Genome recombination fingerprint for characterizing hHRD homologous recombination deficiency and identification method thereof
A technology of homologous recombination and genomics, applied in the fields of genomics, microbial measurement/inspection, biochemical equipment and methods, etc., can solve problems such as undiscovered identification methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
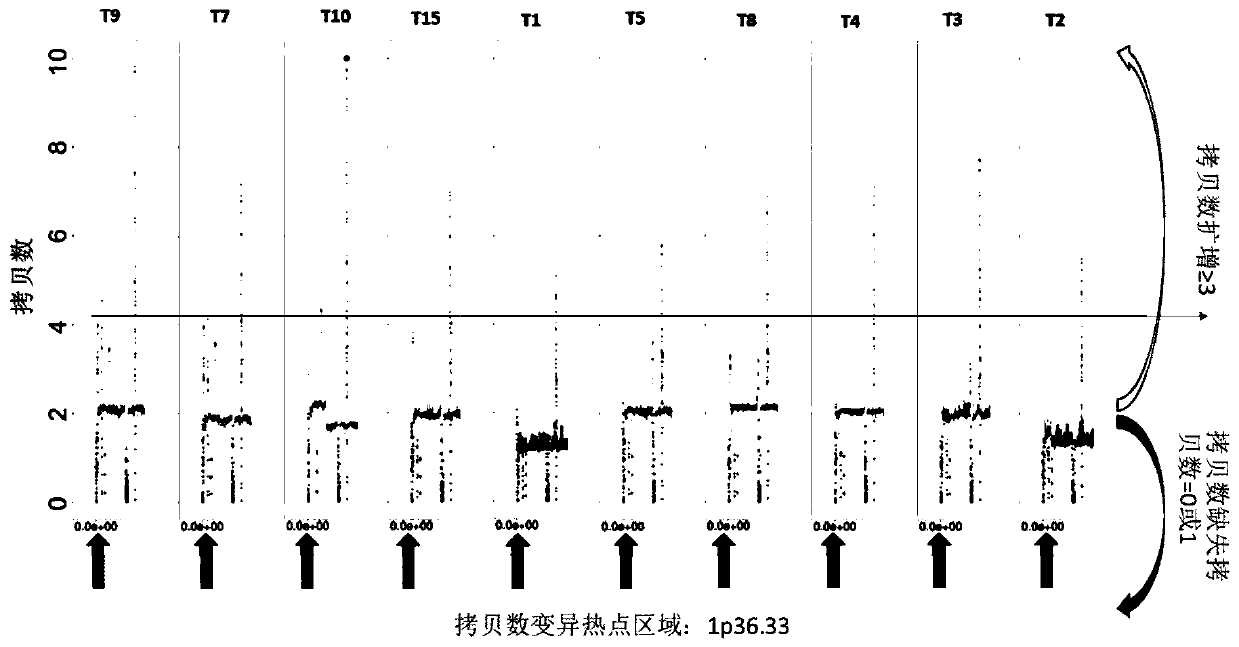
[0048] Example 1 High-frequency site-specific copy number variation in the HBV-related liver cancer genome is a characteristic of hHRD fingerprint Method: Surgical sampling of fresh liver cancer and blood tissue, sampling volume> 1cm 3 , the blood sample was 5 ml of anticoagulated blood, and the blood cells were collected by centrifugation. The samples were quickly frozen in liquid nitrogen and placed in individual sealed packages to prevent contamination between samples, and the packaging containers were marked with basic clinical information. The samples were stored at -80°C and shipped to the sequencing company within two weeks on dry ice for sample quality inspection, nucleic acid extraction and sequencing. After genome purification, use Qubit, 1% AGE to detect the degradation of the sample, the concentration is ≥20ng / ul, the total amount is ≥500ng, the sample has no obvious degradation band and can be used for downstream analysis and sequencing.
[0049] The sequencing p...
Embodiment 2
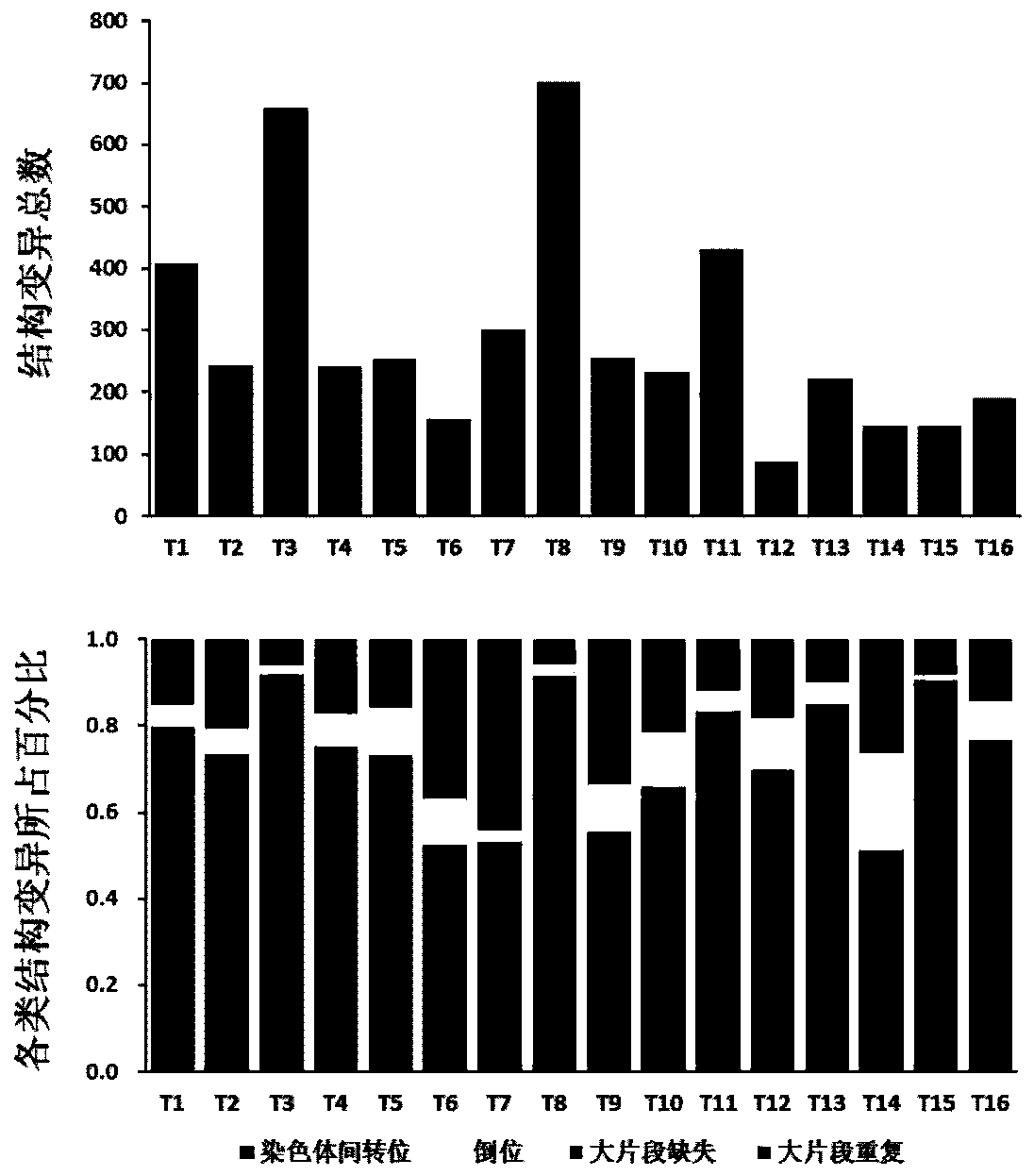
[0056] Example 2 A high proportion of interchromosomal translocation events in HBV-related liver cancer genomes is one of the molecular fingerprints of hHRD
[0057] Methods: Further, the delly software was used to define and count the genome-wide recombination events of HBV-related liver cancer genomes. The software is based on the soft-clipped principle. When deletions, translocations, insertions and other structural variations in the genome cause reads across the deletion site to be compared to the genome, a reads is cut into two segments and matched to different regions (soft-clipped reads) to identify the chromosome structure Variation and integration of foreign sequences. Events with at least three soft-clipped reads covering the breakpoint and one soft-clipped read covering the end of the adapter sequence were screened. SV mutation detection is performed on cancer paired samples (Tumor-Normal), and the SV system mutation data of the tumor itself is obtained.
[0058]...
Embodiment 3
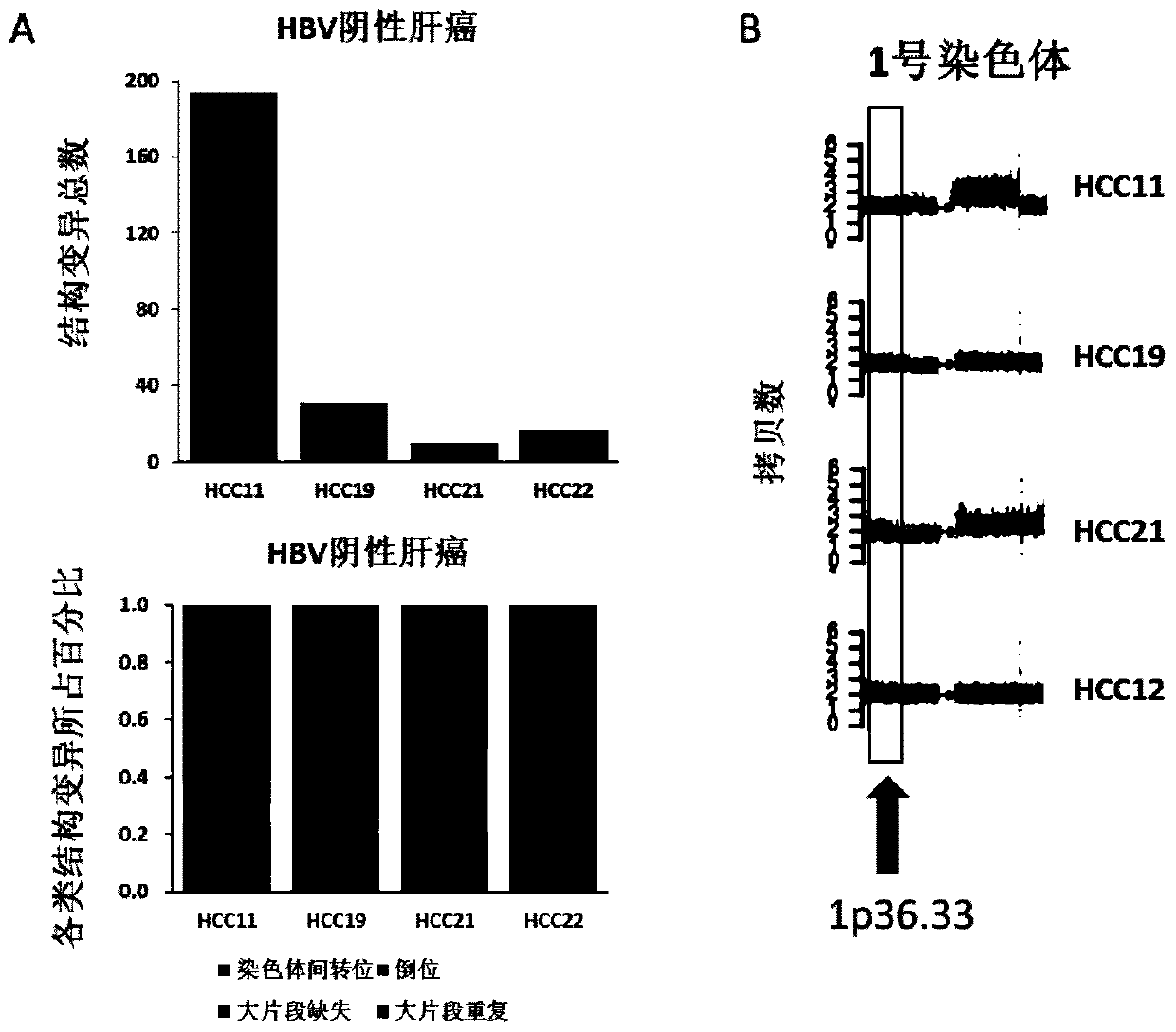
[0059] Example 3 Genome recombination characteristics of HBV-negative liver cancer
[0060] Methods: 4 cases of HBV-negative fresh liver cancer and corresponding individual blood tissues were sampled in the same operation as above, and the whole genome was performed using the illumina2000 sequencer based on the illumina sequencing platform and the Hiseq X-ten paired-end (paired-End) bridge sequencing method Resequencing; delly software was used to analyze the genome recombination characteristics of tumors, and Controlfreec software was used to analyze CNV events.
[0061] Results: A.delly software analysis found that four cases of HBV-negative liver cancer had a low number of genomic structural variations and lacked a high proportion of interchromosomal translocations. The abscissa indicates the sample number, and the ordinate indicates the quantity and composition ratio of the structural variation of each sample. B. Controlfreec software analysis found that four cases of HBV...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com