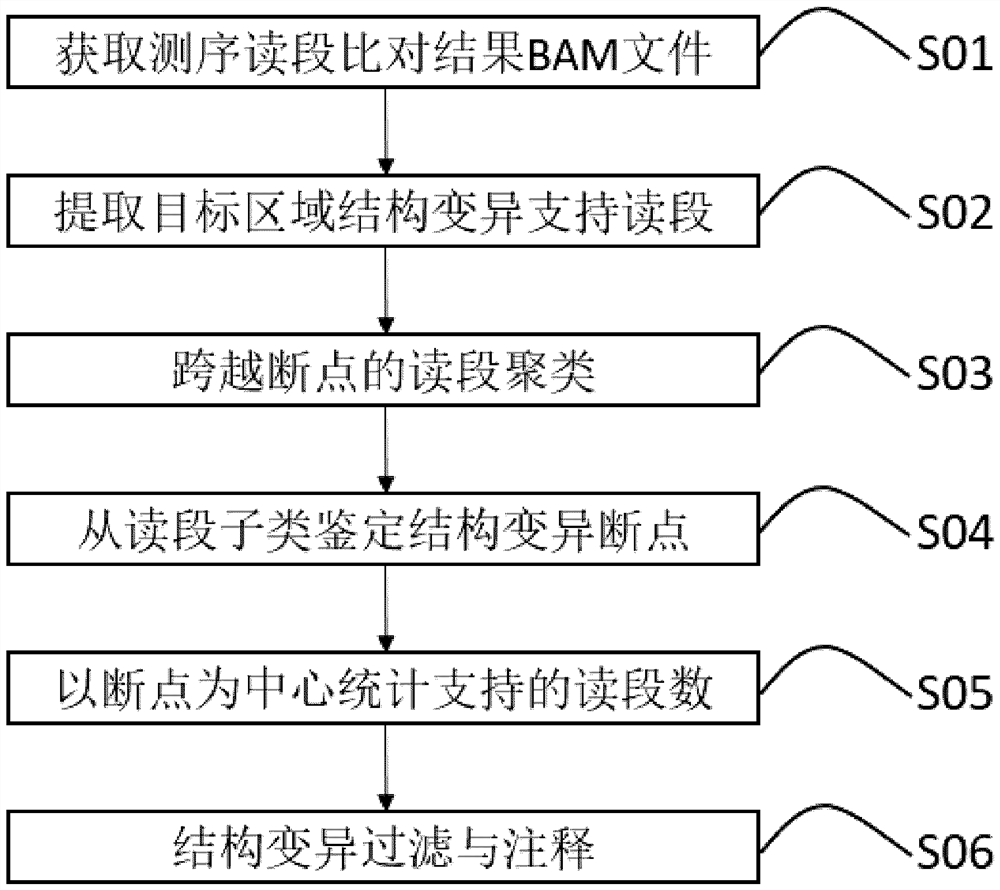
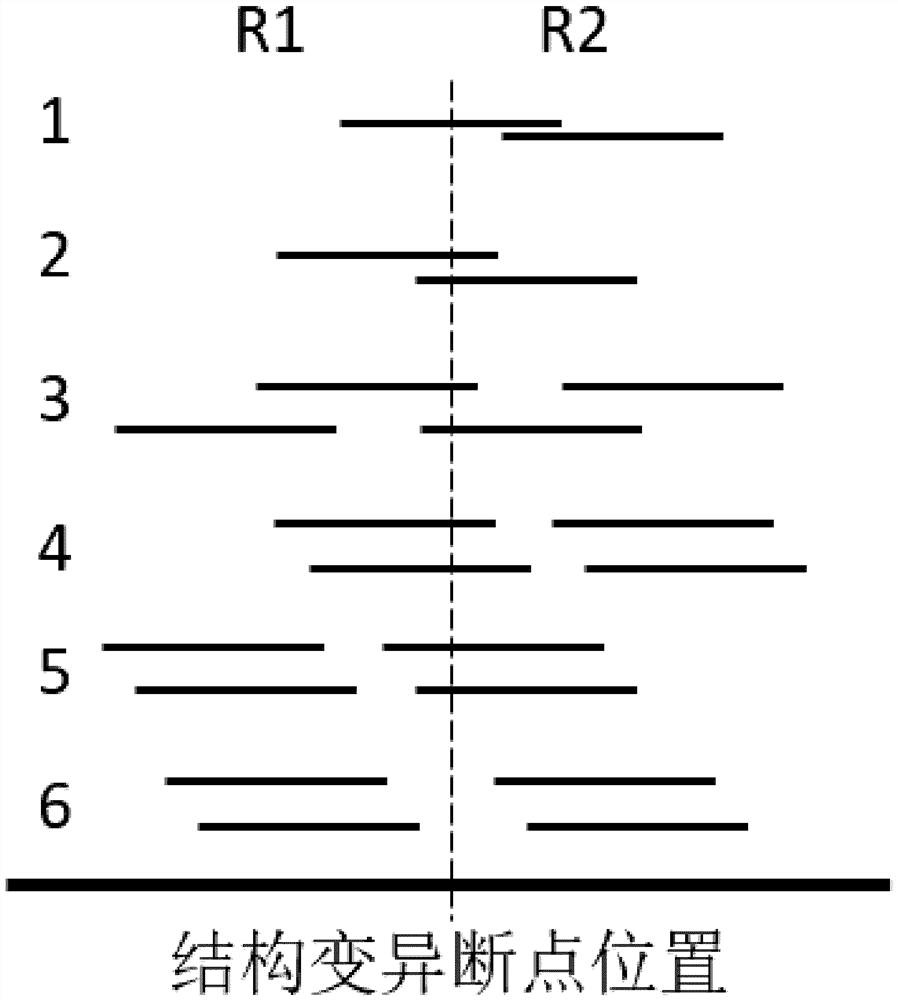
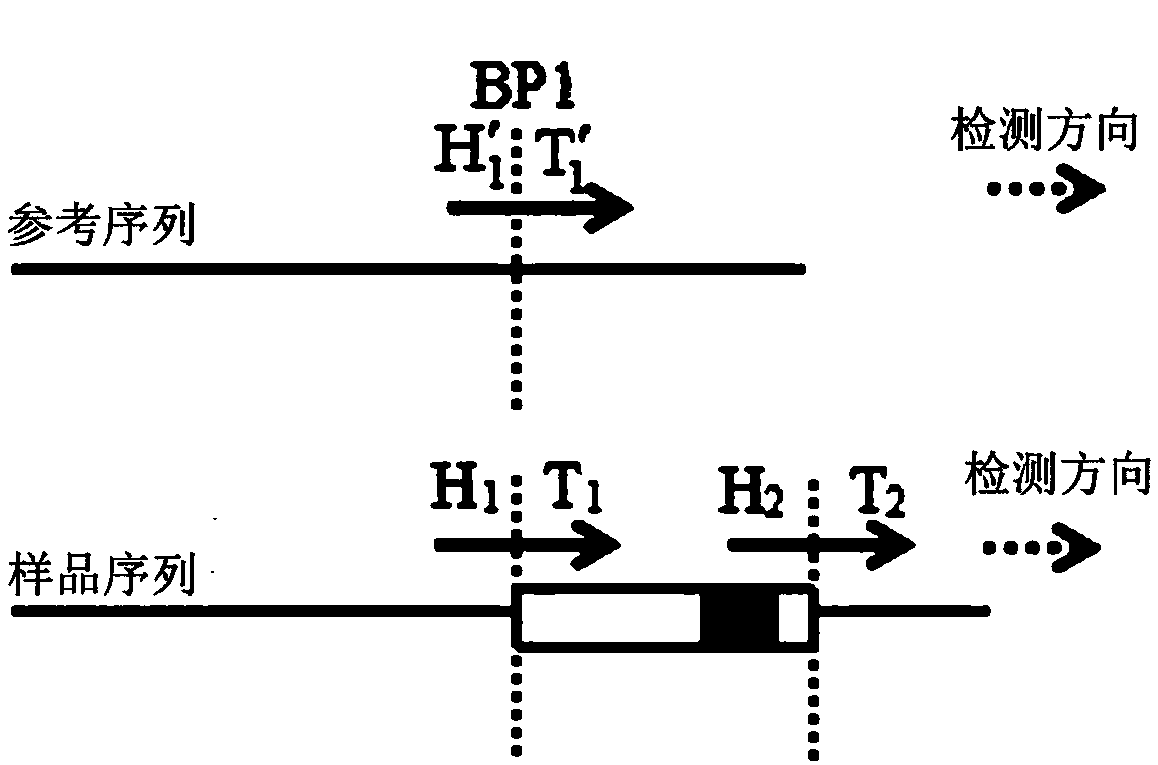
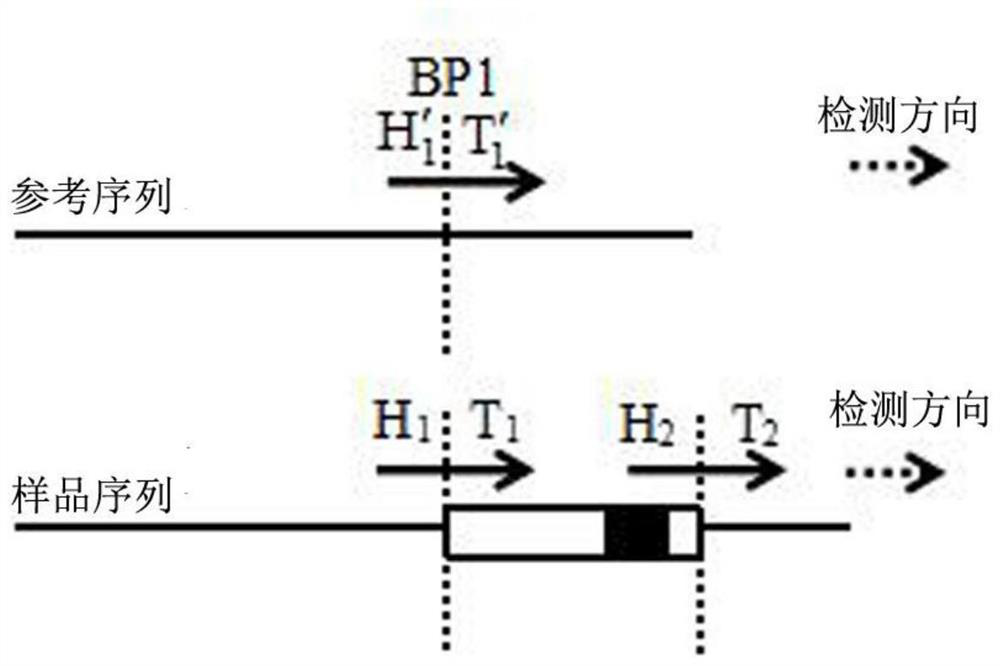
Chromosome structure variation identification method taking breakpoint as center
The invention relates to a chromosome structure variation identification method taking a breakpoint as a center. Specifically, the invention relates to a computer-implemented method for identifying chromosome structural variation by taking a breakpoint as a center. The invention also relates to a computer system or device for implementing the method, and a computer readable medium storing a computer program capable of implementing the method.
Owner:上海思路迪医学检验所有限公司
Method for displaying centromeres and short arms of Larimichthys crocea
The invention discloses a method for displaying centromeres and short arms of large yellow croaker, and belongs to the field of molecular cytogenetics. A molecular marker is a segment of a 28S rDNA sequence cloned from human saliva, and is processed to prepare a probe (named as H-P3K), and the probe is crossed with metaphase chromosomes of Larimichthys crocea to obtain a positive signal. The positive signal in the centromeres and the short arms of every metaphase chromosome can be detected when FISH of the Larimichthys crocea chromosomes is carried out by the probe, the short arm signal intensity of one chromosome is weaker than the centromere signal intensity of the same chromosome, and the centromere signal intensities of different chromosomes are different. The method is used for determining the position of the centromere, analyzing the chromosome karyotype and researching the chromosome structure; and the positive signal can be used as a chromosome identifying and pairing marker.
Owner:JIMEI UNIV
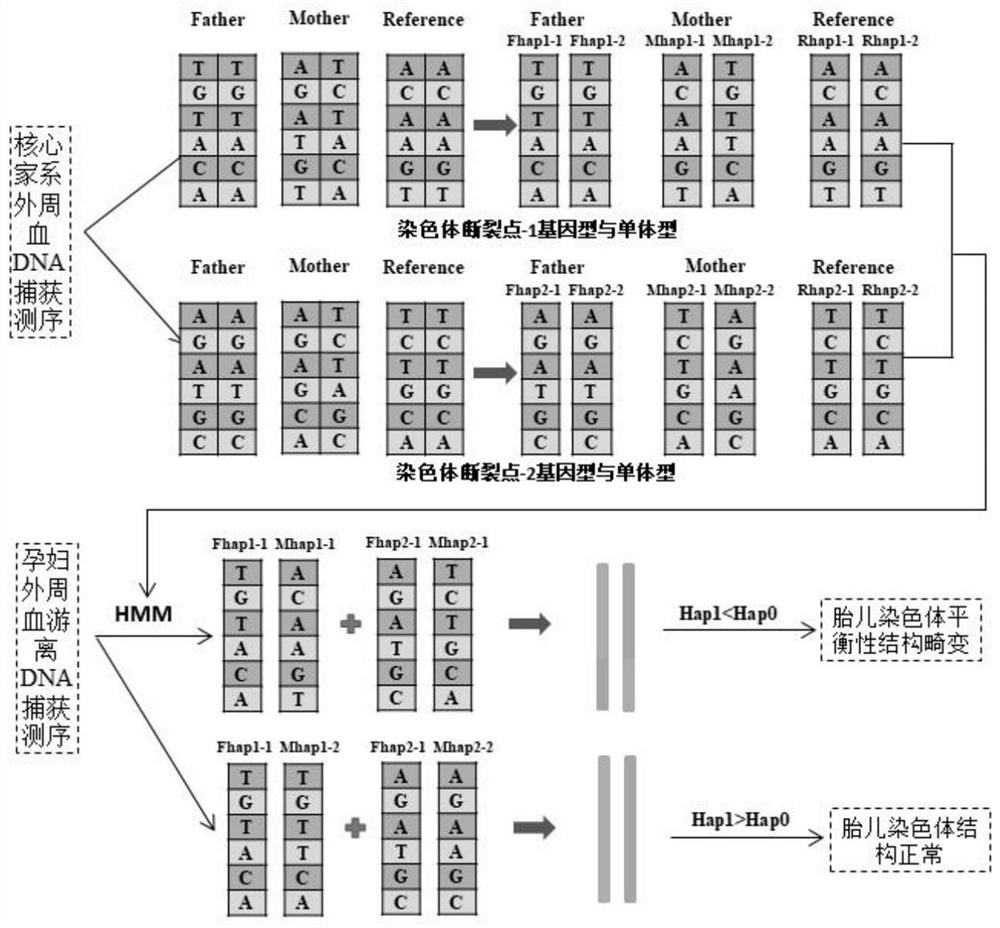
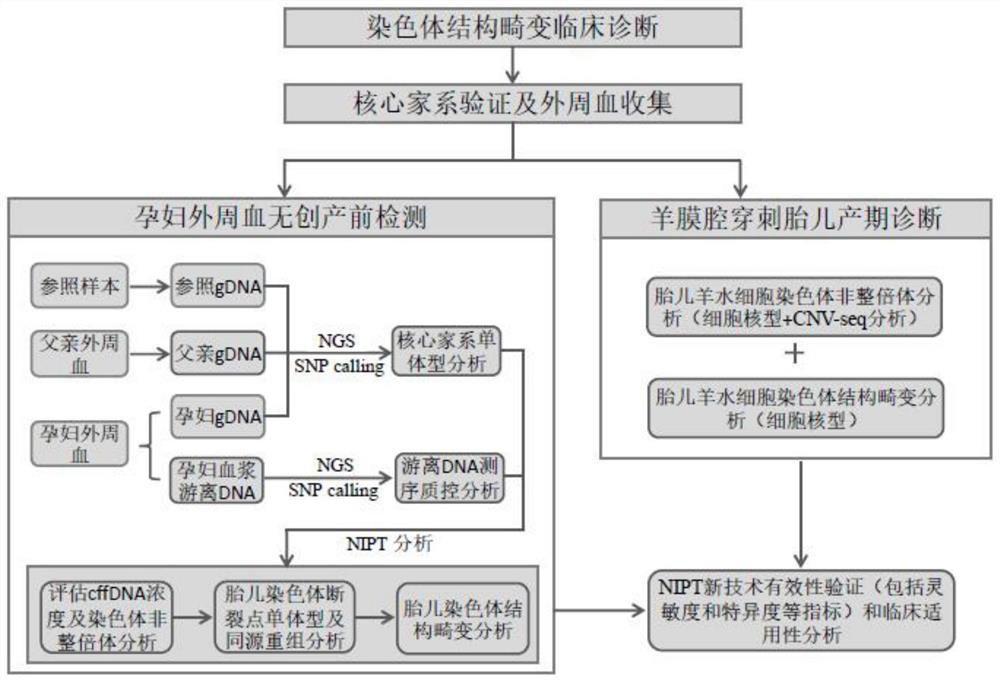
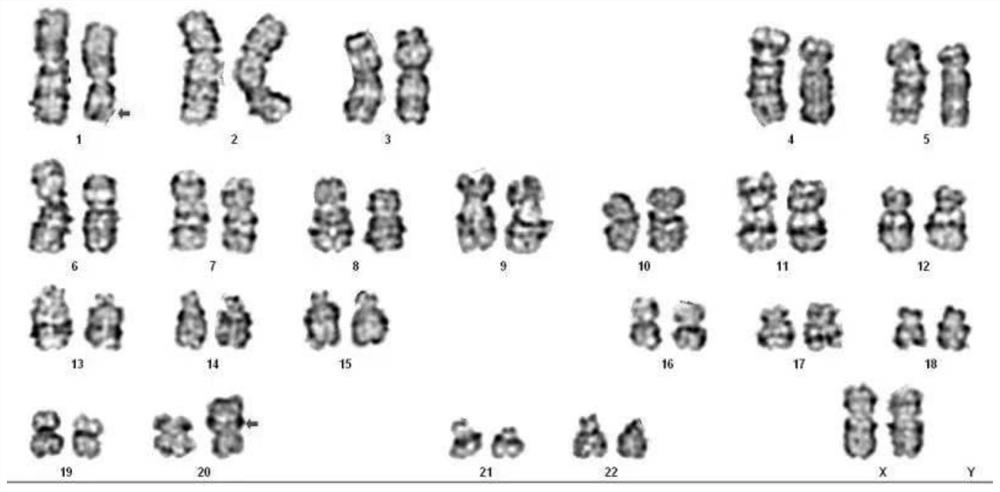
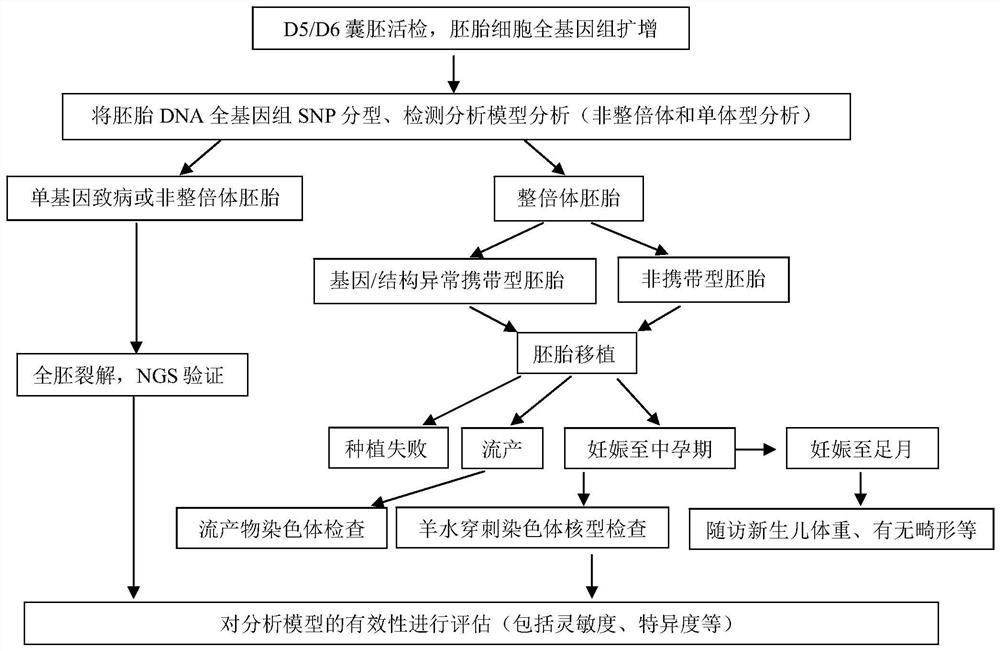
Method for detecting balance structure variation of fetal chromosome through free DNA of peripheral blood of pregnant woman
PendingCN114480667AAvoiding the Difficulty of Directly Detecting BreakpointsHigh-throughput featuresMicrobiological testing/measurementProteomicsFetal structurePhysiology
The invention discloses a method for detecting fetal chromosome balance structure variation through pregnant woman peripheral blood free DNA. The method comprises construction of a core family haplotype model and fetal structure variation detection. The method comprises the following steps: collecting chromosome structure rearrangement variation family carrying Fuffles, carrying out SNP (Single Nucleotide Polymorphism) genotyping on the family, defining haplotypes of structure rearrangement chromosomes and structure normal chromosomes, and constructing a whole genome haplotype model. Free DNA of peripheral blood of a pregnant woman is captured and sequenced, the genotype and haplotype of a fetus in the free DNA are calculated and analyzed according to a hidden Markov model, and embryo chromosome structure rearrangement is predicted by analyzing whether the fetus carries the haplotype near a structure rearrangement chromosome breaking point area or not. According to the invention, detection of fetal balance chromosome structure abnormity through the maternal peripheral blood cfDNA is provided for the first time, and important guidance is provided for prevention and diagnosis of chromosome diseases.
Owner:THE OBSTETRICS & GYNECOLOGY HOSPITAL OF FUDAN UNIV
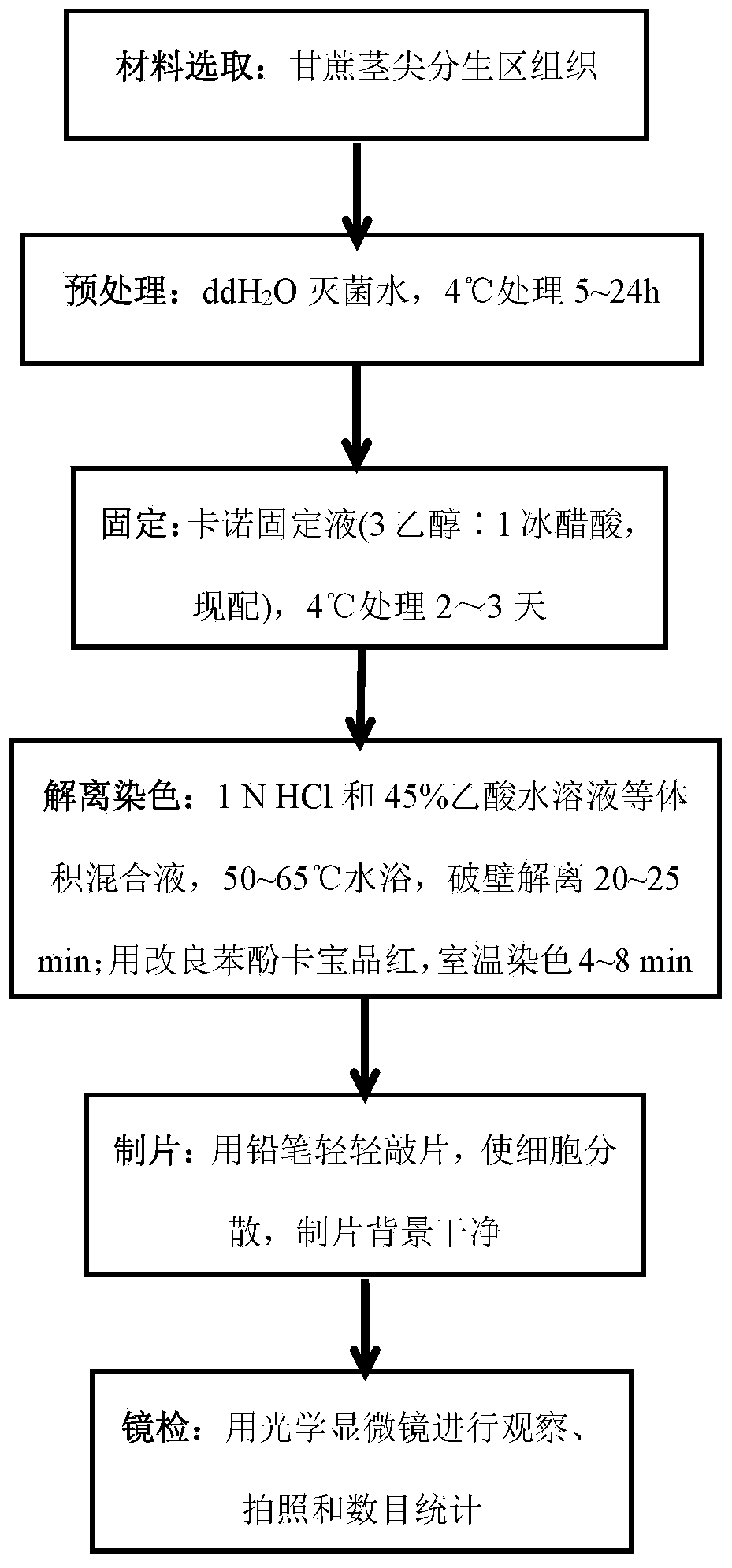
Efficient sugarcane or sugarcane related species stem tip chromosome flaking method
PendingCN111238888AShorten the timeAvoid repeated tedious steps of collection and cleaningPreparing sample for investigationBiotechnologyStaining
The invention provides an efficient sugarcane or sugarcane related species stem tip chromosome flaking method, and belongs to the technical field of cell biology. The invention provides an efficient sugarcane stem tip chromosome flaking method aiming at solving the problems that sugarcane chromosomes are large in number and small in form and ideal division phase metaphase cells are difficult to obtain through root tip flaking. Chromosome flaking is carried out by utilizing stem tip meristematic region tissues of sugarcane and sugarcane related species in a vigorous growth period, the method has the advantages of convenient material taking, large meristematic area tissue sample size, vigorous division, many metaphase cells and clear and dispersed chromosome structure, and the technical keypoints of material selection, pretreatment, fixation, dissociation dyeing, flaking and chromosome morphology microscopic observation are disclosed. The method is simple and convenient, accurate and reliable in result, good in repeatability, easy to operate and short in experiment period and improves the sugarcane chromosome genome analysis efficiency. Technical support is provided for sugarcane chromosome karyotype research, germplasm resource classification identification and protection utilization.
Owner:SUGARCANE RES INST OF YUNNAN ACADEMY OF AGRI SCI
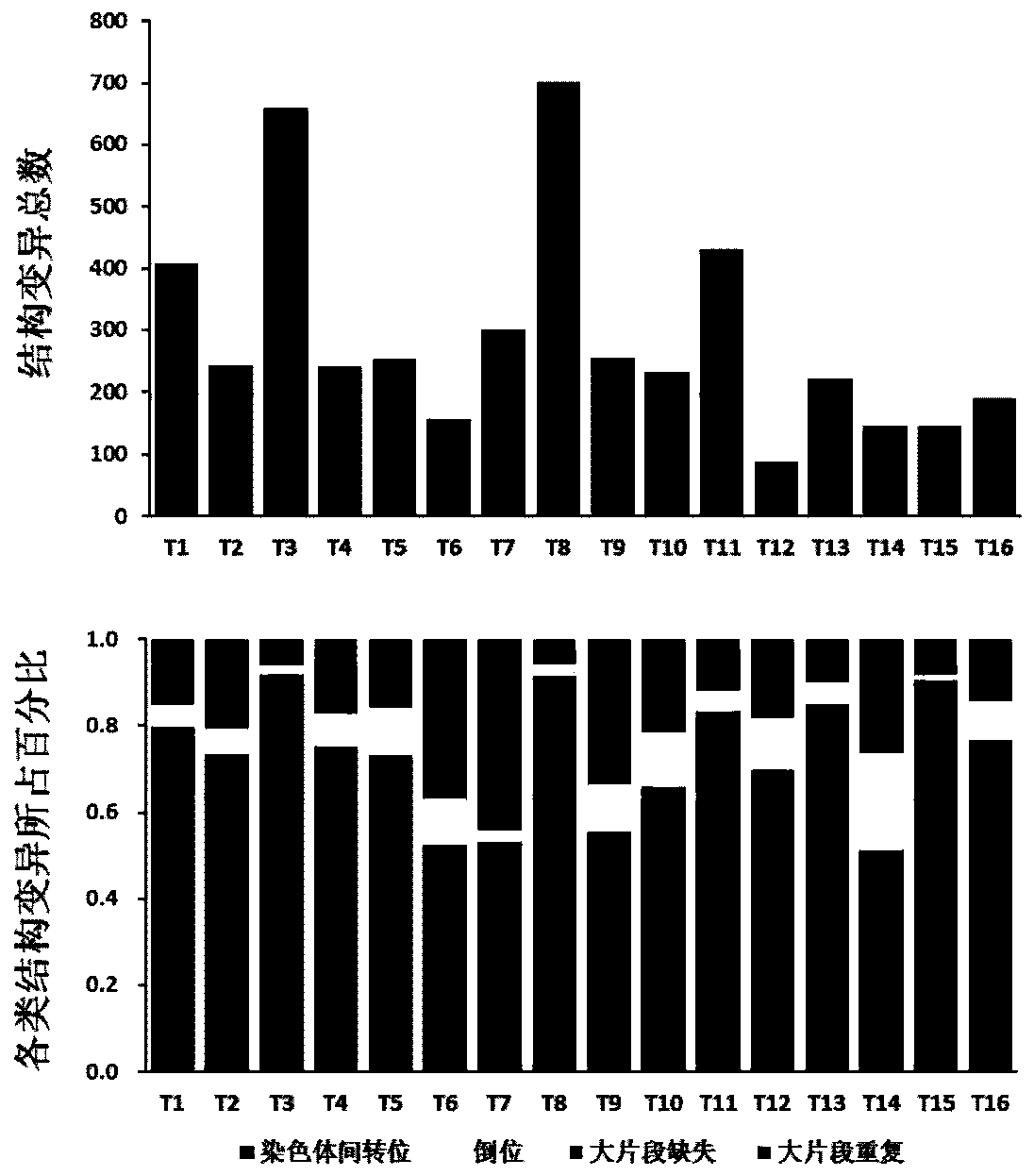
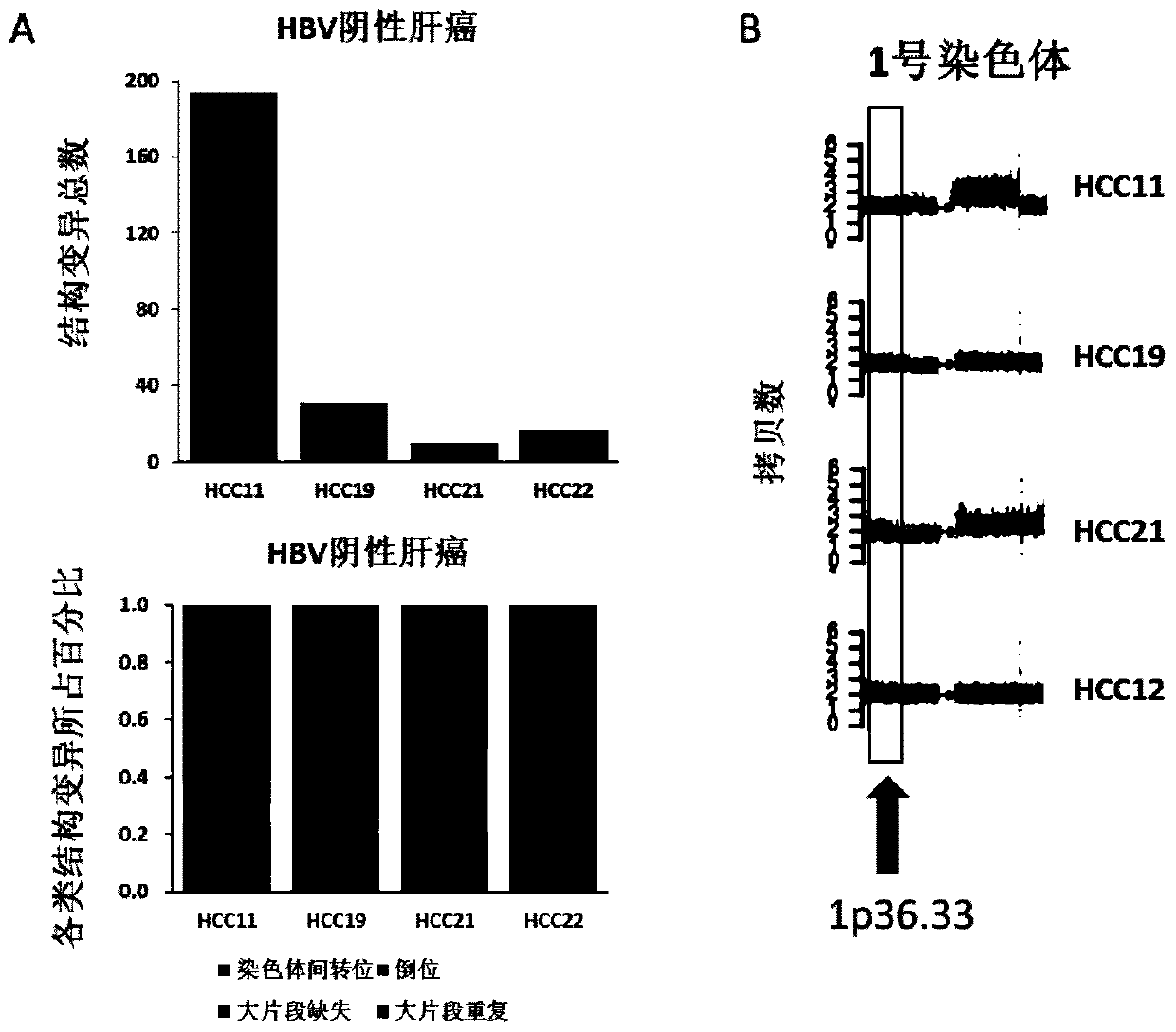
Genome recombination fingerprint for characterizing hHRD homologous recombination deficiency and identification method thereof
The invention relates to a novel method for identifying homologous recombination deficiency and application thereof. More particularly, the present invention relates to a characteristic genome recombination fingerprint which is related to hHRD type homologous recombination repair deficiency and is resolved by using high throughput genome re-sequencing, bioinformatics analysis and statistical correlation analysis. The characteristic hHRD recombination fingerprint is caused by the functional deficiency of a specific homologous recombination repair mechanism (namely CRL4WDR70-H2B mono-ubiquitination pathway), and comprises the total frequency of genome recombination in a single sample, the composition of chromosome structure variation types and site-specific copy number variation. The recombinant fingerprint is used for identifying infectious diseases or tumors with the characteristic mutant fingerprint, and is used for guiding targeted drug treatment of PARP inhibitors. The invention relates to the method and application thereof for diseases including but not limited to breast cancer, ovarian cancer, endometrial (like) cancer, ovarian clear cell cancer, prostate cancer, pancreatic cancer, skin cancer and gastric cancer with such characteristics, as well as hepatitis B virus infection or related liver fibrosis cirrhosis, liver cancer and cholangiocarcinoma.
Owner:成都吉诺迈尔生物科技有限公司 +1
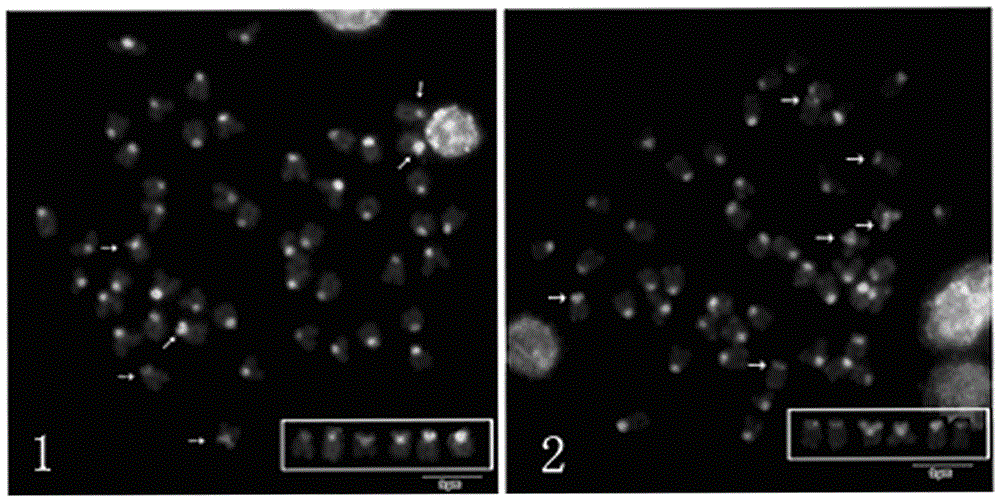
Classifier model training method and device for detecting chromosome structure abnormality
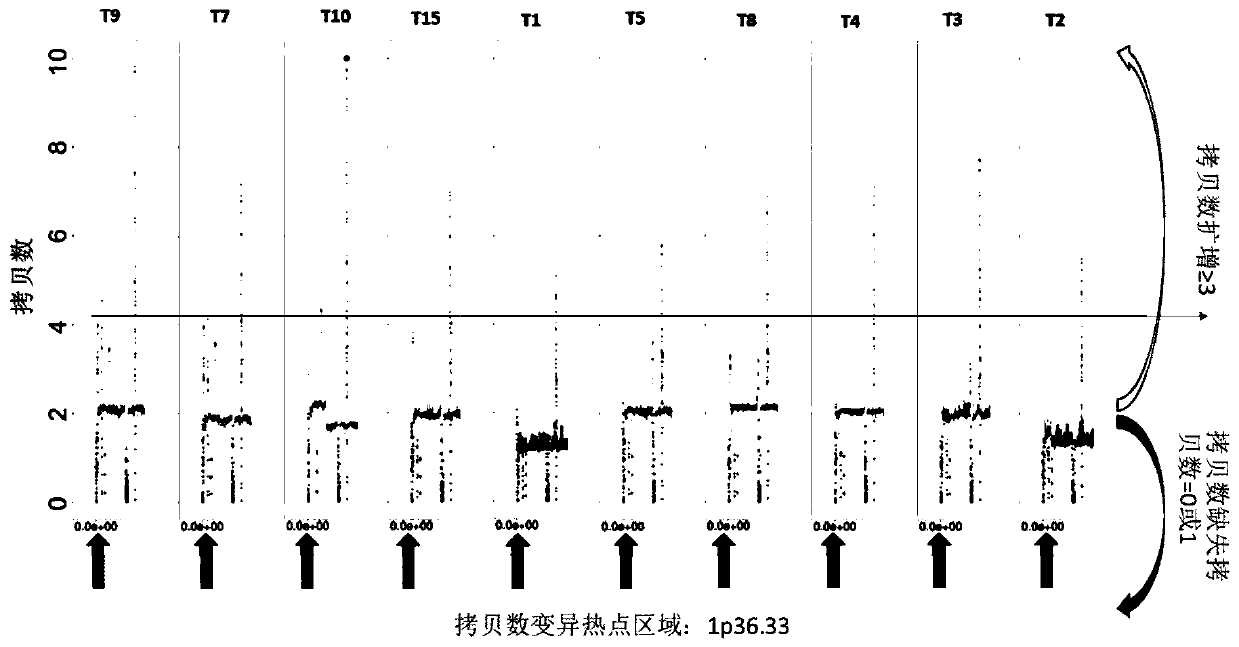
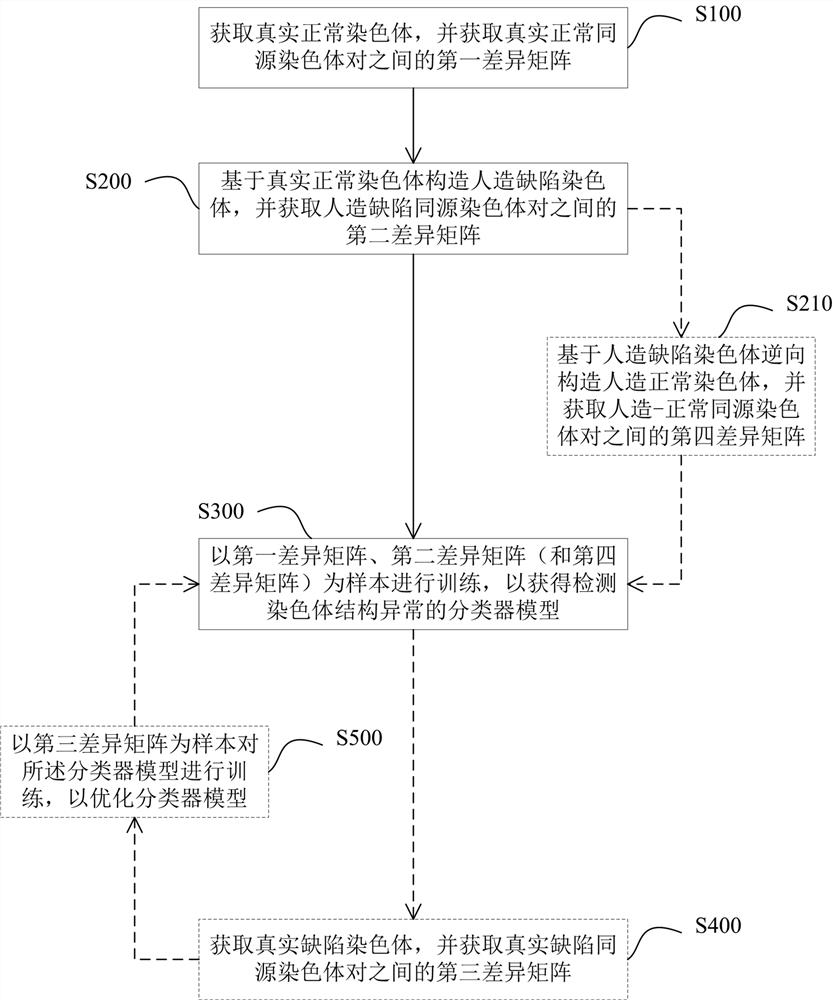
ActiveCN114841294AAutomated screeningCharacter and pattern recognitionPattern recognitionHomologous chromosome
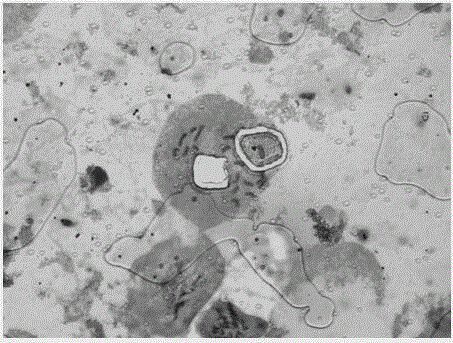
The invention provides a classifier model training method and device for detecting chromosome structure abnormality. The training method comprises the steps that real normal chromosomes are obtained, a first difference matrix between real normal homologous chromosome pairs is obtained, and two homologous chromosomes in the real normal homologous chromosome pairs are both real normal chromosomes; artificial defect chromosomes are constructed based on the real normal chromosomes, a second difference matrix between the artificial defect homologous chromosome pairs is obtained, and at least one of two homologous chromosomes in the artificial defect homologous chromosome pairs is an artificial defect chromosome; training at least by taking the first difference matrix and the second difference matrix as samples to obtain a classifier model for detecting the chromosome structure abnormality; and judging whether the to-be-diagnosed user has chromosome abnormality based on the classifier model. According to the method, abundant structure abnormal chromosomes of different types are artificially constructed, so that sufficient and balanced samples are provided for classifier model training.
Owner:HANGZHOU DIAGENS BIOTECH CO LTD
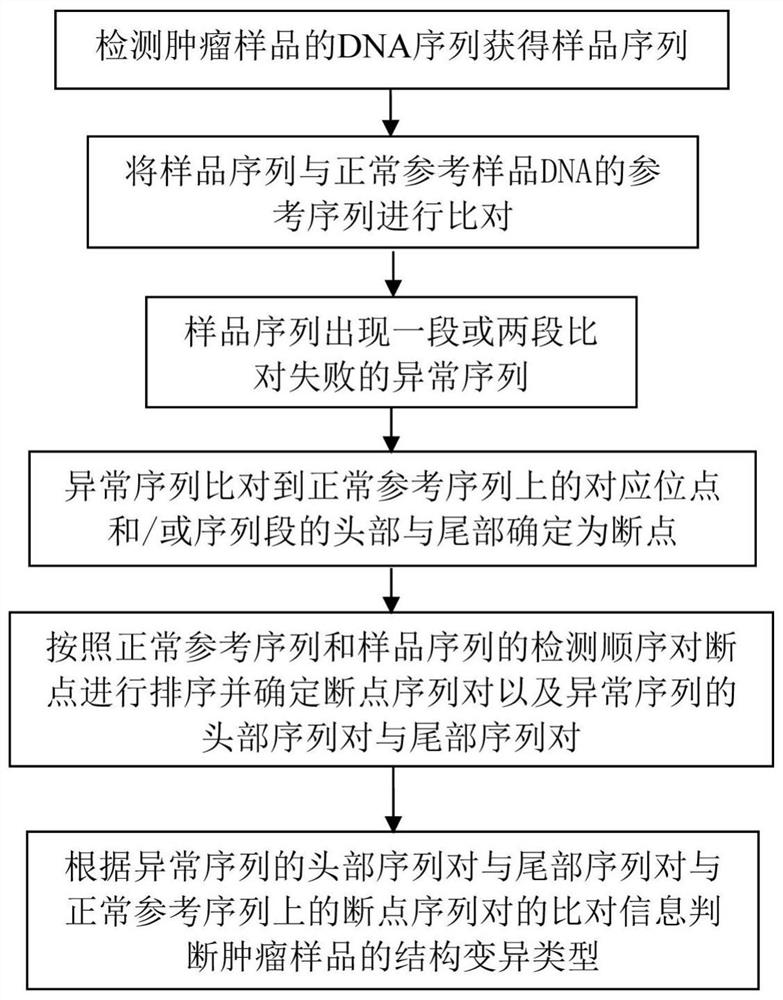
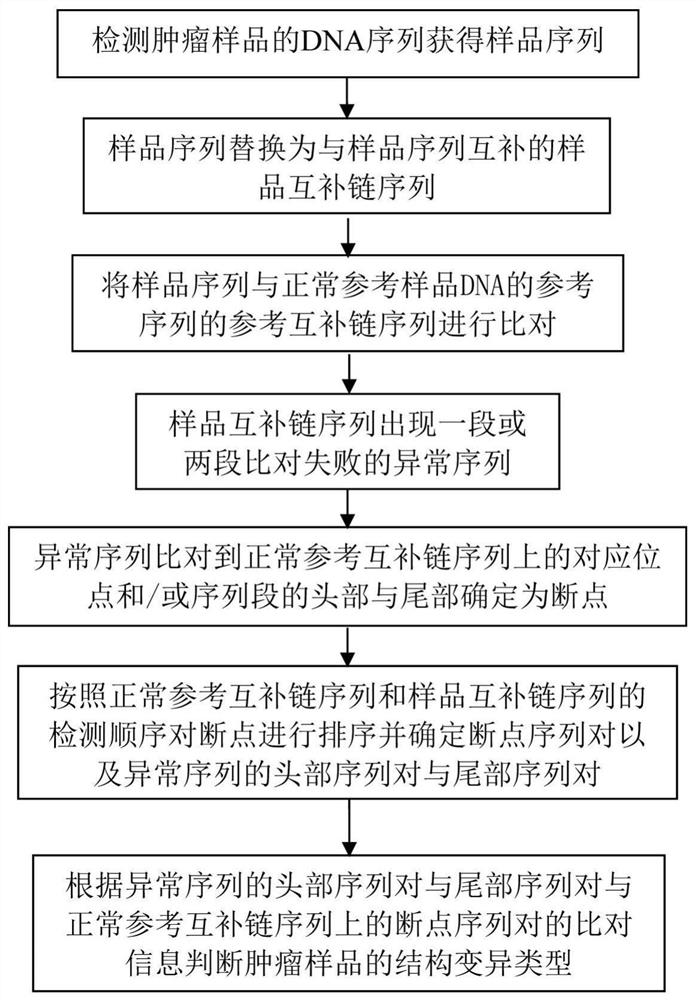
DNA (desoxyribonucleic acid) complex structure variation diagnosis method
ActiveCN108171011AImproving the Accuracy of Structural Variation DetectionAvoid incomplete acquisitionProteomicsGenomicsDiagnosis methodsA-DNA
The invention discloses a DNA (desoxyribonucleic acid) complex structure variation diagnosis method. 16 chromosome structure variation types are designed, and a corresponding judgment model is designed by aiming at each variation type. Comparison information is directly obtained in a sequencing data comparison file. Compared with the prior art, the method omits sequence re-splicing and re-comparing parts, DNA complex structure variation detection can be quickly finished, and time cost is saved. Meanwhile, since information required for deducing the variation type is fully obtained in the comparison, the problem that re-comparison information can not be completely obtained and wrong information is obtained in a traditional process can be avoided, and chromosome structure variation detectionaccuracy is improved.
Owner:志诺维思(北京)基因科技有限公司
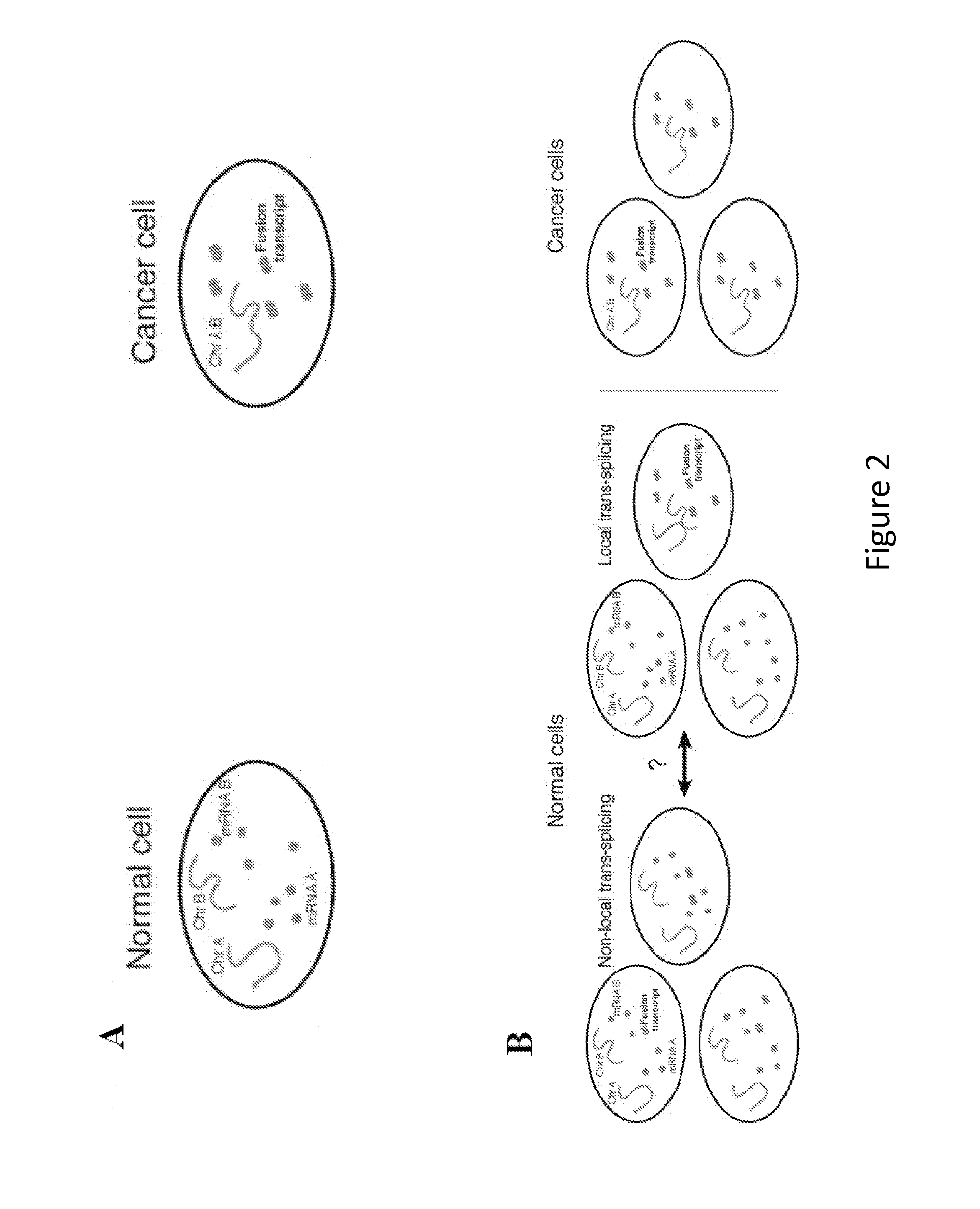
DNA conformation (loop structures) in normal and abnormal gene expression
ActiveUS20180080081A1Easy to detectEarly stageMicrobiological testing/measurementChromosome StructuresBiology
Method of detection or diagnosis of abnormal gene expression in an individual comprising determining in a sample from the individual the presence or absence of a chromosome structure in which two separate regions of the gene have been brought into close proximity, to thereby detect or diagnose whether the individual has abnormal gene expression.
Owner:OXFORD UNIV INNOVATION LTD
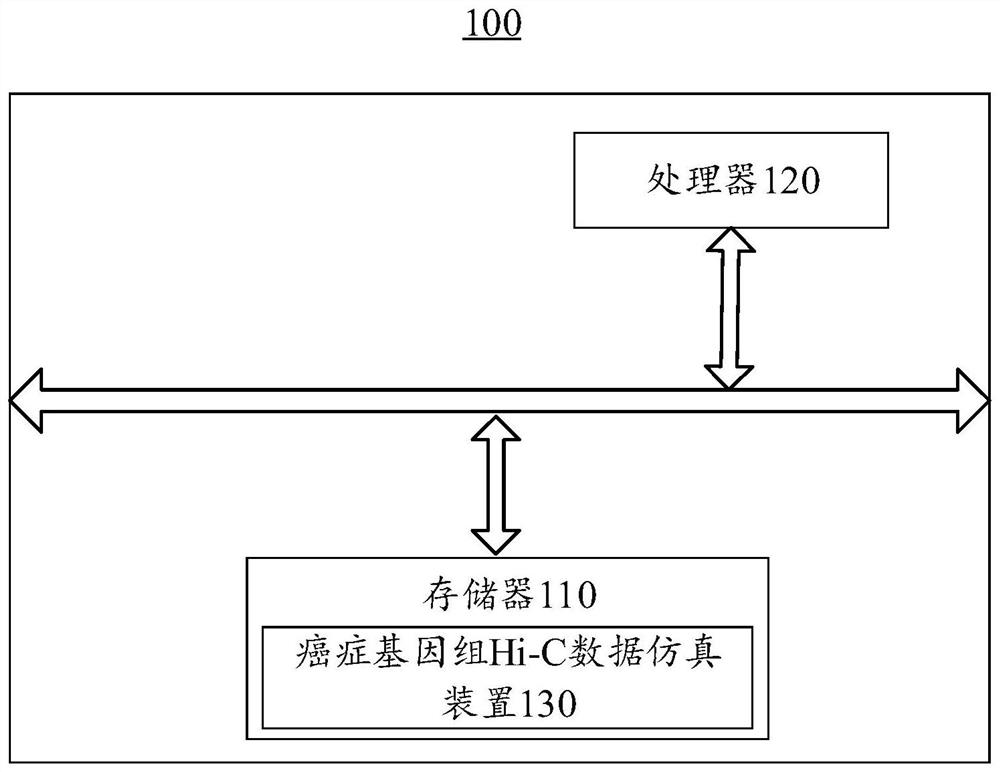
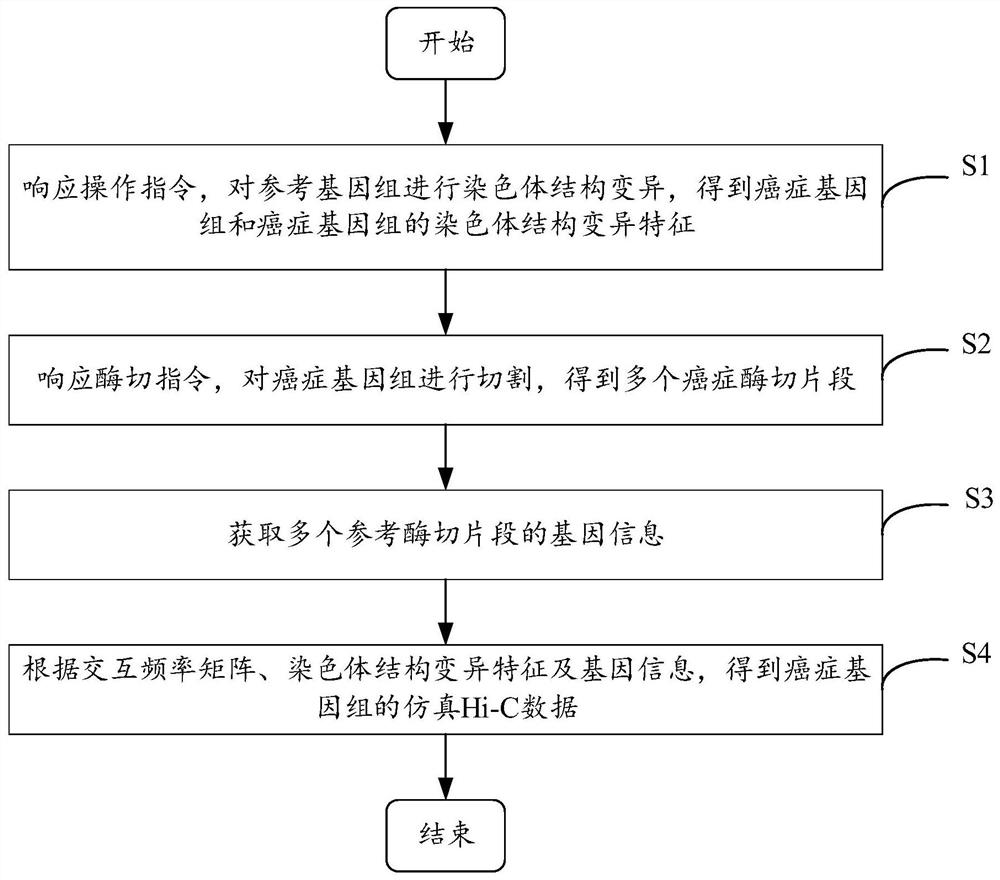
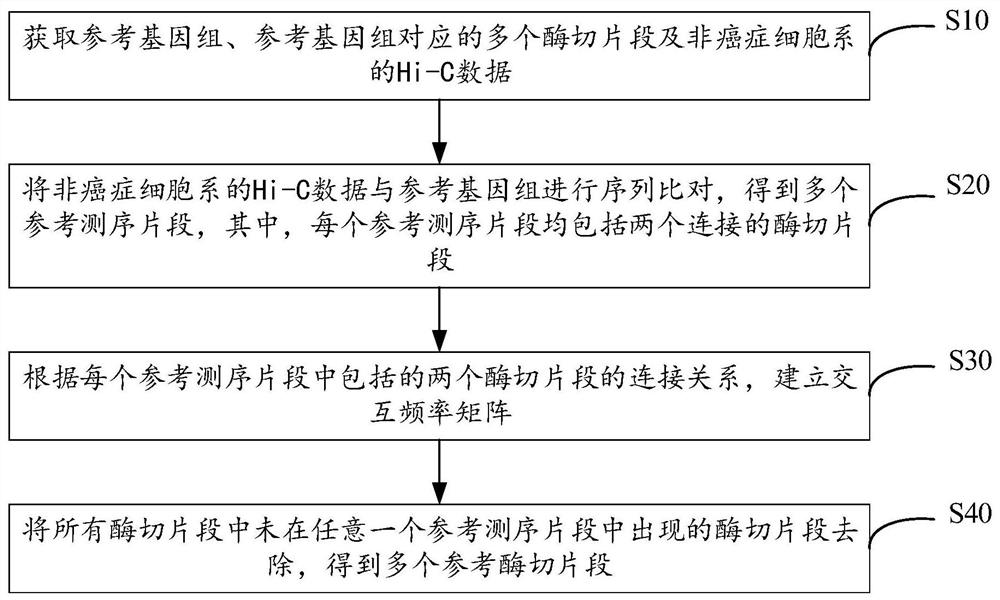
Cancer genome Hi-C data simulation method and device and electronic equipment
The embodiment of the invention provides a cancer genome Hi-C data simulation method and device and electronic equipment, and relates to the technical field of genomics. According to the method, a specified variation mode can be adopted to simulate chromosome structure variation of a cancer genome and the interaction frequency matrix and the gene information of the reference enzyme digestion fragment are taken as templates; the simulated Hi-C data of the cancer genome is obtained in combination with the chromosome structure variation characteristics, various variation conditions of the chromosome are simulated, and the interaction characteristics of different positions of the cancer genome can be reflected, so that the accuracy of simulating the Hi-C data of the cancer genome is improved.
Owner:ACADEMY OF MILITARY MEDICAL SCI
DNA conformation (loop structures) in normal and abnormal gene expression
ActiveUS9777327B2Easy to detectEarly stageMicrobiological testing/measurementChromosome StructuresBiology
Owner:OXFORD UNIV INNOVATION LTD
System and method for resource allocation of semiconductor testing industry
InactiveUS8271311B2Efficient developmentFacilitate decision-makingGenetic modelsDigital computer detailsChromosome StructuresComputer science
A system and a method for resource allocation in the semiconductor testing industry are provided. In the system, an industry characteristic conversion module is used to transform the industry characteristic obtained from an input module into a chromosome structure. Next, an artificial intelligence evolution module proceeds to find an optimal solution by handling the chromosome structure until a candidate solution having a maximum final total profit converges to a value.
Owner:NAT TAIWAN UNIV OF SCI & TECH
Plant single chromosome laser microdissection separation method
InactiveCN106318897AEvenly distributedReduce crossoverPreparing sample for investigationPlant cellsAdhesion processNitrous oxide gas
The invention provides a plant single chromosome laser microdissection separation method. According to the method, firstly, a plant organ requiring single chromosome separation is put into nitrous oxide gas to be pretreated; then, the pre-treated plant organ is subjected to immobilization treatment and enzymolysis treatment; the plant organ subjected to enzymolysis is crushed and is prepared into cell suspension; the cell suspension is dripped onto a glass slide or a coverslip to obtain a chromosome mounting chip; dye liquid is dripped into the prepared chromosome mounting chip, and then, drying is performed; microscope is used for observing the dried chromosome to determine the target chromosome to be separated; a laser capture micro cutting process is used for separating the target chromosome. The chromosome dispersion degree of the prepared chromosome mounting chip is high, so that the later stage laser cutting operation difficulty is reduced; the damage to the chromosome in the ultraviolet laser cutting and infrared laser adhesion process is reduced; the single chromosome structure obtained after the separation is not damaged; the later-stage experiment study requirements are met.
Owner:THE AFFILIATED HOSPITAL OF SOUTHWEST MEDICAL UNIV +1
Method and device for detecting chromosome structure abnormality based on deep learning
ActiveCN114842472AImprove screening efficiencyGet detailed and accurateAcquiring/recognising microscopic objectsRecognition of DNA microarray patternStainingAlgorithm
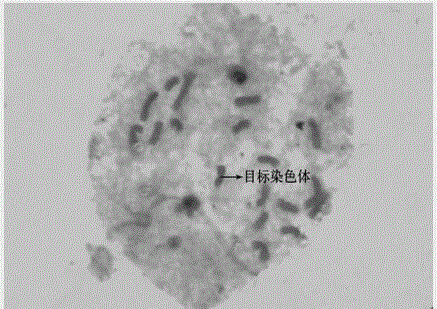
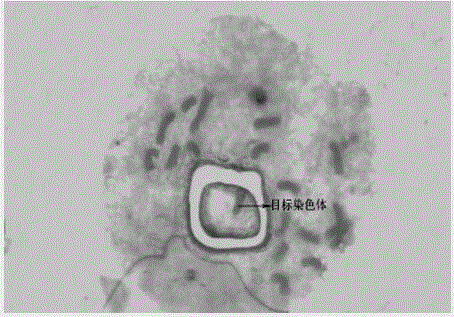
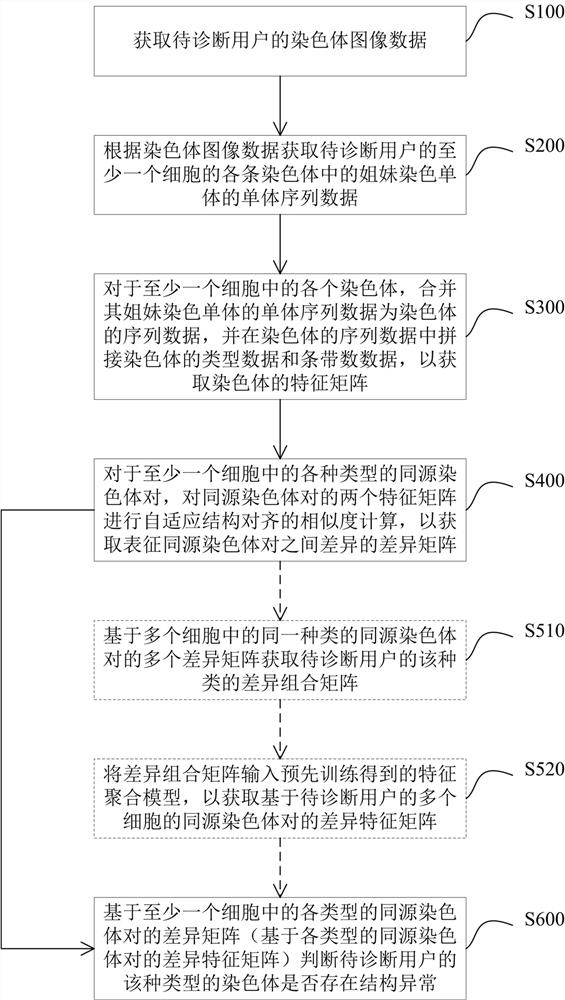
The invention provides a chromosome structure anomaly detection method and device based on deep learning. The detection method comprises the following steps: acquiring chromosome image data of a to-be-diagnosed user; according to the chromosome image data, obtaining a feature matrix of each chromosome through monomer sequence data, type data and stripe number data of sister dyeing monomers of each chromosome; obtaining a difference matrix representing the difference between the homologous chromosome pairs based on the two feature matrixes of the homologous chromosomes; and at least based on the difference matrix of the homologous chromosome pairs of various types in the at least one cell, judging whether the chromosome of the type of the user to be diagnosed has structural abnormality or not. According to the method, the chromosomes are represented through the feature matrix, and the difference between the homologous chromosome pairs is represented through the difference matrix, so that whether the chromosome structure abnormality exists in the user or not can be judged according to the difference matrix through deep learning, and the screening efficiency of the chromosome structure abnormality can be greatly improved.
Owner:HANGZHOU DIAGENS BIOTECH CO LTD
Method for simultaneously identifying embryo chromosome structure abnormality and virulence gene carrying state
ActiveCN113436680ANo pre-experimentVersatilityMicrobiological testing/measurementProteomicsMedicineEmbryo
The invention discloses a method for simultaneously identifying embryo chromosome structure abnormality and pathogenic gene carrying state. The present invention is in the field of genetic diagnosis and human assisted reproduction. On the basis of a family whole genome haplotype analysis model, a comprehensive general technical method is constructed, the method can detect the pathogenic gene of the embryo only through one-time detection, and also can detect the chromosome aneuploid and chromosome structure abnormality of the embryo. The method provides guidance for accurate embryo diagnosis of genetic illness patients, and has important clinical significance.
Owner:THE OBSTETRICS & GYNECOLOGY HOSPITAL OF FUDAN UNIV +1
A dna complex structure variation detection method
ActiveCN108171011BImproving the Accuracy of Structural Variation DetectionSave time and costProteomicsGenomicsGeneticsA-DNA
The invention discloses a DNA (desoxyribonucleic acid) complex structure variation diagnosis method. 16 chromosome structure variation types are designed, and a corresponding judgment model is designed by aiming at each variation type. Comparison information is directly obtained in a sequencing data comparison file. Compared with the prior art, the method omits sequence re-splicing and re-comparing parts, DNA complex structure variation detection can be quickly finished, and time cost is saved. Meanwhile, since information required for deducing the variation type is fully obtained in the comparison, the problem that re-comparison information can not be completely obtained and wrong information is obtained in a traditional process can be avoided, and chromosome structure variation detectionaccuracy is improved.
Owner:志诺维思(北京)基因科技有限公司
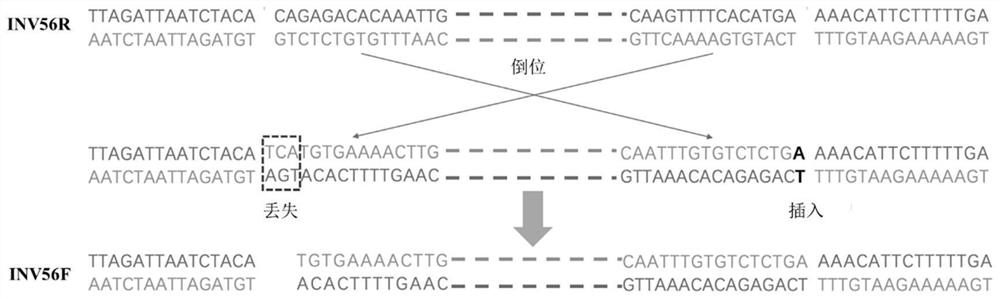
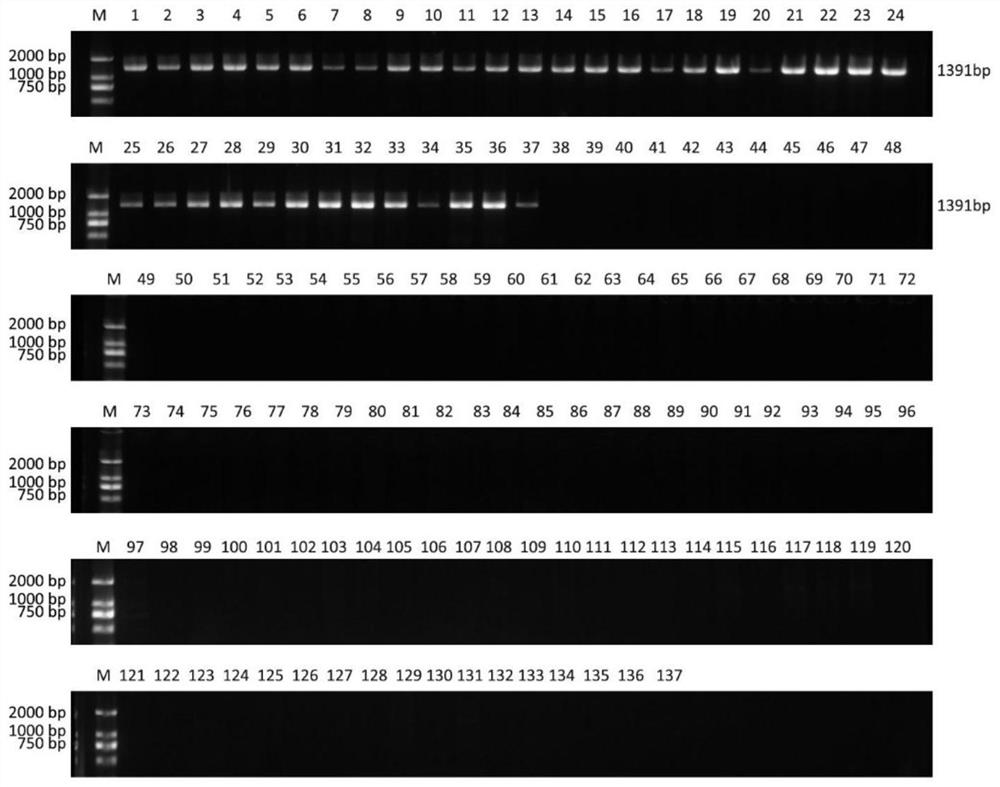
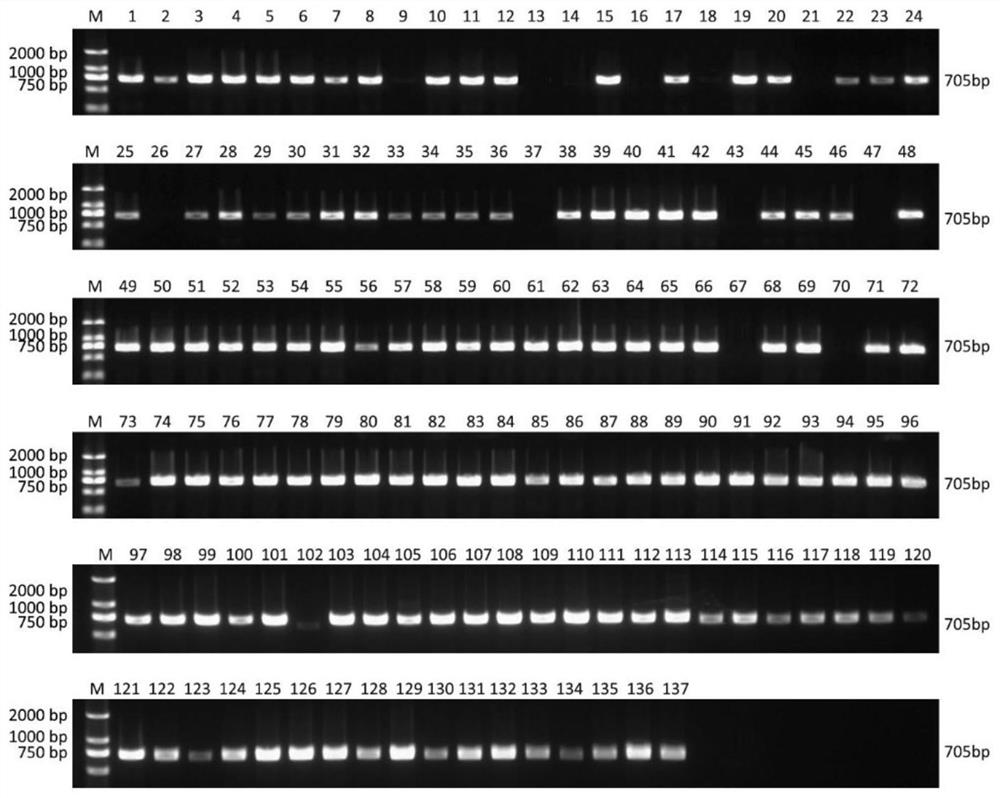
A method for early detection of peach fruit shape based on chromosome structure variation
ActiveCN111944918BStrong specificityEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationGenomic DNARapid identification
The invention relates to a method for early identification of cultivated peach fruit shape based on PCR technology. The PCR detection of the present invention uses 4 pairs of primers, including 1 pair of primers (P1) for identifying INV56F (Flat), and 3 pairs of primers (P2 / 3 / 4) for identifying INV56R (Round). The identification method is to use the peach genomic DNA to be tested as a template, first use P1 and P2 to amplify respectively, and detect the amplified products. If two specific amplification bands of 1391bp and 705bp are obtained, the fruit shape is flat; if only If there is a 705bp specific amplification band, the fruit shape is round; if no 705bp band is obtained, continue to amplify with the P3 primer pair, if a 981bp specific amplification product band is obtained, the fruit shape is round, otherwise Continue to amplify with the P4 primer; if a 651bp specific amplification product band is obtained, the fruit shape is round, otherwise the fruit shape cannot be judged temporarily. The method has high primer specificity, and the setting of control primers can effectively prevent the occurrence of false negatives, and has the characteristics of simple operation, rapid identification, and accurate results.
Owner:BEIJING AGRO BIOTECH RES CENT
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com