Chromosome structure variation molecular marker related to tomato fruit color, specific primers and application thereof
A specific primer and chromosome technology, applied in the field of plant genetic engineering and molecular biology, can solve the problem of uneven accumulation of anthocyanins, and achieve the effect of accelerating the breeding process, reducing the investment of planting area and human and material resources, and improving the breeding efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Test material
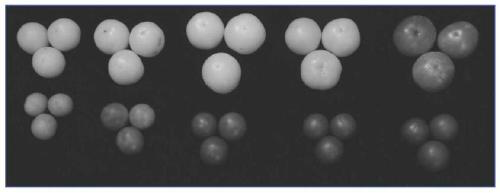
[0035] The ginkgo tomato (hereinafter referred to as wf mutant) used in this experiment is a natural mutant, the fruit is yellow-white when immature, and red when ripe; Ailsa Craig (AC) is a normal tomato, green when immature, and red when ripe . The F1 generation was obtained by crossing AC as the female parent, the wf mutant was obtained by crossing the male parent, and the F2 was obtained by selfing F1.
[0036] Identification and marker development of chromosomal structural variation in wf mutants
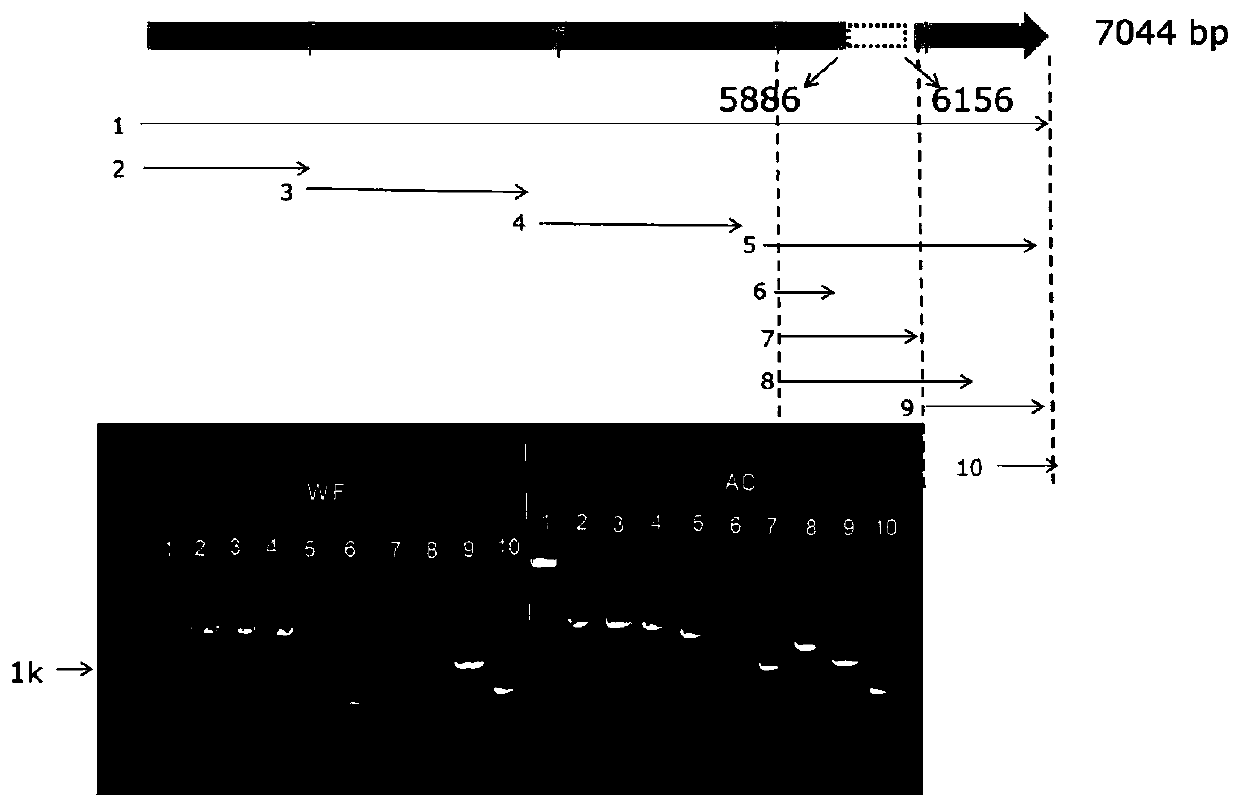
[0037] According to the similar phenotype between the wf mutant and the luteescent2 mutant, we first performed PCR detection on the luteescent2 gene in the wf mutant, and found that no band could be amplified when the full length of the luteescent2 gene in the wf mutant was amplified by PCR , so we amplified the luteescent2 gene in segments, and found that there may be a mutation from the 5886th base to the 6156th base after the start codon of the lutee...
Embodiment 2
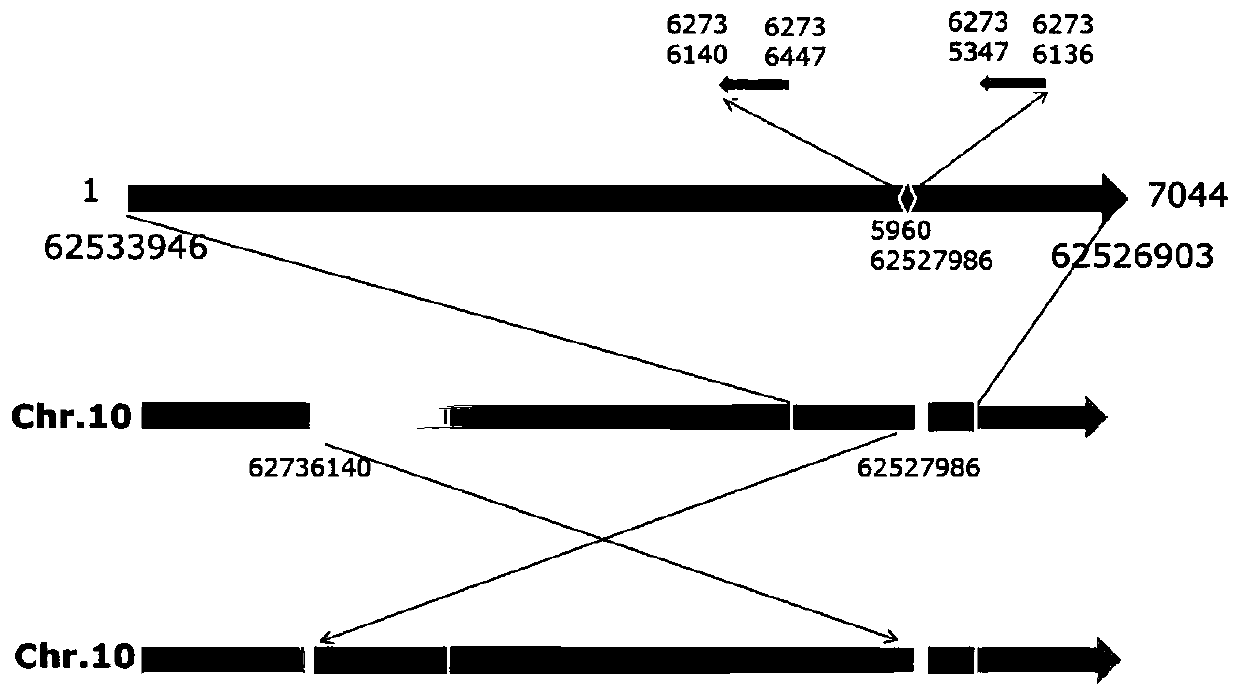
[0041] 5. On the basis of Example 1, we first designed primer SH1 with the sequence of about 1000 bp at both ends of the wf mutant inversion site 62527986 bases as a template, so that it can amplify to about 1000 bp band in the wf mutant ( For the specific sequence, refer to the sequence table (SEQ ID NO: 5), no band can be amplified in AC; the primer SH2 is designed with the sequence of about 1500 bp at both ends of the normal tomato 62527986 base as a template, so that it can amplify about 1500 bp in AC. A 1500bp band (see SEQ ID NO: 6 in the sequence listing for the specific sequence), but no band can be amplified in the wf mutant.
[0042] Using primers SH1 and SH2 to carry out molecular detection on wf mutant, Ailsa Craig (AC), F1 generation, F2 generation plants, the results showed that in AC, SH1 could not amplify any band, and SH2 could amplify a 1548bp band ; In the wf mutant, SH1 can be amplified to a 986bp band, SH2 can not be amplified to any band; in the F1 genera...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com