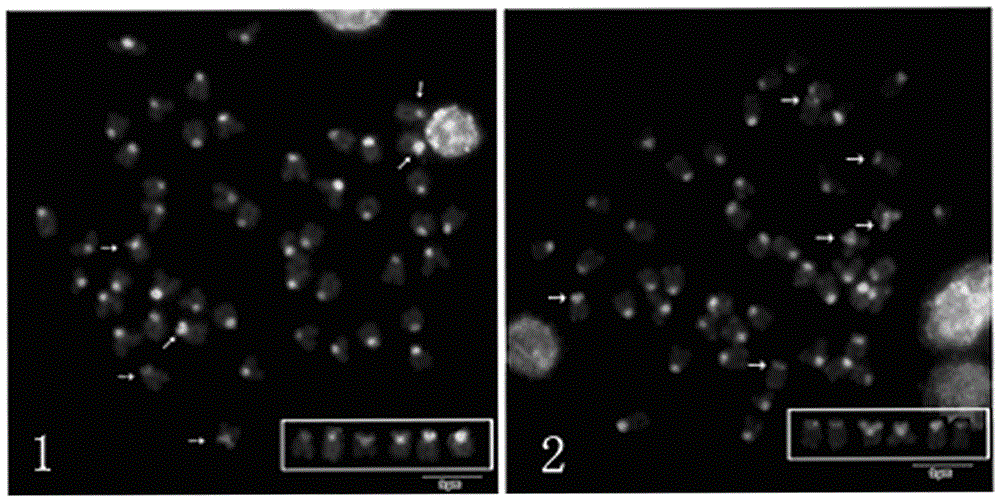
Method for displaying centromeres and short arms of Larimichthys crocea
A centromere and large yellow croaker technology, applied in the field of molecular cytogenetics, can solve the problems of unsuitability for genomics and functional gene research, poor quality of stained images, and difficulty in identifying chromosomes and fragments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0030] (1) Chromosome preparation
[0031] Inject 8mg Codonopsis Codonopsis juice / 20μgBSA mixture at the base of the pectoral fins 20 hours in advance at the base of the pectoral fins, and temporarily raise them in seawater at 25°C; inject the same reagent with the same method 5 hours in advance; inject the same reagent at the base of the pectoral fins at the rate of 2.5 hours in advance Inject 0.5 μg / g colchicine at the base; take the head kidney cells, use 0.075MKCl hypotonicity for 40 minutes, and fix with Carnoy’s fixative solution for 3 times; before dropping the tablets, replace the Carnoy’s fixative solution with a volume ratio of methanol / acetic acid = 2:1 mixed solution; put a layer of water vapor on the surface of the slide, drop the cell suspension at 1cm from the slide, and let it dry naturally in the air;
[0032] (2) Extraction of human genomic DNA
[0033] Use the tissue genome DNA extraction kit (Shanghai Jierui) to extract DNA from human saliva; before breakf...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com