A method for early detection of peach fruit shape based on chromosome structure variation
A fruit and purpose technology, applied in biochemical equipment and methods, microbiological determination/testing, DNA/RNA fragments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1: Primer Design for Detecting Peach Fruit Shape
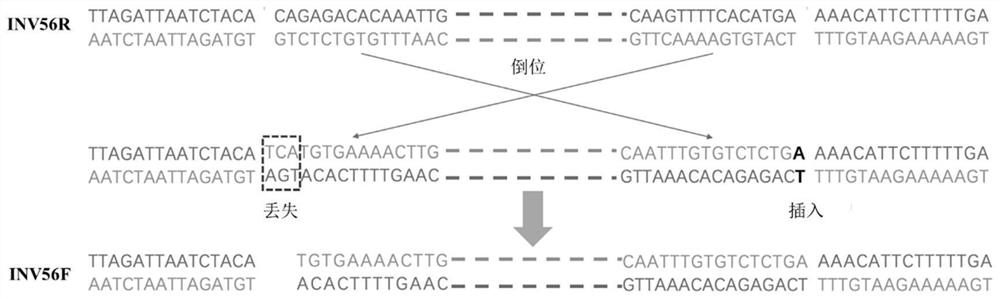
[0035] With the help of the advanced third-generation sequencing platform Pacbio, the present invention completed the genome sequencing of Ruiyoupan 1, and successfully assembled the first draft genome of the flat peach. By comparing the sequence with the existing peach reference genome Lovell (circle), screening the chromosome structure variation, and combining the resequencing data of 149 cultivated peaches, it was found that a chromosomal inversion structure variation located on chromosome 6 was related to the round and flat shape of peach fruit The shape is closely related, and the structural variant was named INV56 (Scaffold_6:27,959,880-29,634,101). Among them, the structural state that exists specifically in flat peach is called INV56F (flat shape, see figure 1 ), another state is INV56R (round, see figure 1 ). Because the locus is heterozygous in flat peaches, the genotype of flat peaches is INV56F / R;...
Embodiment 2
[0039] Embodiment 2: application embodiment
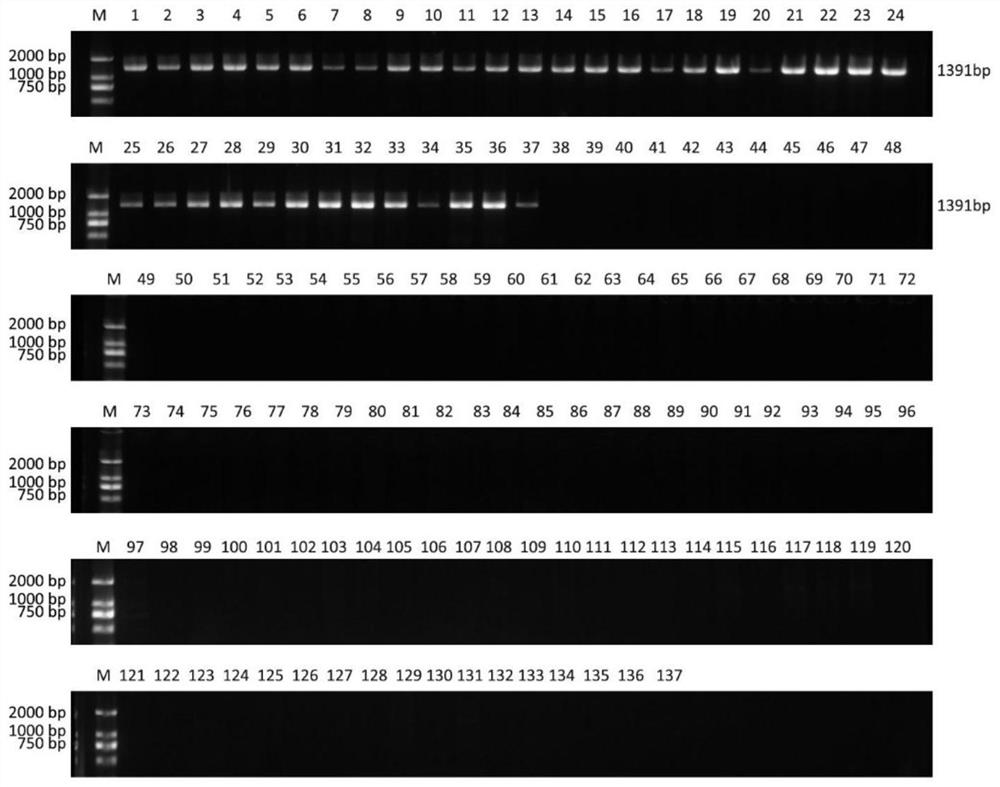
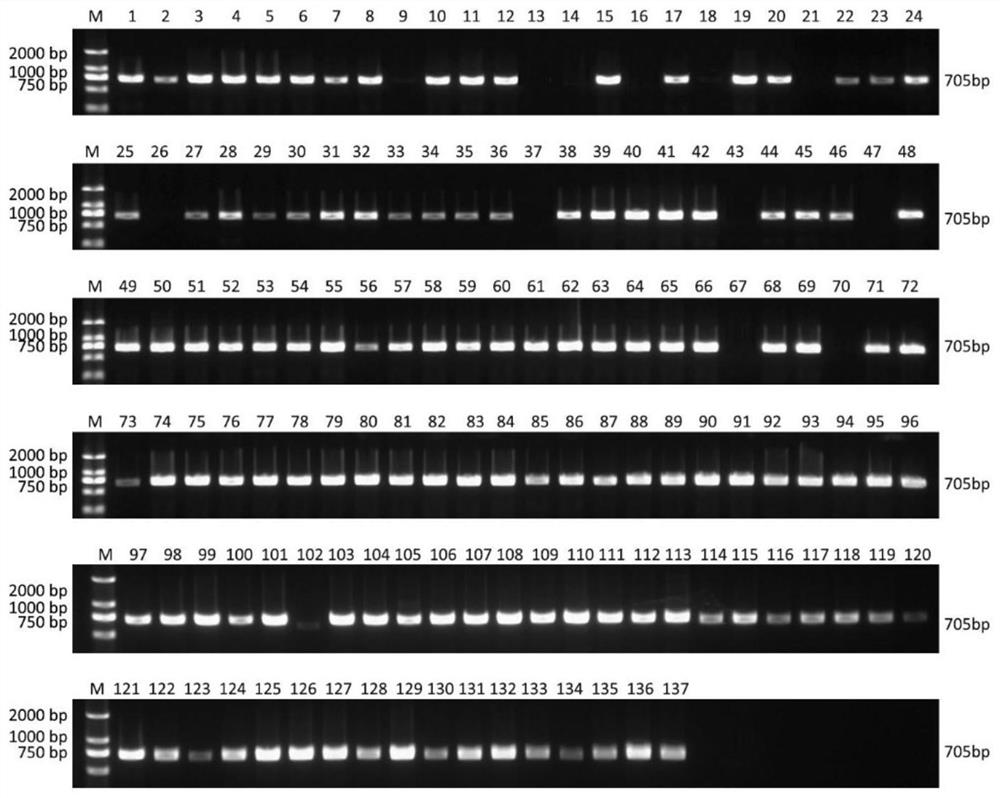
[0040] 137 young leaves of cultivated peaches were collected, including 37 flat peaches and 100 round peaches (Table 2). Genomic DNA was extracted using a plant DNA extraction kit (Tiangen Biochemical Technology (Beijing) Co., Ltd.), and then quantified using sterile Bacteria water was configured as a 20ng / μL template solution. The configuration and reaction conditions of the PCR reaction solution are as follows:
[0041] Composition of PCR reaction solution:
[0042]
[0043]
[0044] PCR reaction conditions:
[0045]
[0046] In the above experimental process, first use the P1 and P2 primer pair for amplification detection. If no specific amplification band is obtained with the P2 primer, continue to use the P3 primer pair for amplification detection. If there are still samples without amplified bands , continue to use the P4 primer pair for amplification detection.
[0047] The detection of the amplification product...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com