DNA (desoxyribonucleic acid) complex structure variation diagnosis method
A diagnostic method and technology of complex structure, applied in the field of gene detection of tumor cells, can solve the problems of complex structure mutation diagnosis and chromosomal structure mutation technology, such as slow speed and inability, to save time and cost, avoid obtaining wrong information, and improve the effect of accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
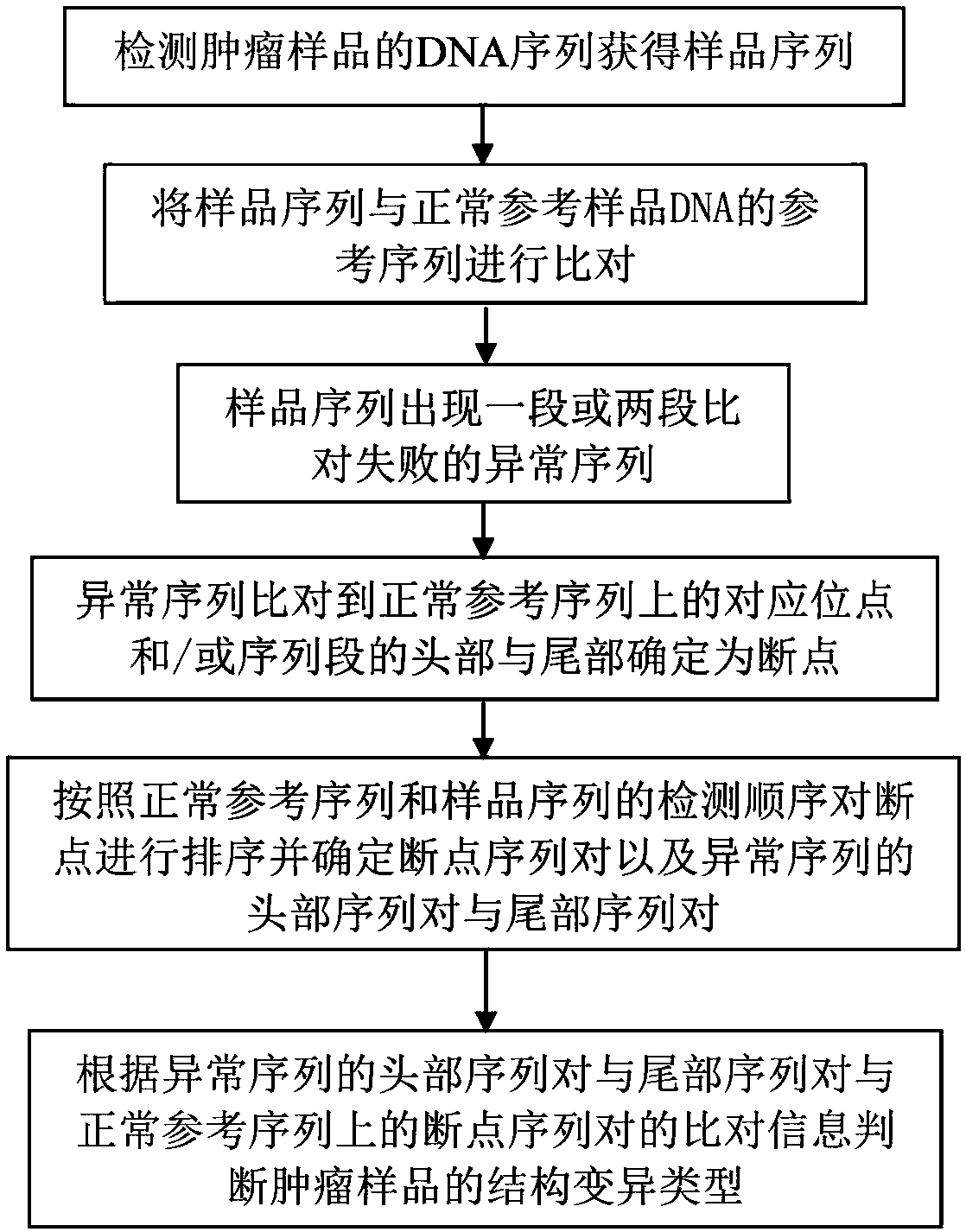
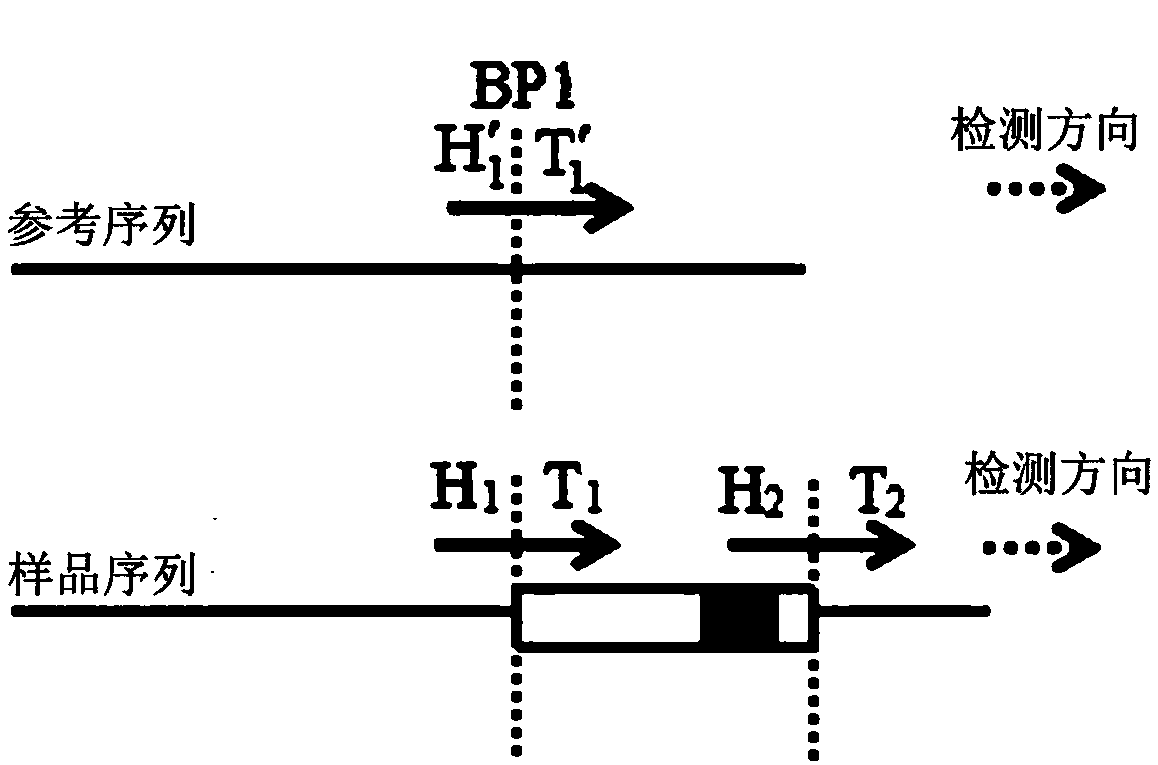
[0037] Such as figure 1 As shown, a DNA complex structure mutation diagnosis method disclosed in this embodiment includes: detecting the DNA sequence of the tumor sample to obtain the sample sequence; comparing the sample sequence with the reference sequence of the normal reference sample DNA; image 3 As shown, when the sample sequence is compared with the reference sequence of the normal reference sample DNA, there is an abnormal sequence in the sample sequence that fails to be aligned; the corresponding position of the abnormal sequence compared to the normal reference sequence is determined as the breakpoint BP1; according to the normal reference sequence Sort the breakpoints with the detection order of the sample sequence and determine the breakpoint sequence pair (H′ 1 , T 1 ′) and the head sequence pair (H 1 , T 1 ) And the tail sequence pair (H 2 , T 2 ); the head point H of the head sequence pair of the abnormal sequence on the sample sequence 1 The tail point T of the ta...
Embodiment 2
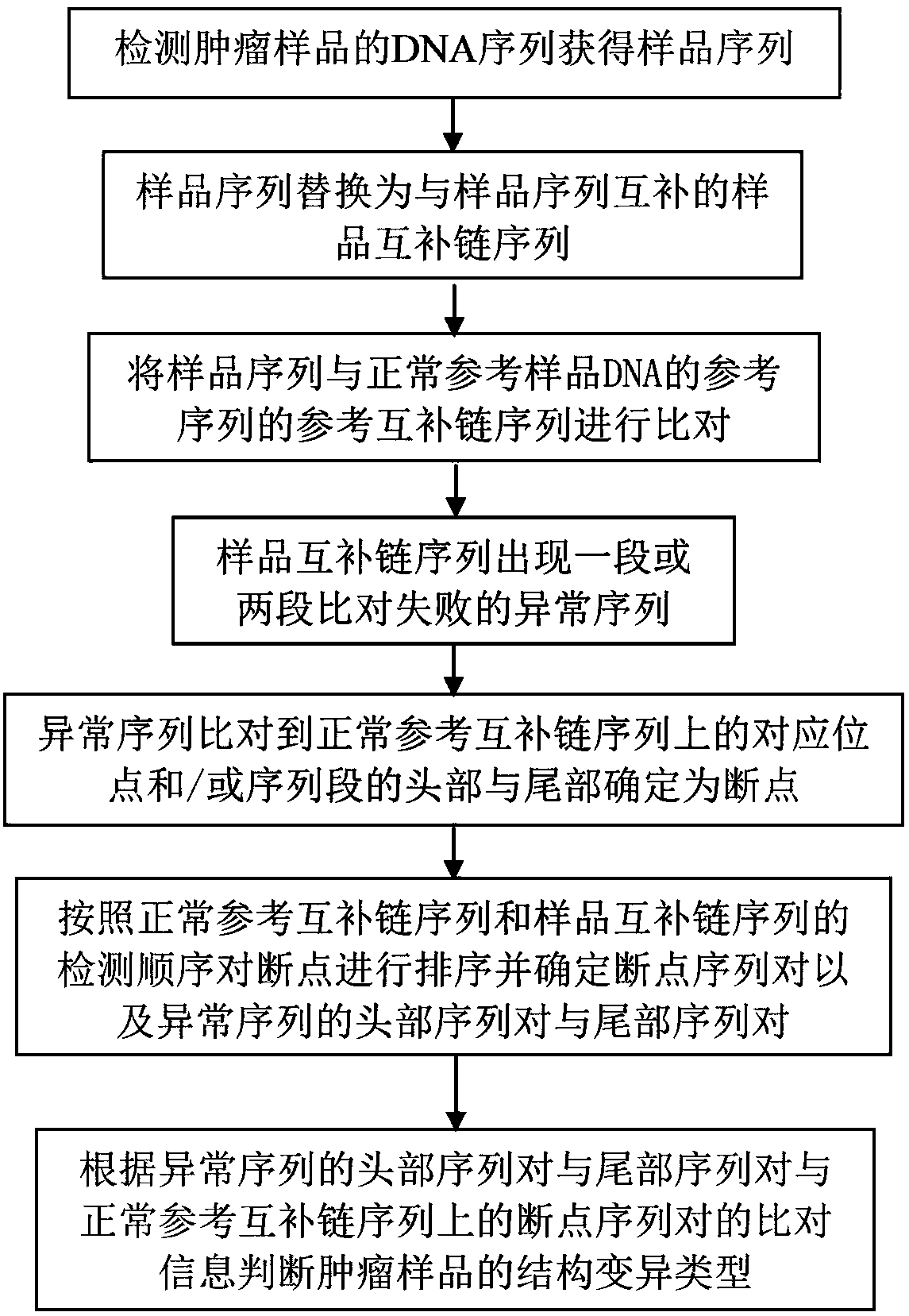
[0039] Such as figure 1 As shown, a DNA complex structure mutation diagnosis method disclosed in this embodiment includes: detecting the DNA sequence of the tumor sample to obtain the sample sequence; comparing the sample sequence with the reference sequence of the normal reference sample DNA; Figure 4 As shown, when the sample sequence is compared with the reference sequence of the normal reference sample DNA, there is a missing sequence in the sample sequence; this missing sequence is compared to the head and tail of the sequence segment to determine the breakpoint; according to the normal reference sequence and the sample sequence Sort the breakpoints BP1 and BP2 and determine the breakpoint sequence pair (H′ 1 , T 1 ′) and (H′ 2 , T 2 ′) and the sequence pair (H 1 , T 1 ); the head point H of the corresponding position of the missing sequence on the sample sequence 1 With tail point T 1 Align respectively to the head point H′ of the head breakpoint sequence pair on the normal...
Embodiment 3
[0041] Such as figure 1 As shown, a DNA complex structure mutation diagnosis method disclosed in this embodiment includes: detecting the DNA sequence of the tumor sample to obtain the sample sequence; comparing the sample sequence with the reference sequence of the normal reference sample DNA; Figure 5 As shown, when the sample sequence is compared with the reference sequence of the normal reference sample DNA, there is an abnormal sequence that fails the alignment; the head and tail of the sequence segment where the abnormal sequence is compared to the normal reference sequence are determined as breakpoints; Sequence the breakpoints BP1 and BP2 according to the detection sequence of the normal reference sequence and the sample sequence, and determine the breakpoint sequence pair (H′ 1 , T 1 ′) and (H′ 2 , T 2 ′) and the head sequence pair (H 1 , T 1 ) And the tail sequence pair (H 2 , T 2 ); the head point H of the head sequence pair of the abnormal sequence on the sample sequen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com