Patents
Literature
51 results about "Specific chromosome" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
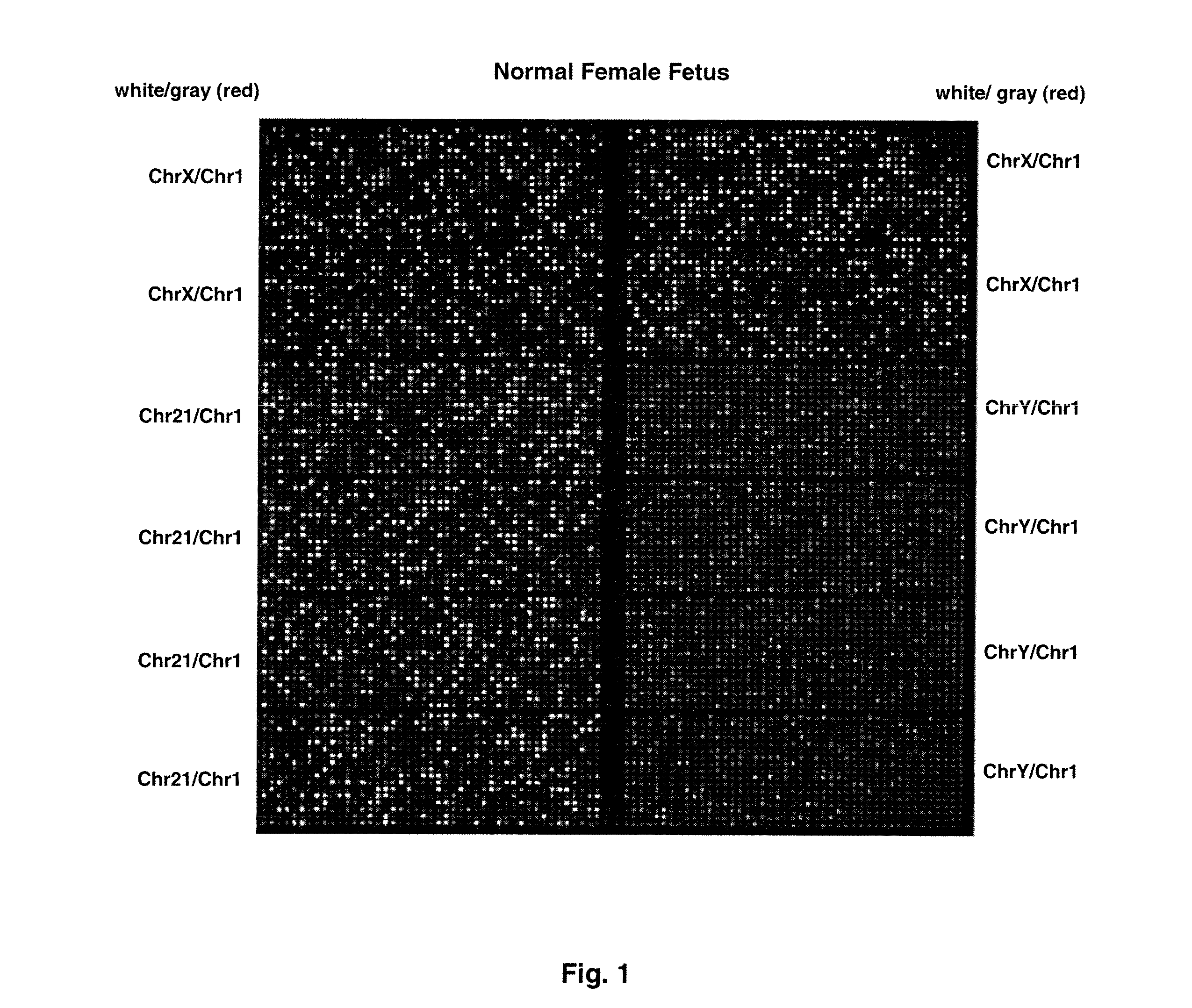
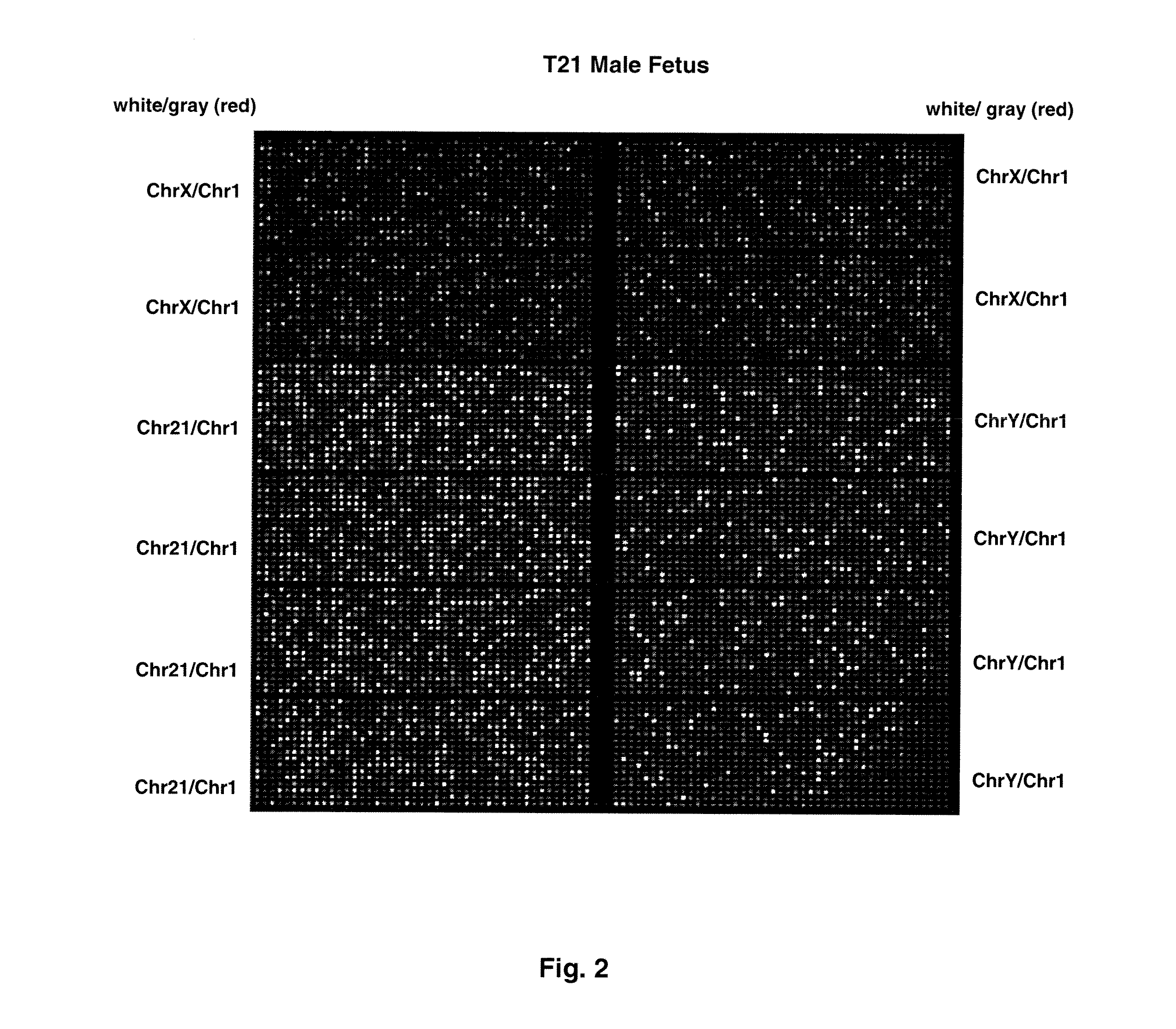
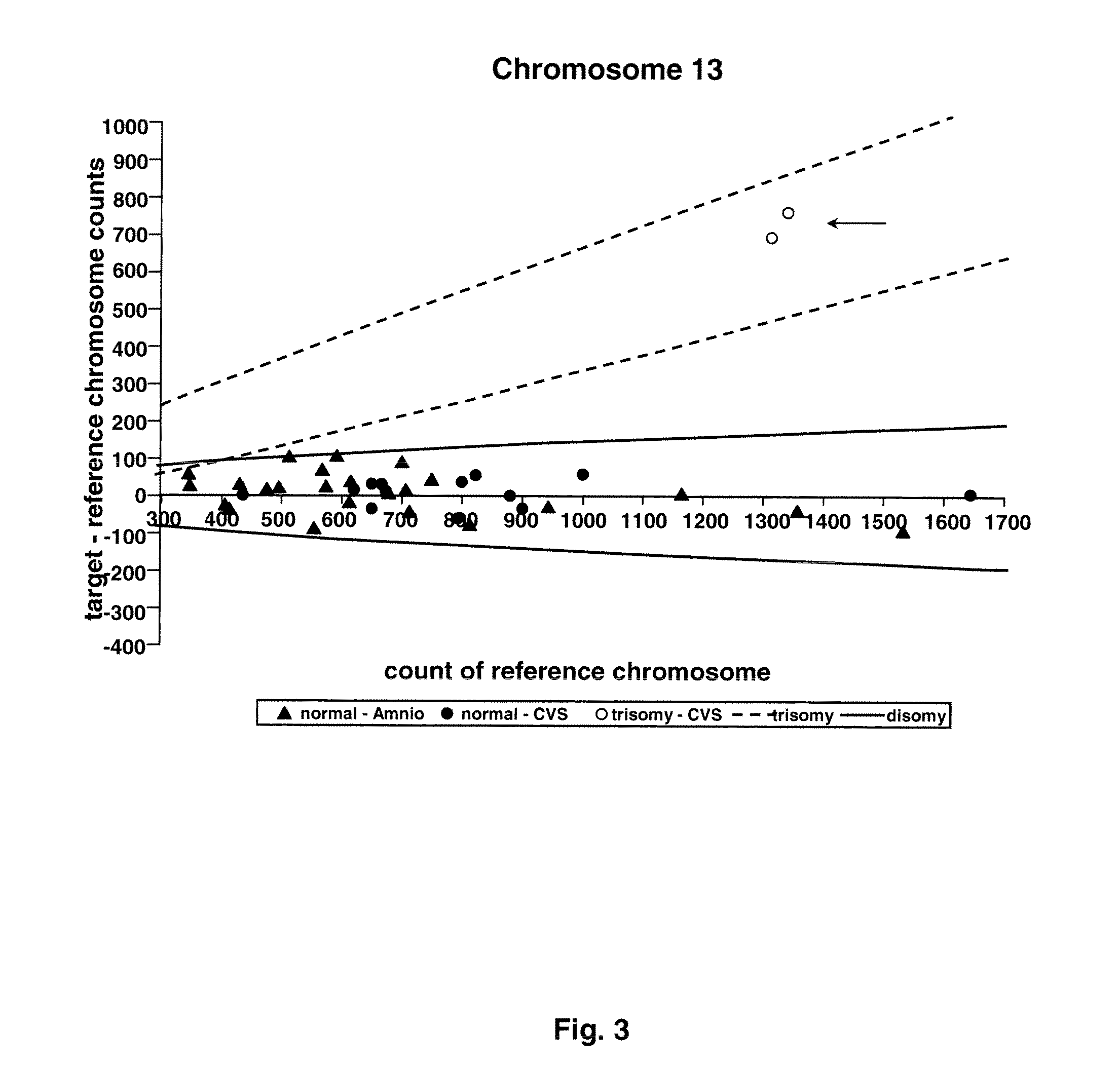
Direct Molecular Diagnosis of Fetal Aneuploidy
ActiveUS20110151442A1Addressing slow performanceSugar derivativesMicrobiological testing/measurementSpecific chromosomeFetal tissues
Methods and materials for detection of aneuploidy and other chromosomal abnormalities using fetal tissue are disclosed. Results can be obtained rapidly, without cell culture. The method uses digital PCR for amplification and detection of single target sequences, allowing an accurate count of a specific chromosome or chromosomal region. Specific polynucleic acid primers and probes are disclosed for chromosomes 1, 13, 18, 21, X and Y. These polynucleic acid sequences are chosen to be essentially invariant between individuals, so the test is not dependent on sequence differences between fetus and mother.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
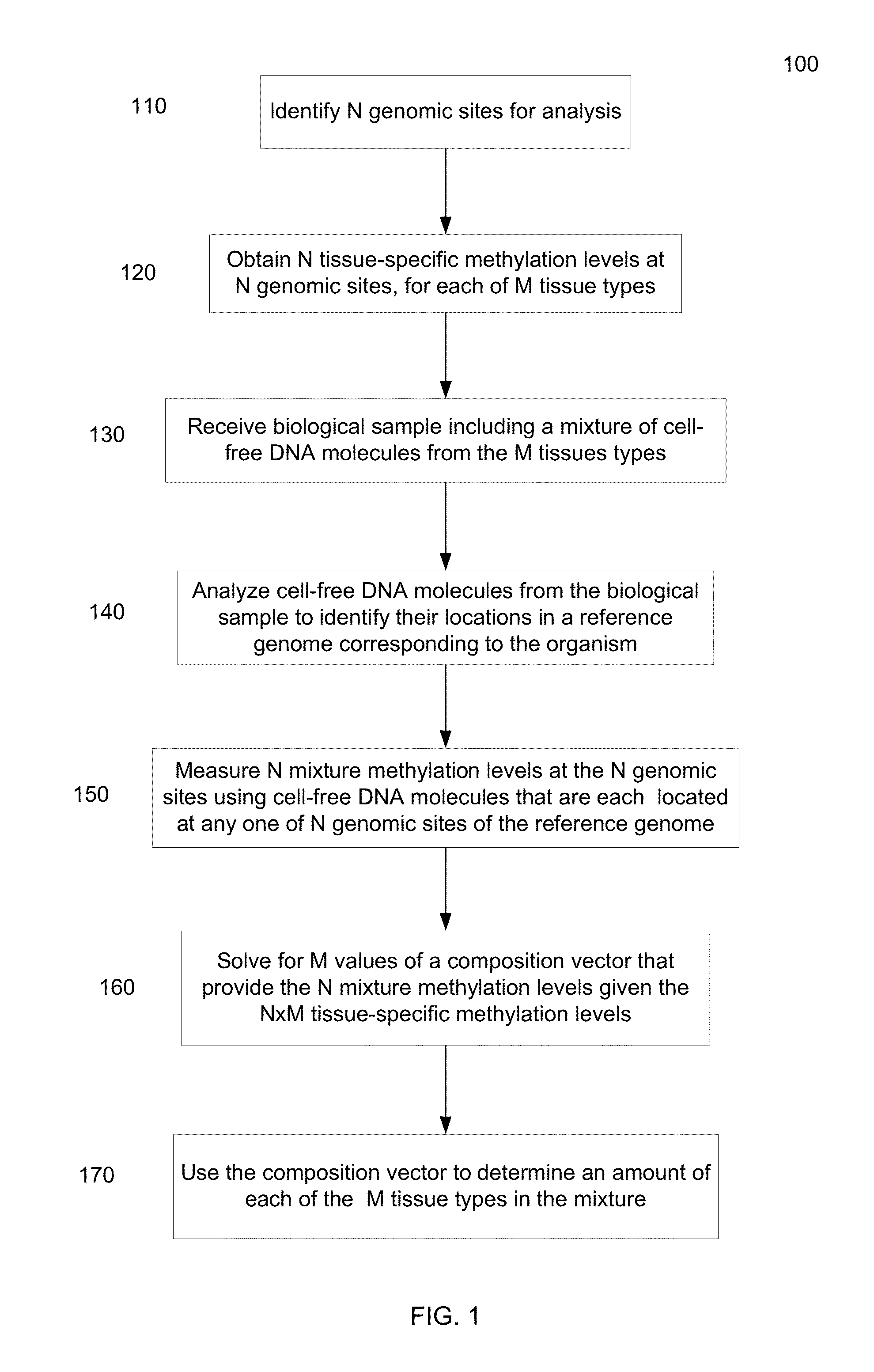
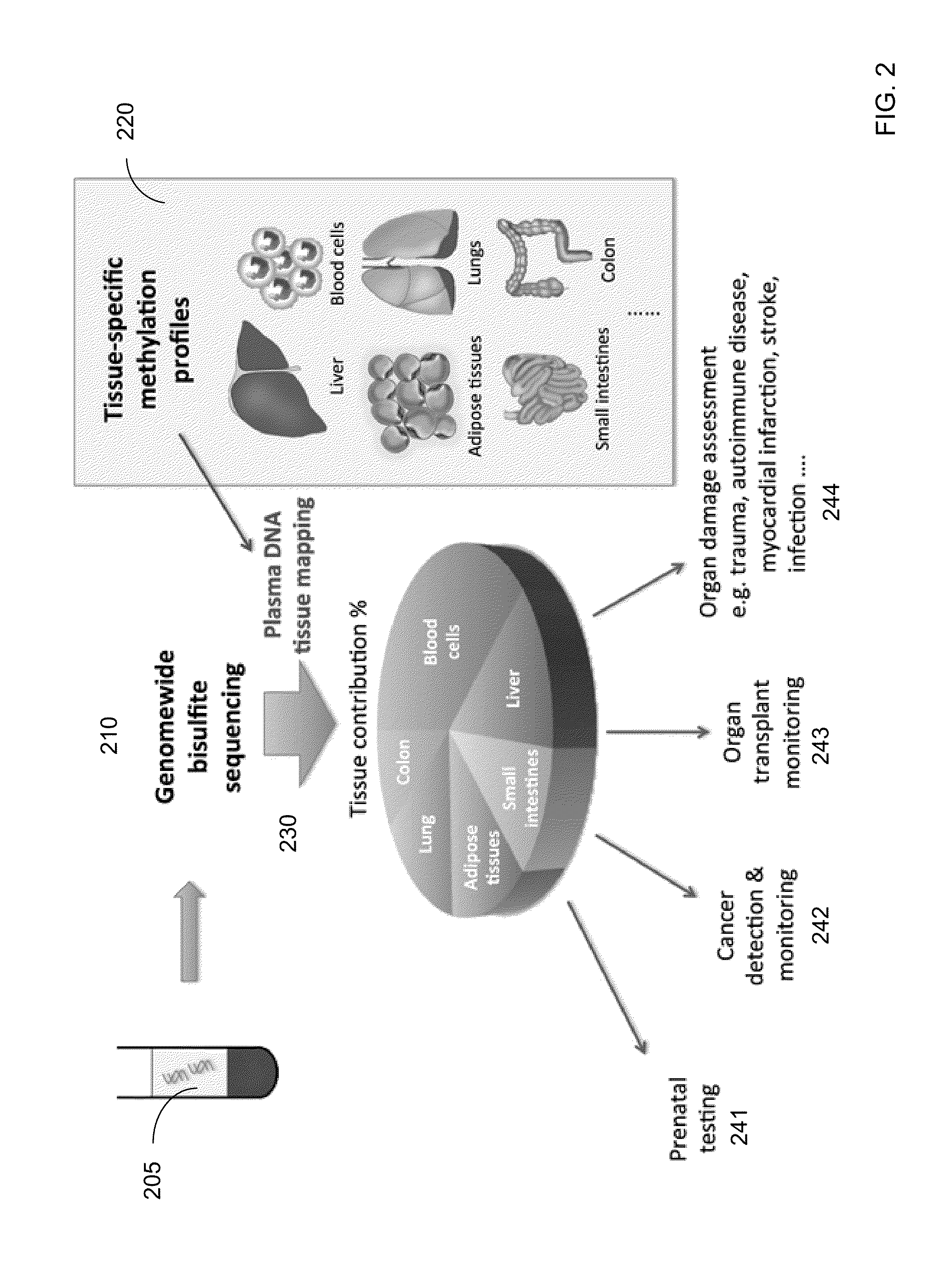
Methylation pattern analysis of tissues in a DNA mixture
ActiveUS20160017419A1Improve accuracyMicrobiological testing/measurementLibrary screeningAbnormal tissue growthSpecific chromosome
The contributions of different tissues to a DNA mixture are determined using methylation levels at particular genomic sites. Tissue-specific methylation levels of M tissue types can be used to deconvolve mixture methylation levels measured in the DNA mixture, to determine fraction contributions of each of the M tissue types. Various types of genomic sites can be chosen to have particular properties across tissue types and across individuals, so as to provide increased accuracy in determining contributions of the various tissue types. The fractional contributions can be used to detect abnormal contributions of a particular tissue, indicating a disease state for the tissue. A differential in fractional contributions for different sizes of DNA fragments can also be used to identify a diseased state of a particular tissue. A sequence imbalance for a particular chromosomal region can be detected in a particular tissue, e.g., identifying a location of a tumor.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
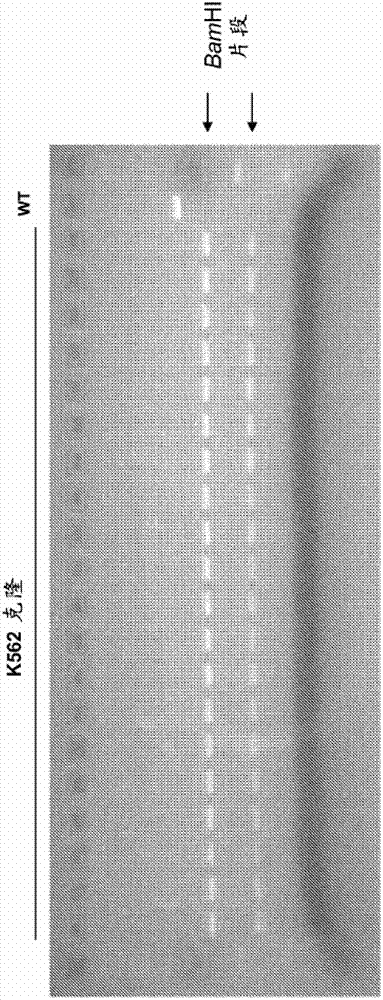
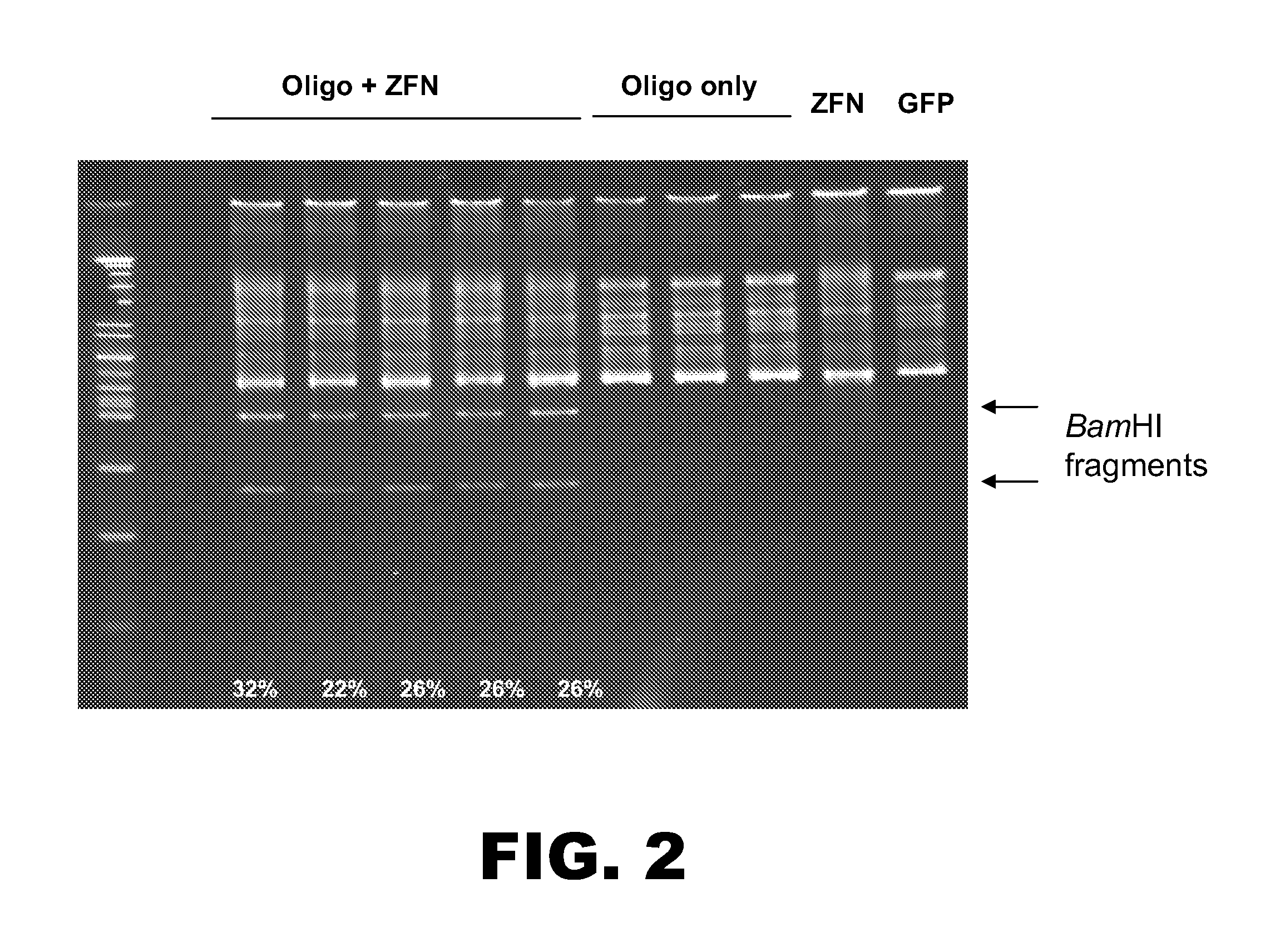
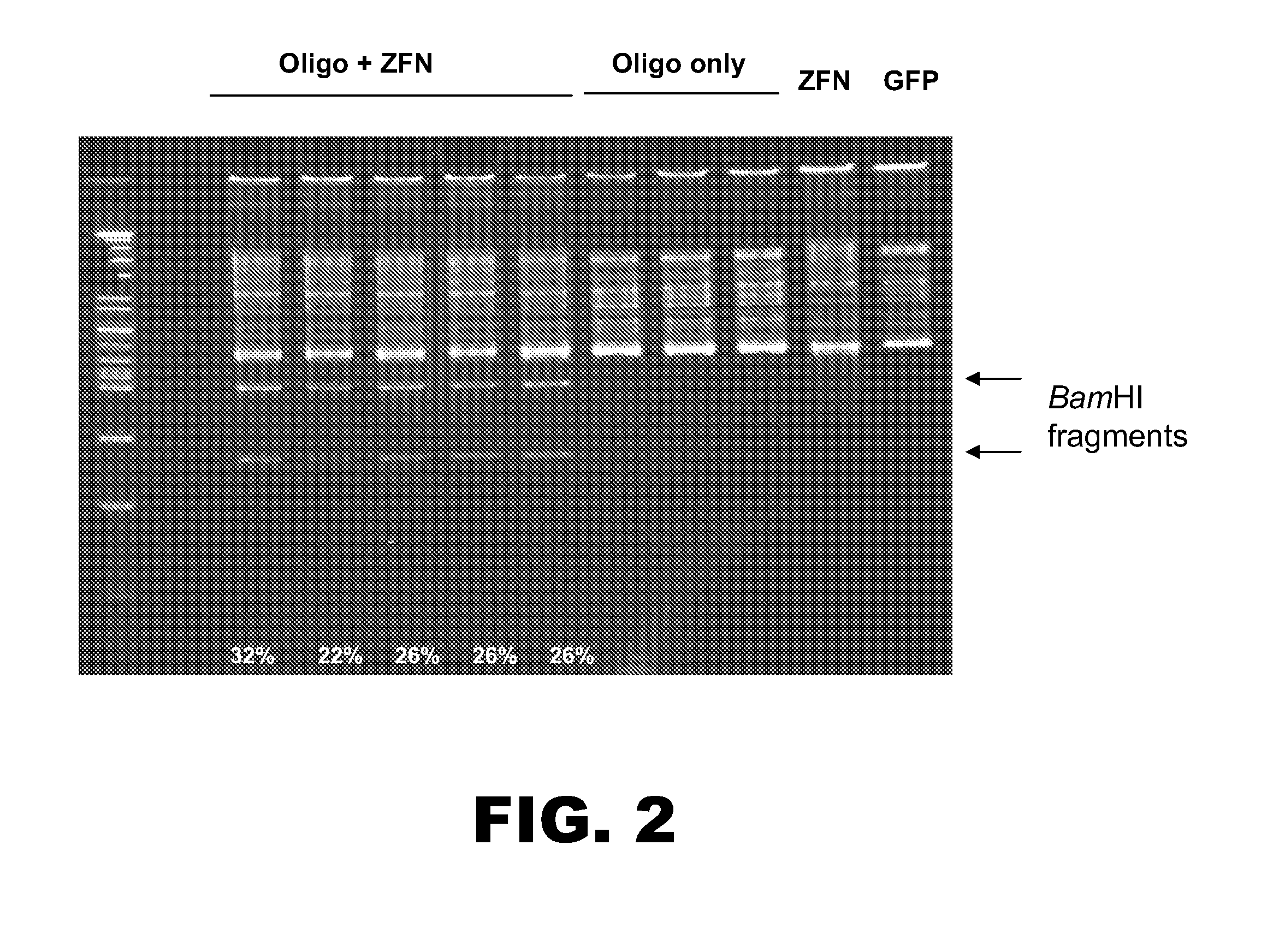
Rabbit genome editing with zinc finger nucleases
The present invention provides a genetically modified rabbit or cell comprising at least one edited chromosomal sequence. In particular, the chromosomal sequence is edited using a zinc finger nuclease-mediated editing process. The disclosure also provides zinc finger nucleases that target specific chromosomal sequences in the rabbit genome.
Owner:SIGMA ALDRICH CO LLC
System, Method and computer program product for integrated analysis and visualization of genomic data
Described is a system for analysis and visualization of genomic data. The system allows a user to select at least one individual sample. The sample has chromosomal data representing a genome with a chromosome and also includes chromosomal measurements of at least one event at a particular location on the chromosome. A frequency of event is generated based on the selected sample. The frequency of event is a frequency of occurrence of the event in the selected sample. At least one annotation can be selected that includes chromosomal region specific information as related to the chromosome. Finally, the chromosomal data, the annotation, and the frequency of event on a display can all be simultaneously displayed, thereby allowing a user to view chromosomal region specific information with respect to a particular chromosomal event.
Owner:BIODISCOVERY
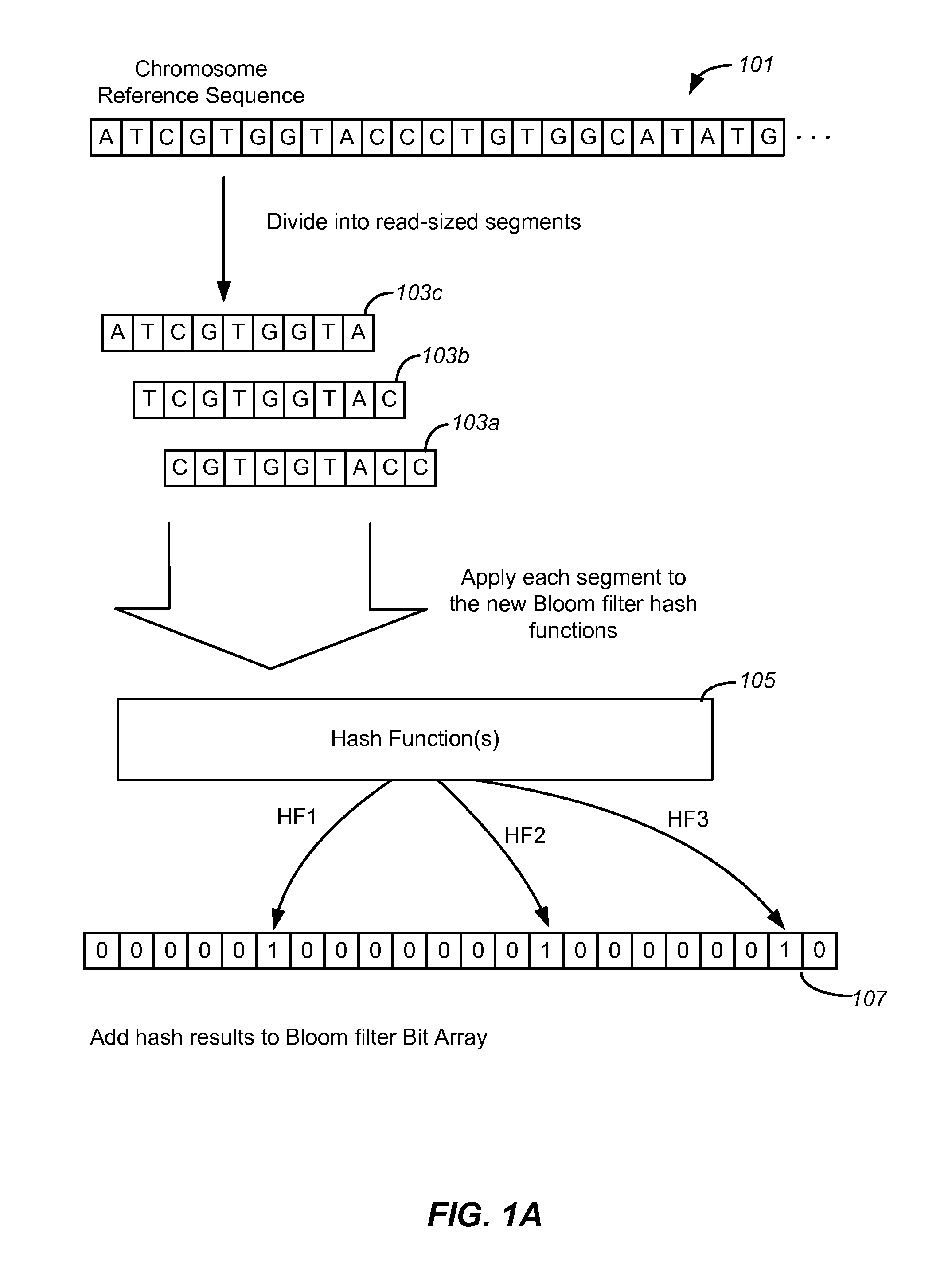
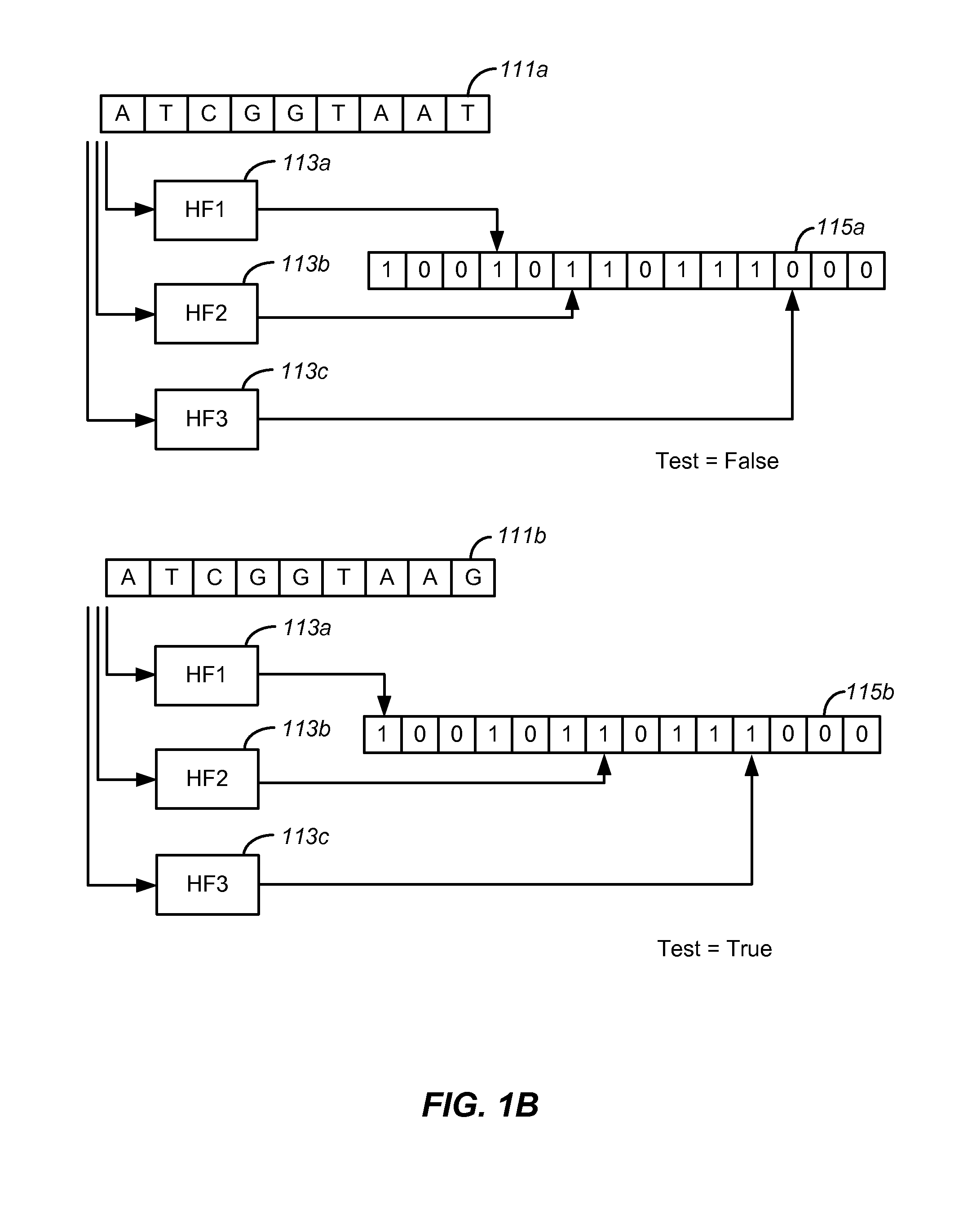
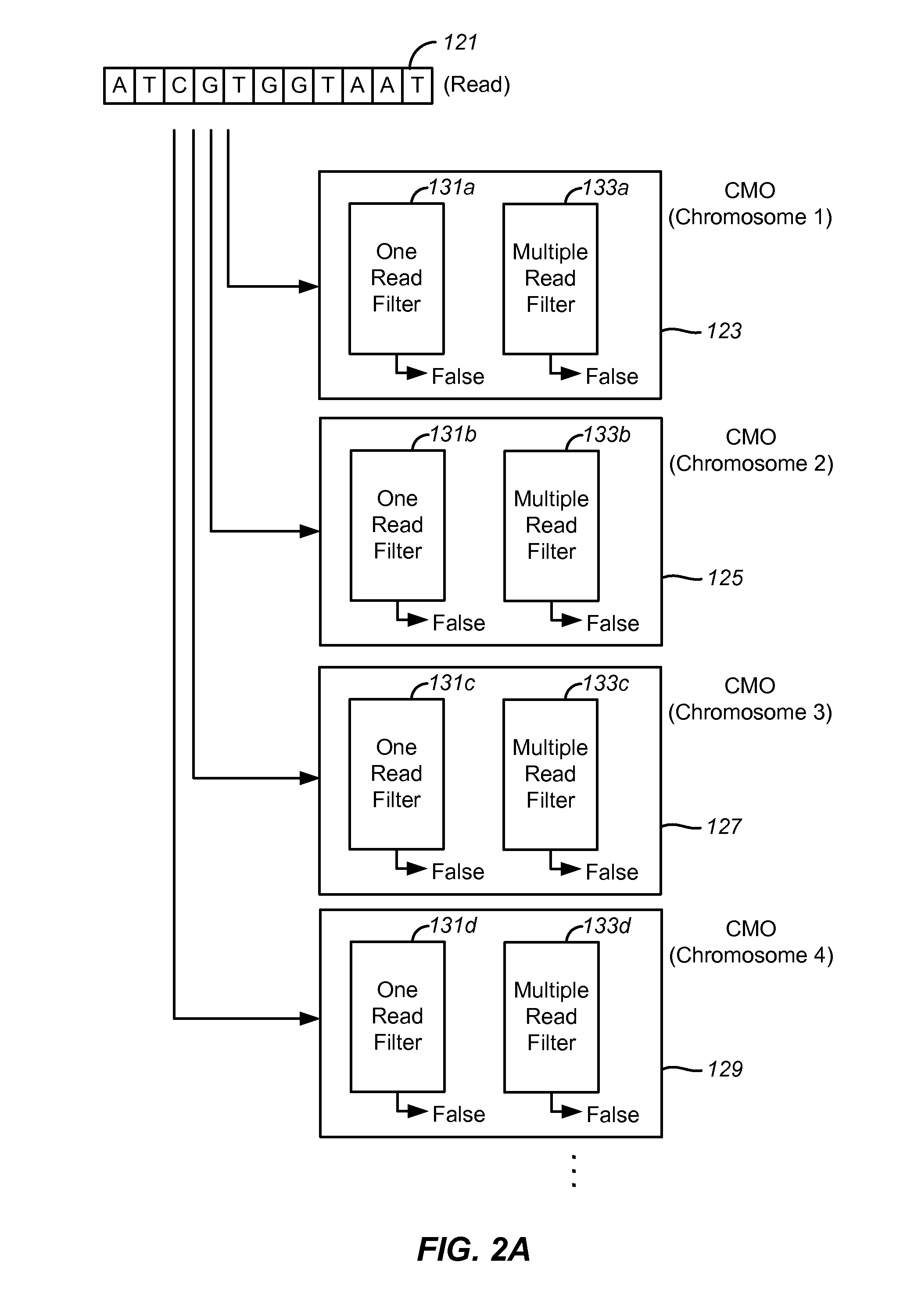
Set membership testers for aligning nucleic acid samples
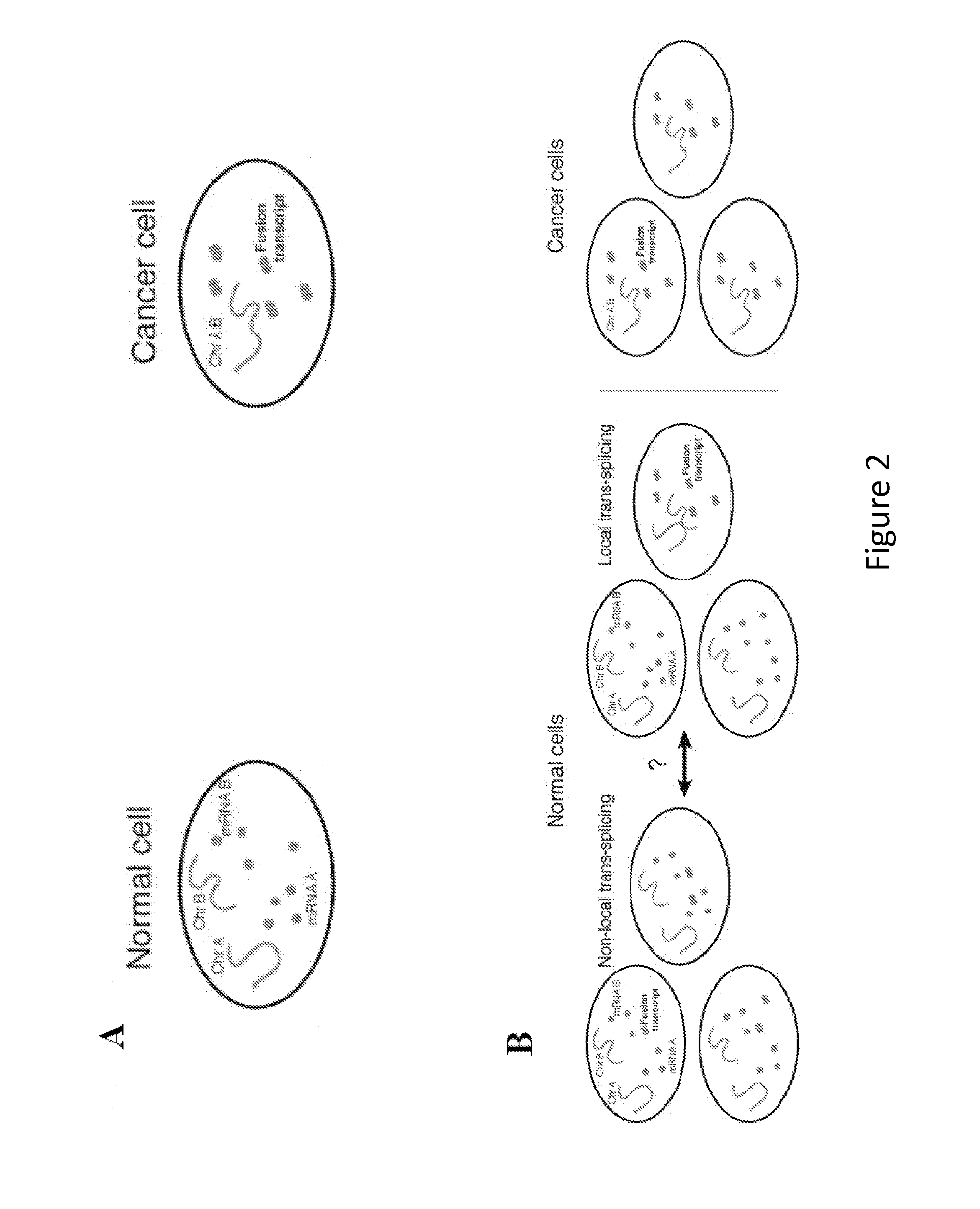
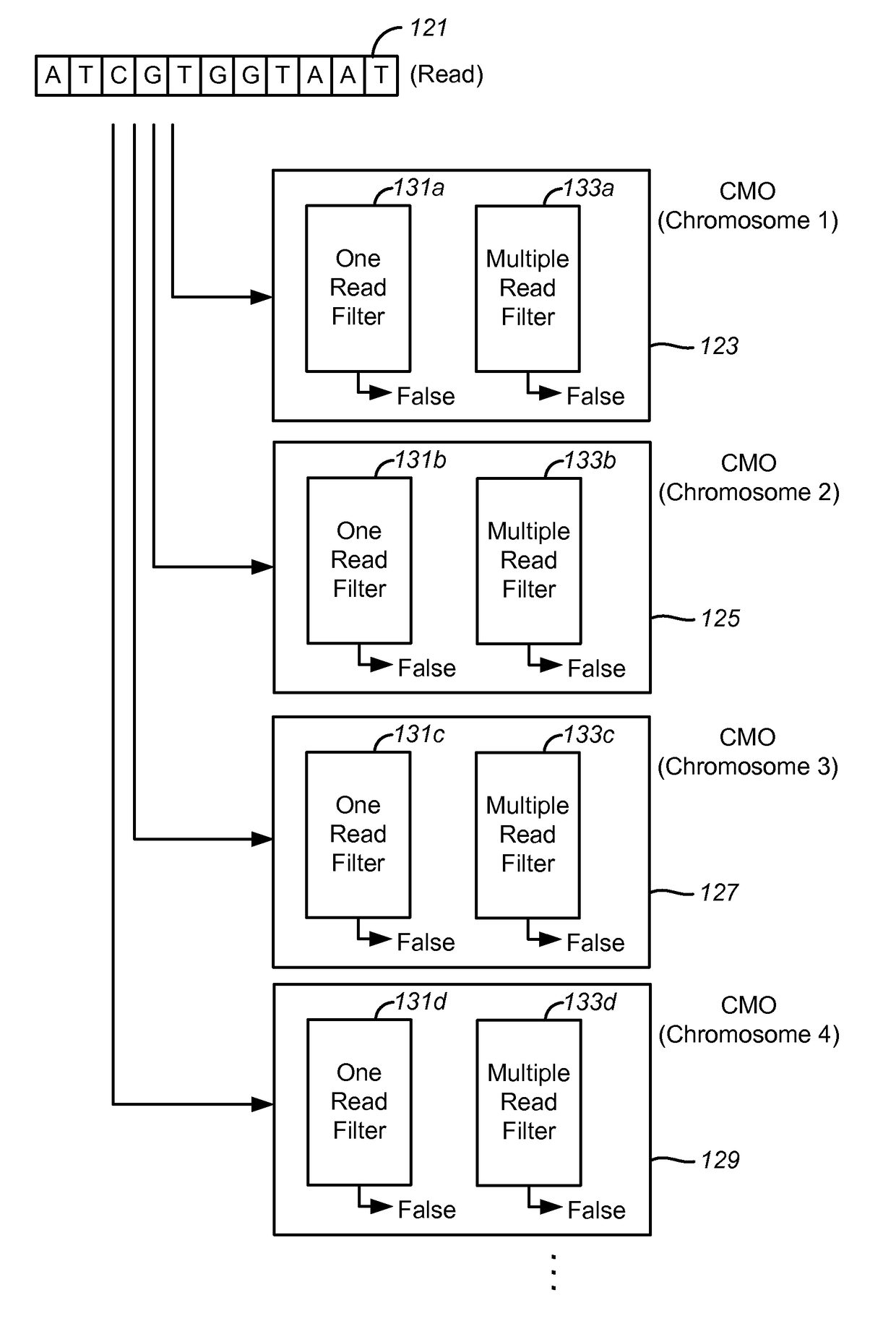
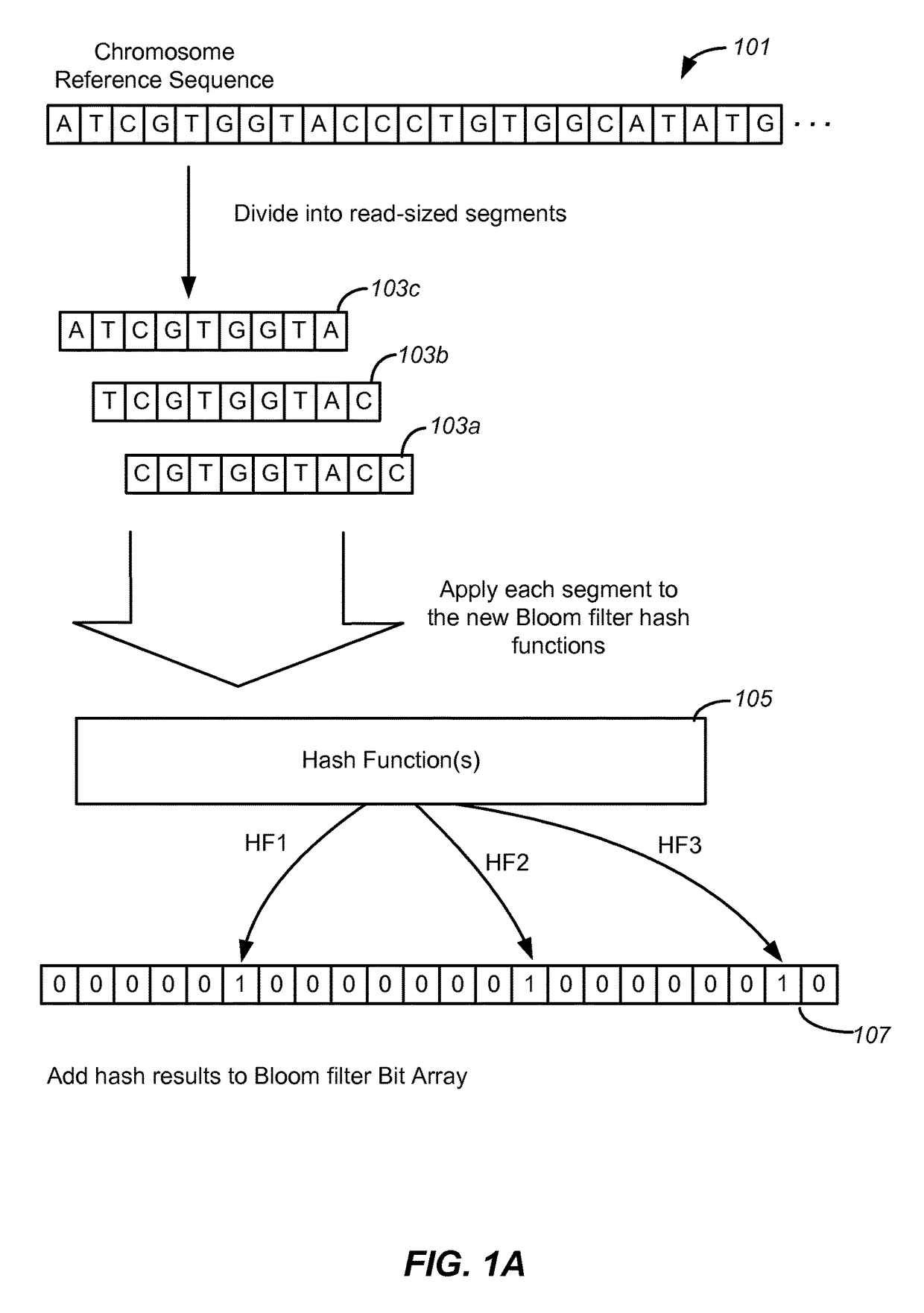
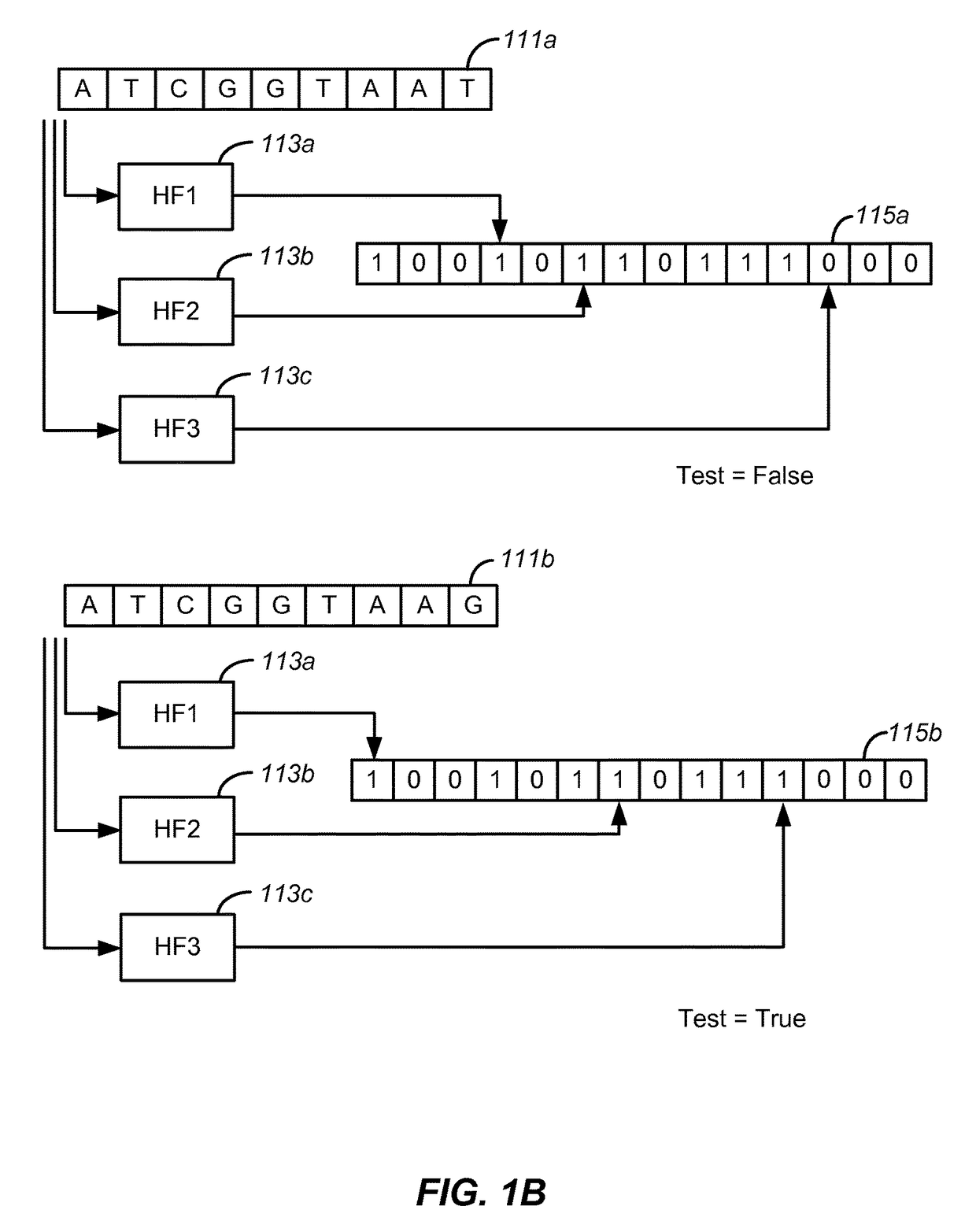
Disclosed are methods and tools for rapidly aligning reads to a reference sequence. These methods and tools employ Bloom filters or similar set membership testers to perform the alignment. The reads may be short sequences of nucleic acids or other biological molecules and the reference sequences may be sequences of genomes, chromosomes, etc. The Bloom filters include a collection of hash functions, a bit array, and associated logic for applying reads to the filter. Each filter, and there may be multiple of these used in a particular application, is used to determine whether an applied read is present in a reference sequence. Each Bloom filter is associated with a single reference sequence such as the sequence of a particular chromosome. In one example, chromosomal abundance is determined by aligning reads from a sequencer to multiple chromosomes, each having an associated Bloom filter or other set membership tester.
Owner:VERINATA HEALTH INC
Method for generating cloned animals using chromosome shuffling
InactiveUS20020174449A1Population uniformImprove quality controlNew breed animal cellsFermentationSpecific chromosomeMammal
The present invention concerns the use of chromosomal replacement techniques in the context of producing cloned and transgenic animals, in order to correct chromosome abnormalities or alter autosomal genotypes, and provide for novel breeding pairs by replacing the sex chromosome in animals to be cloned. Replacement of a sex chromosome, or an X or Y chromosome, will result in animals that are autosomally isogenic and sexually non-isogenic (AISN), with "autosomally isogenic" meaning that the paired sets of autosomes (non-sex chromosomes) in each animal are isogenic or identical. Also included in the invention are animals that are both "autosomally" and "allelically" isogenic whereby each particular pair of chromosomes is internally isogenic or identical within a single animal as well as between animals. Such animals are particularly useful in generating a line of cloned mammals using sexual reproduction, without having to undergo nuclear transfer in order to propagate cloned animals.
Owner:ADVANCED CELL TECH INC
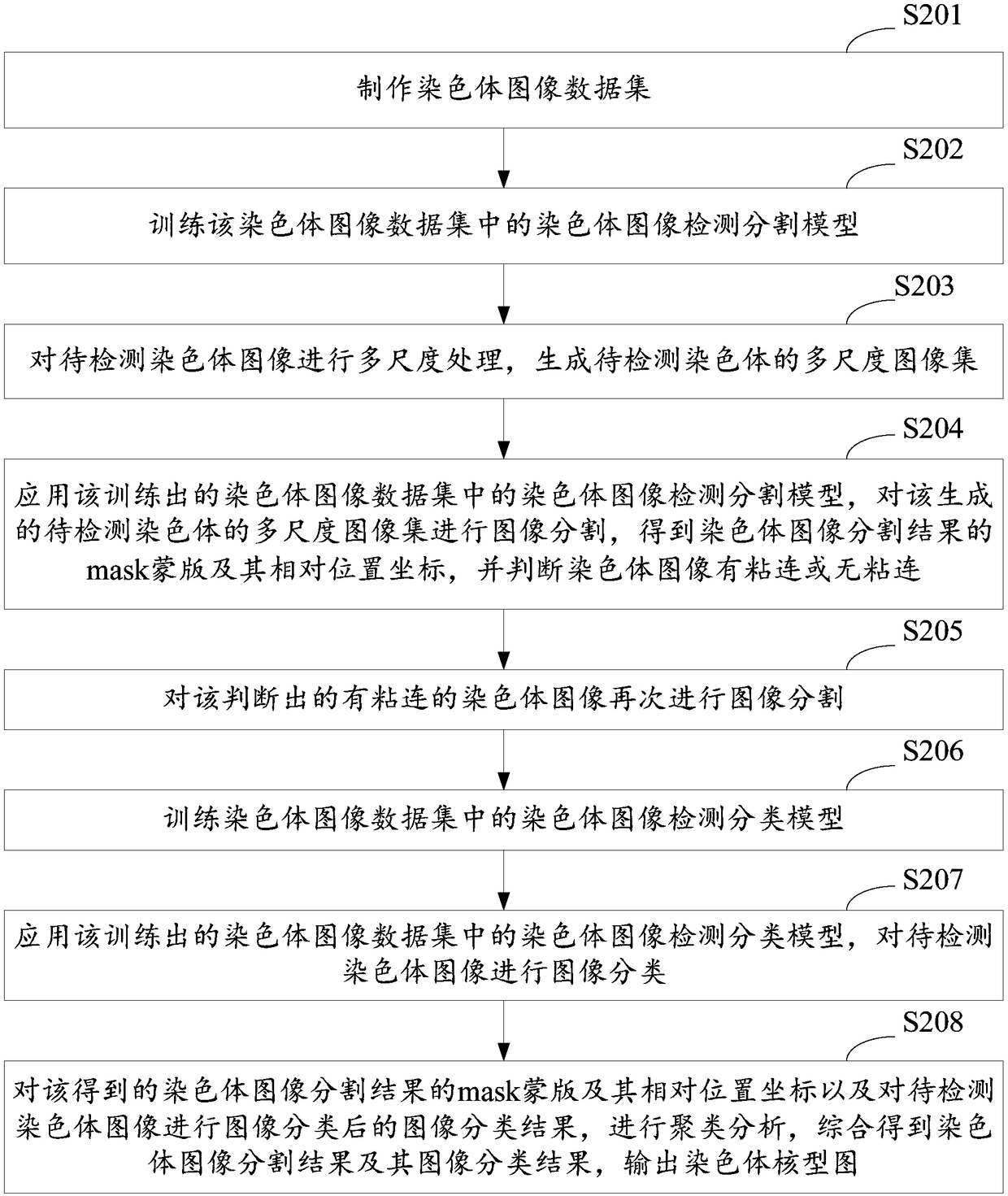
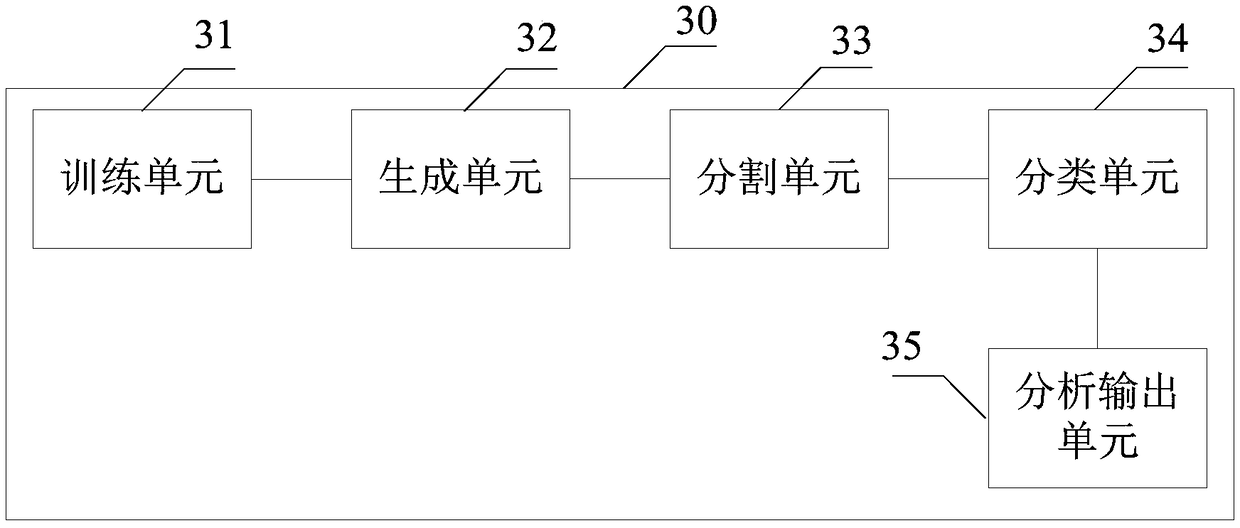
A method and system for automatic chromosome analysis based on depth learning
ActiveCN109344874ATaking into account the overall shapeTaking into account featuresImage enhancementImage analysisPattern recognitionChromosome localisation
The invention discloses a chromosome automatic analysis method and a chromosome automatic analysis system based on depth learning, which can adopt multi-level processing, stratify the independent formand overlapping form of chromosomes, and perform cluster analysis on chromosome position coordinates, classification labels and classification confidence to output a karyotype map. In this manner, the method of chromosome segmentation based on depth learning can be adopted, independent of specific chromosomal morphological patterns, The method has high generalization ability, can adopt chromosomeclassification method based on depth learning, take into account the global morphology and banding characteristics of chromosomes, improve classification accuracy, can adopt multi-scale processing, more fully utilize the detected images, and effectively improve the segmentation effect in the case of chromosome overlap and adhesion.
Owner:HUAQIAO UNIVERSITY
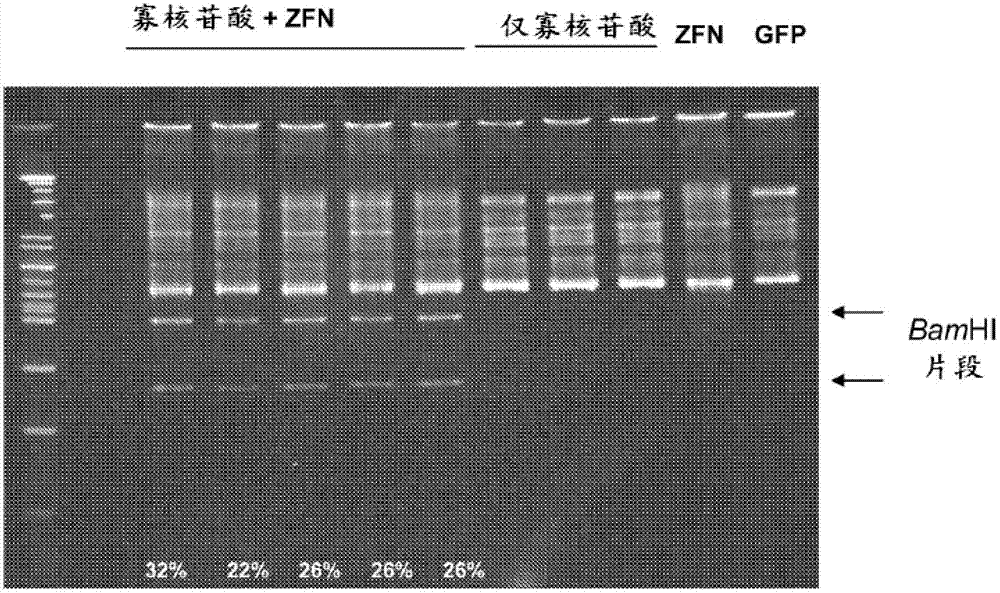
Feline genome editing with zinc finger nucleases
The present invention provides a genetically modified feline or cell comprising at least one edited chromosomal sequence. In particular, the chromosomal sequence is edited using a zinc finger nuclease-mediated editing process. The disclosure also provides zinc finger nucleases that target specific chromosomal sequences in the feline genome.
Owner:SIGMA ALDRICH CO LLC
Silkworm genome editing with zinc finger nucleases
The present invention provides a genetically modified silkworm or cell comprising at least one edited chromosomal sequence. In particular, the chromosomal sequence is edited using a zinc finger nuclease-mediated editing process. The disclosure also provides zinc finger nucleases that target specific chromosomal sequences in the silkworm genome.
Owner:SIGMA ALDRICH CO LLC
Canine genome editing with zinc finger nucleases
The present invention provides a genetically modified canine or cell comprising at least one edited chromosomal sequence. In particular, the chromosomal sequence is edited using a zinc finger nuclease-mediated editing process. The disclosure also provides zinc finger nucleases that target specific chromosomal sequences in the canine genome.
Owner:SIGMA ALDRICH CO LLC
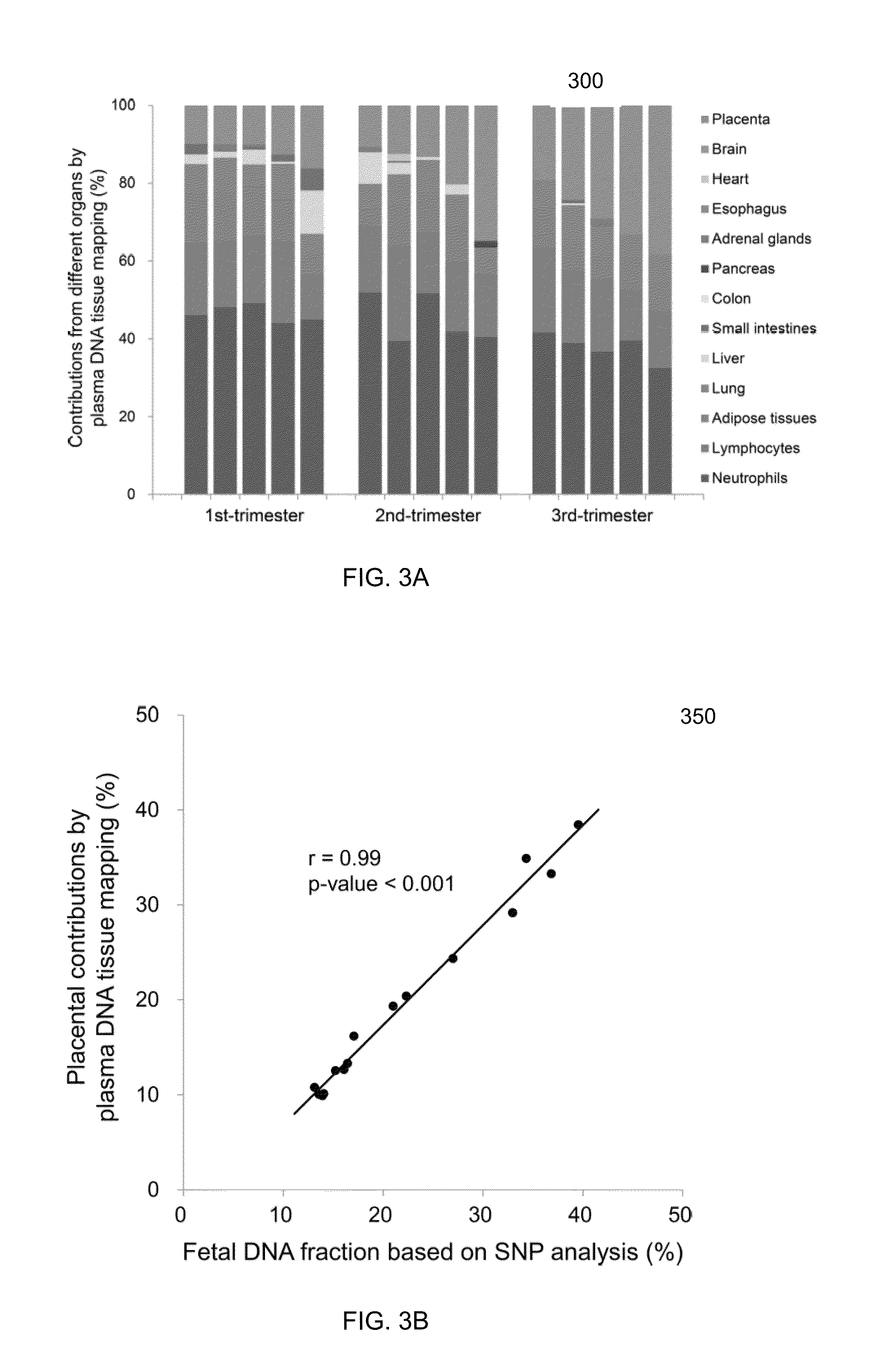
Molecular testing of multiple pregnancies
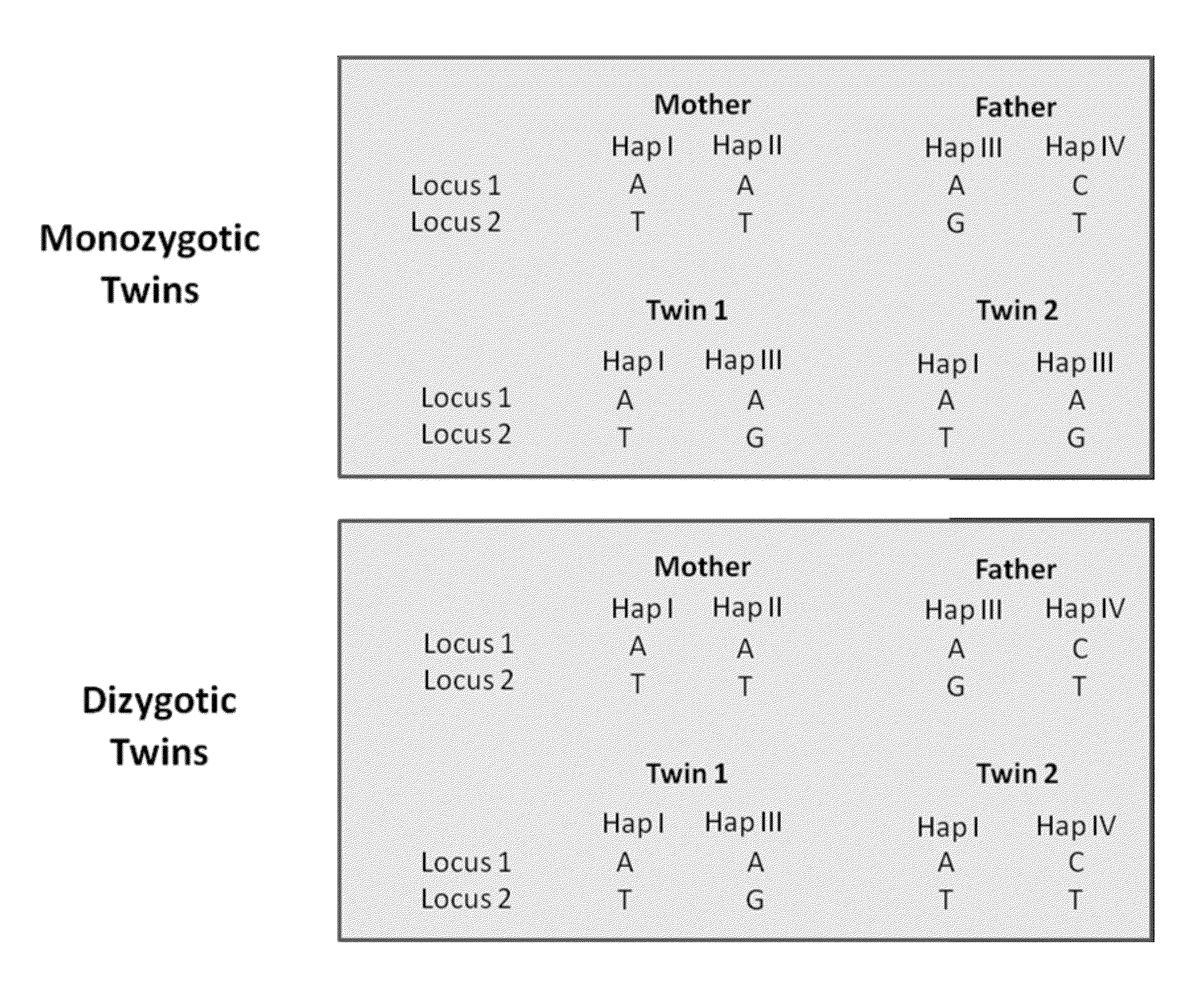
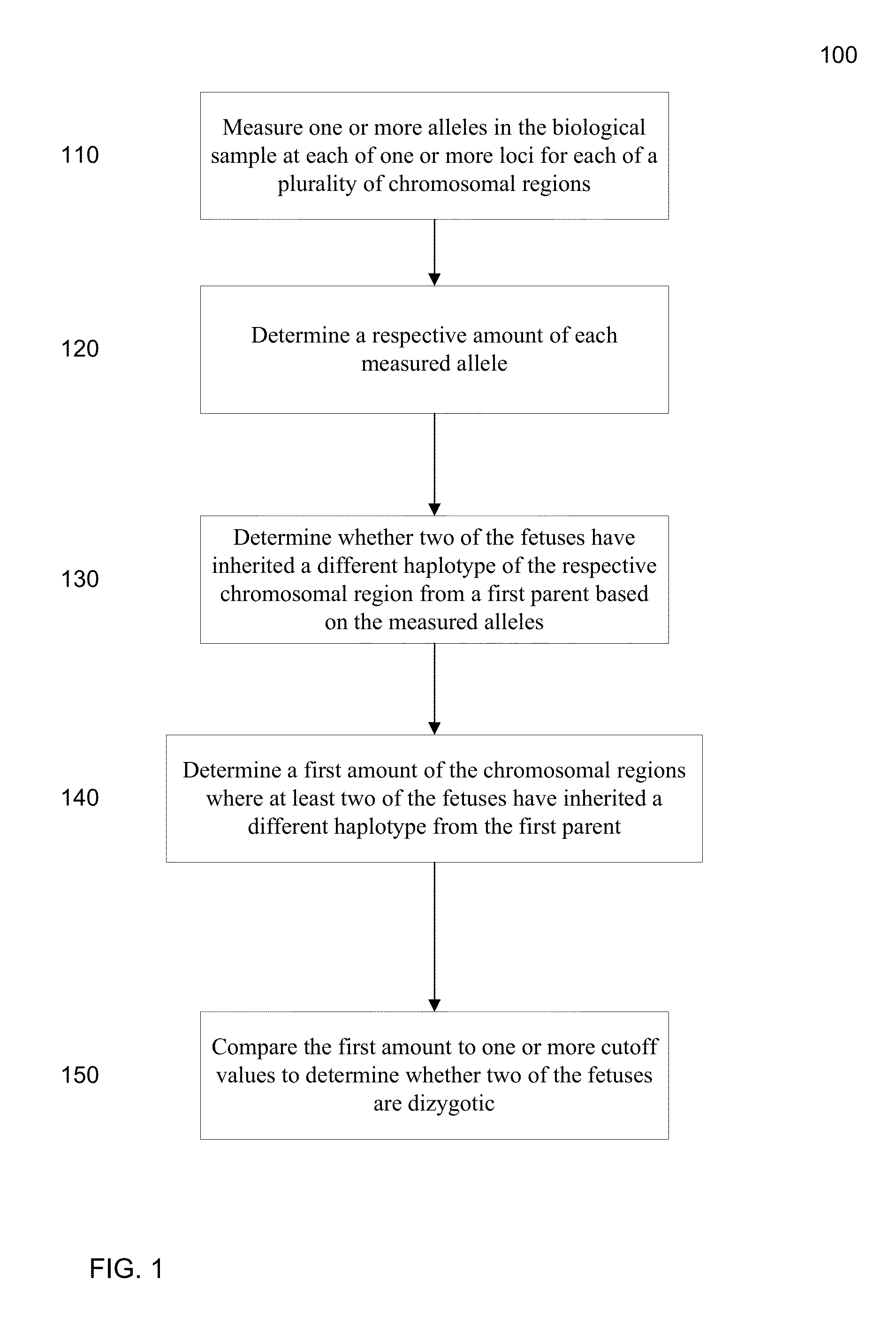
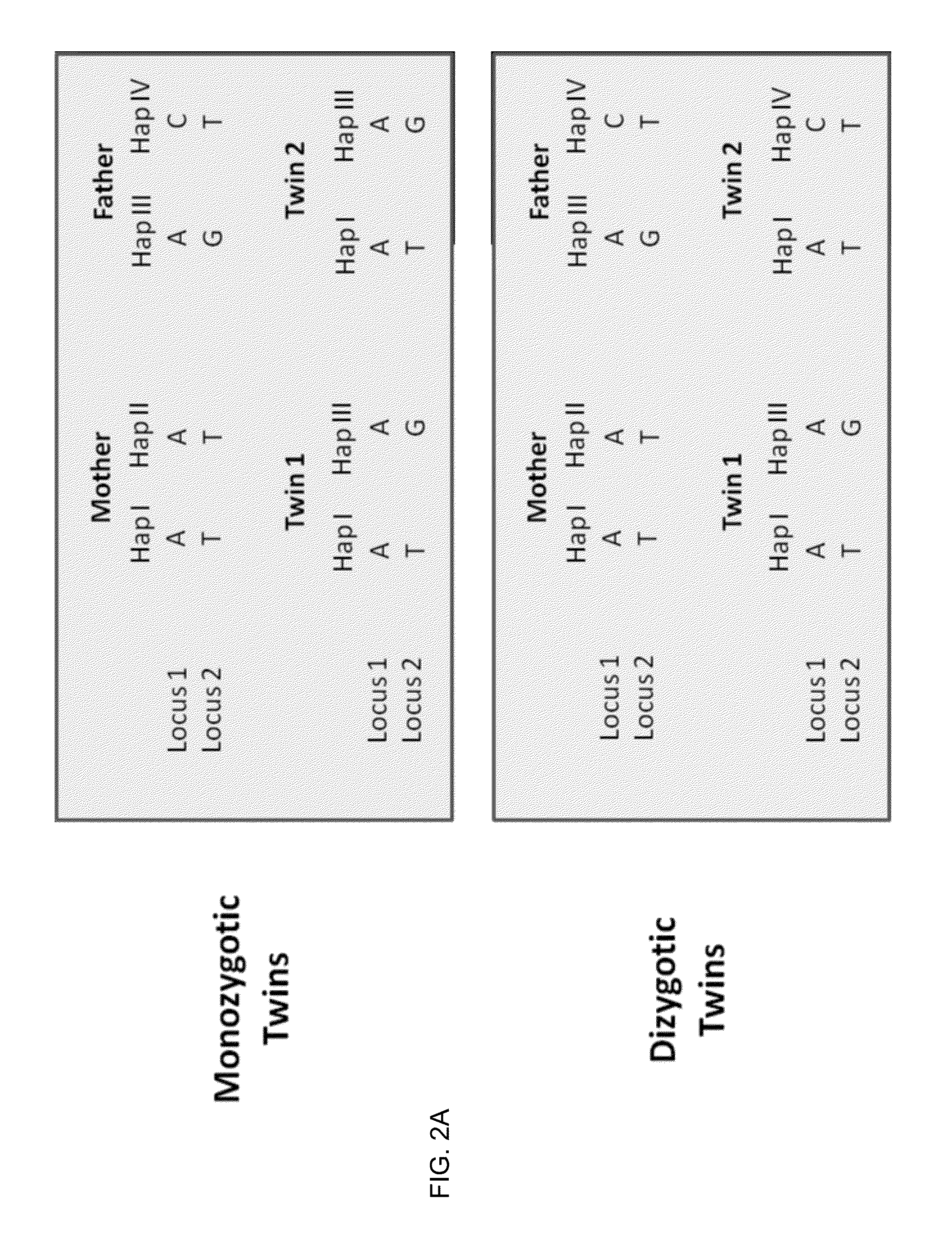
ActiveUS20130059733A1Non invasiveMicrobiological testing/measurementLibrary screeningSpecific chromosomeGenetic divergence
Methods, systems, and apparatus are provided for determining zygosity of a multiple-fetus pregnancy using a biological sample taken from the mother. The fetal and maternal DNA in the sample (e.g. plasma) can be analyzed for a particular chromosomal region to identify genetic differences in the fetuses. For example, a normalized parameter for the measure of a primary or secondary allele can show variances for different chromosomal regions when fetuses are dizygotic. Such a variance can be determined relative to an expected value if the fetuses were genetically identical. Statistical methods are provided for analyzing the variation of the normalized parameters to determine fetal DNA concentration and the maternal-fetal mixed genotype at various loci. Parental genotype and haplotype information can also be used to identify inheritance of different parental haplotypes to indicate genetic differences among the fetuses.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
Genome editing method using attachment carrier for encoding targeted endonuclease and kit
InactiveCN104450785AMicrobiological testing/measurementVector-based foreign material introductionEucaryotic cellSpecific chromosome
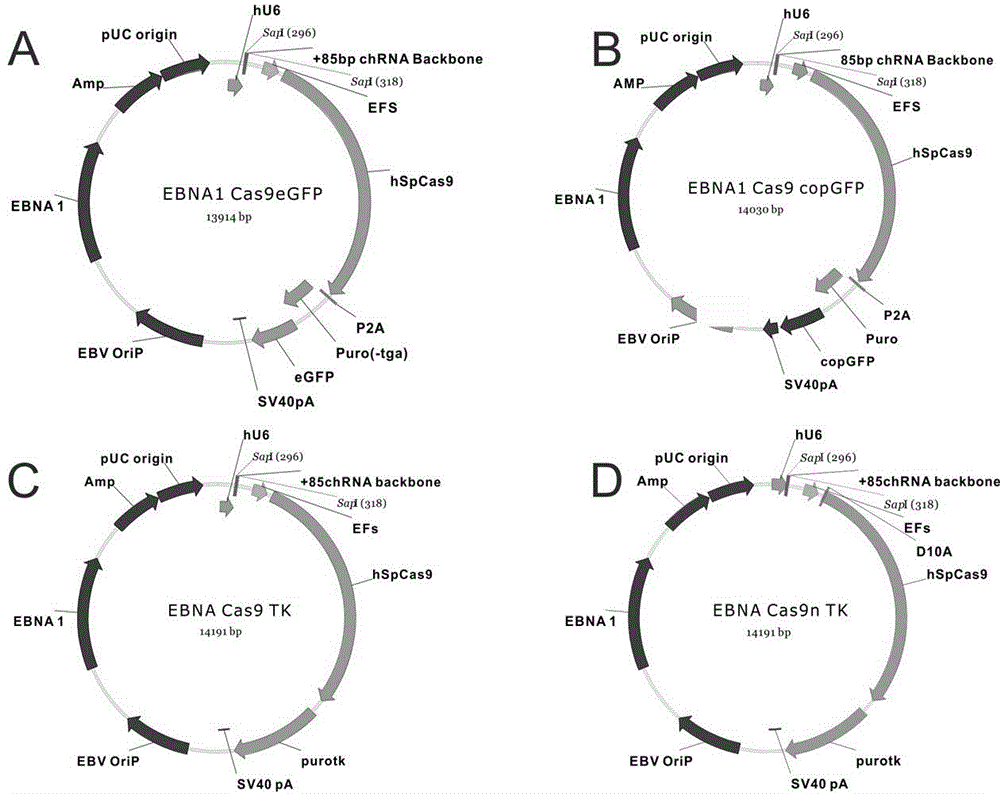
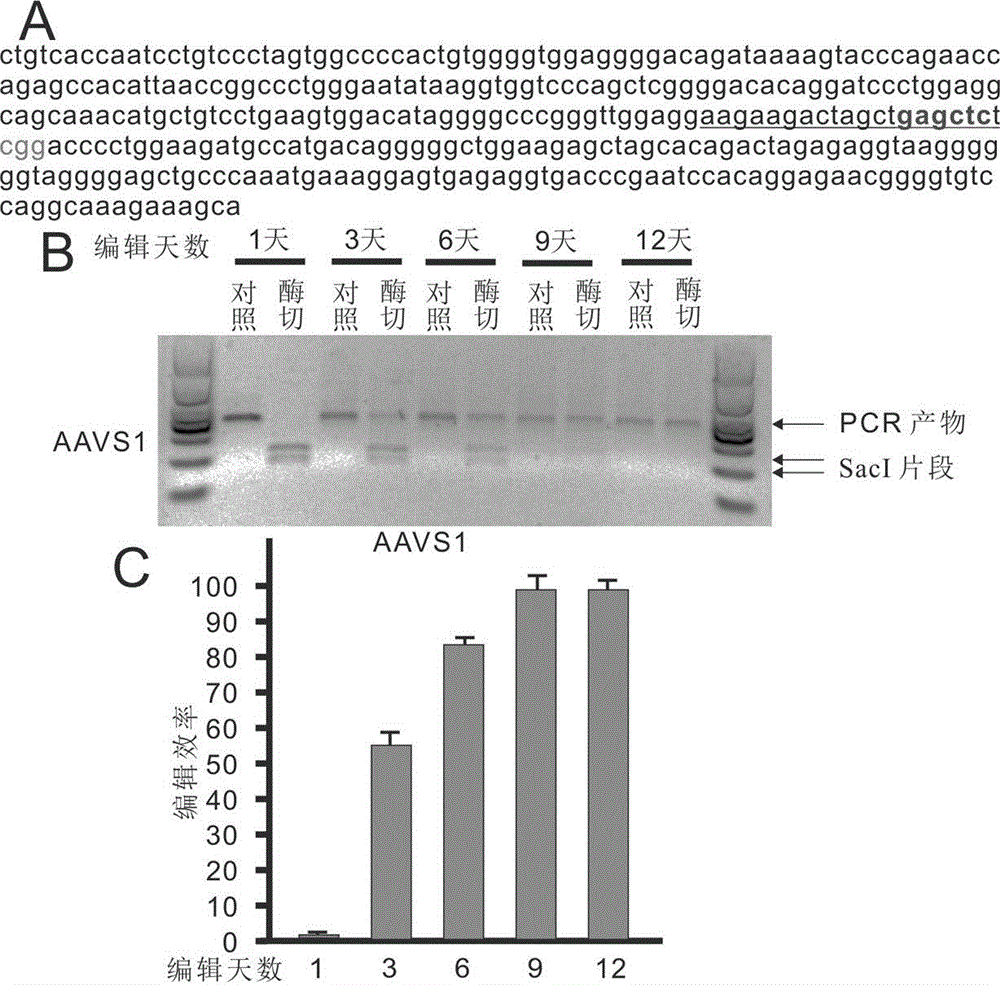
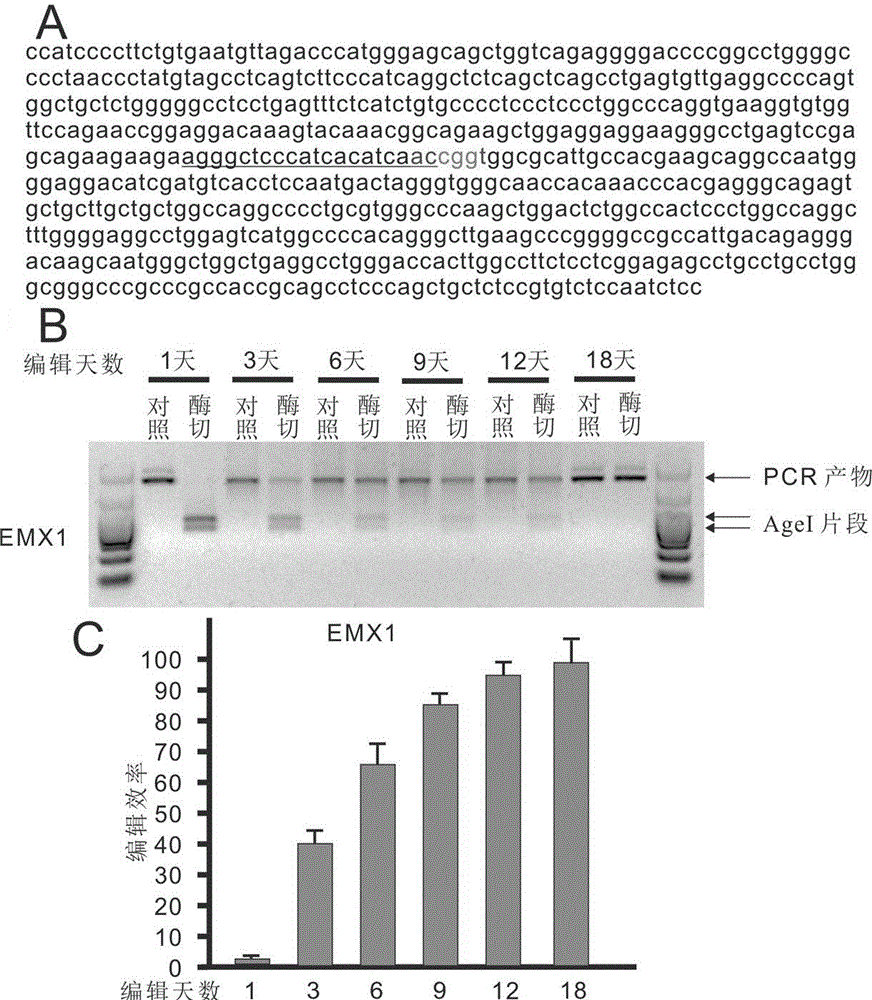
The invention belongs to the technical field of bio-genetic engineering and in particular relates to a genome editing method using an attachment carrier for encoding targeted endonuclease and a kit. According to the genome editing method, the attachment carrier is utilized to express the targeted endonuclease, and the attachment carrier can realize nonconformable and stable gene expression and simultaneously expresses a screening gene; cells without getting plasmids can be removed by virtue of drug screening; and when edition is finished, a drug is removed, the cells can gradually lose the attachment carriers, and therefore, an editing cell line without containing a foreign gene is obtained. With the adoption of the genome editing method, the editing time is prolonged, and the gene editing efficiency is improved. The invention also provides the kit for editing a specific chromosome sequence of the edited cells. The genome editing method and the kit are suitable for genome edition of various eucaryotic cells.
Owner:FUDAN UNIV
Genome editing using targeting endonucleases and single-stranded nucleic acids
Owner:SIGMA ALDRICH CO LLC
Oligonucleotide probe and acquisition method thereof
ActiveCN106566876ASimple designLow costMicrobiological testing/measurementHybridisationRepetitive SequencesIn situ hybridisation
The invention relates to a method for acquisition of an oligonucleotide probe. The method comprises the following steps: downloading genomic sequence of crop species from a public database, filtering and finding out tandem repeat sequences, screening preliminarily found repetitive sequences, comparing residual tandem repeat sequence and known probe sequence, removing already used probe sequence, comparing residual screened tandem repeat sequences, carrying out probe design and synthesizing and verifying the designed probe sequence so as to obtain effective oligonucleotide sequence. According to the invention, cost is low and efficiency is high. The probe can be used for non-degenerated fluorescence in situ hybridization (ND-FISH) analysis of crop chromosome, determination of distribution of tandem repeat sequences represented by the probe on chromosome, understanding of the structure characteristics of chromosome and establishment of specific landmark of chromosome. Thus, specific chromosome or chromosome region of crops is identified, and the developed new oligonucleotide probe has specificity of chromosome or chromosome region.
Owner:SICHUAN AGRI UNIV
Genome editing using targeting endonucleases and single-stranded nucleic acids
Owner:SIGMA ALDRICH CO LLC
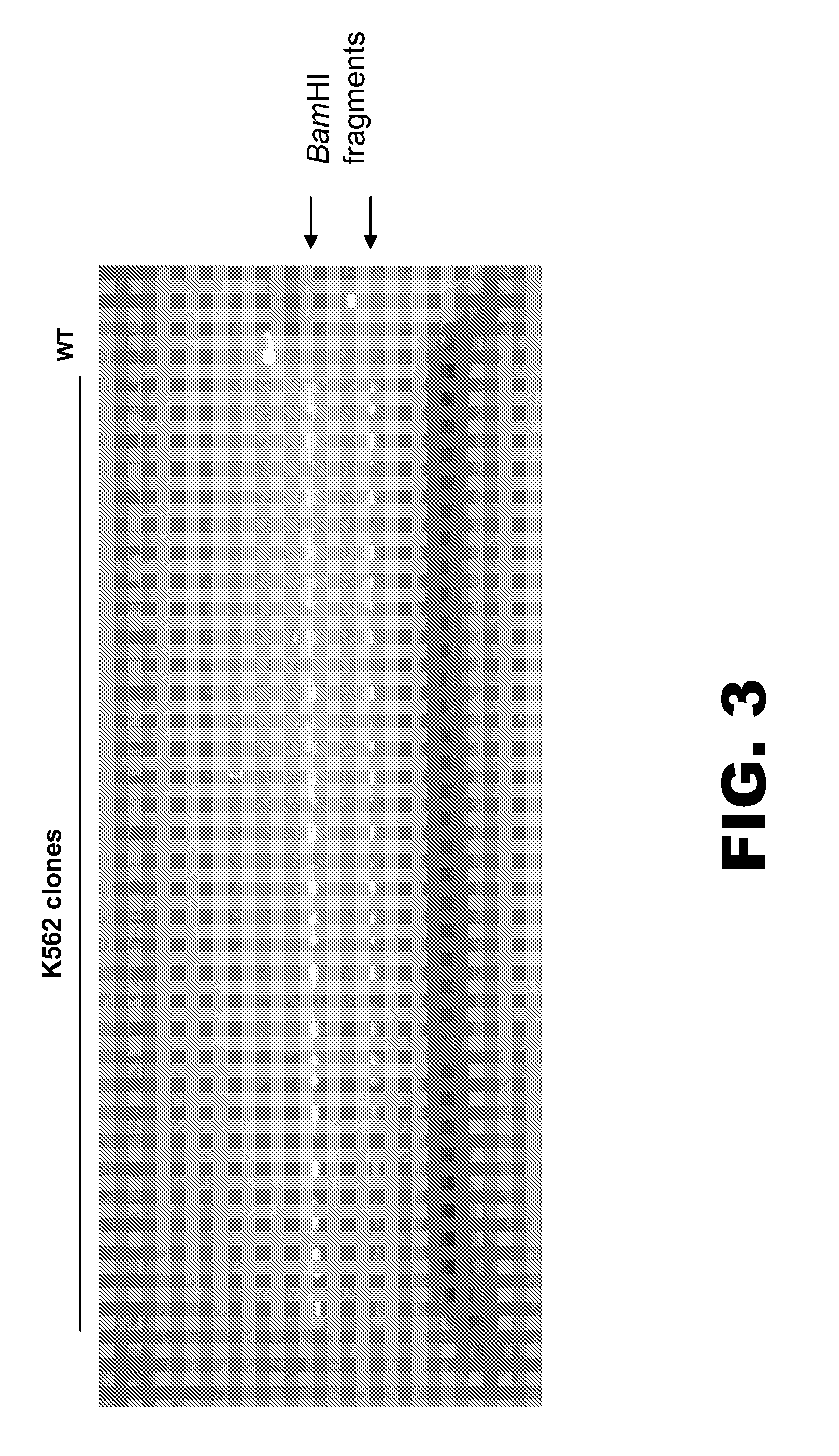
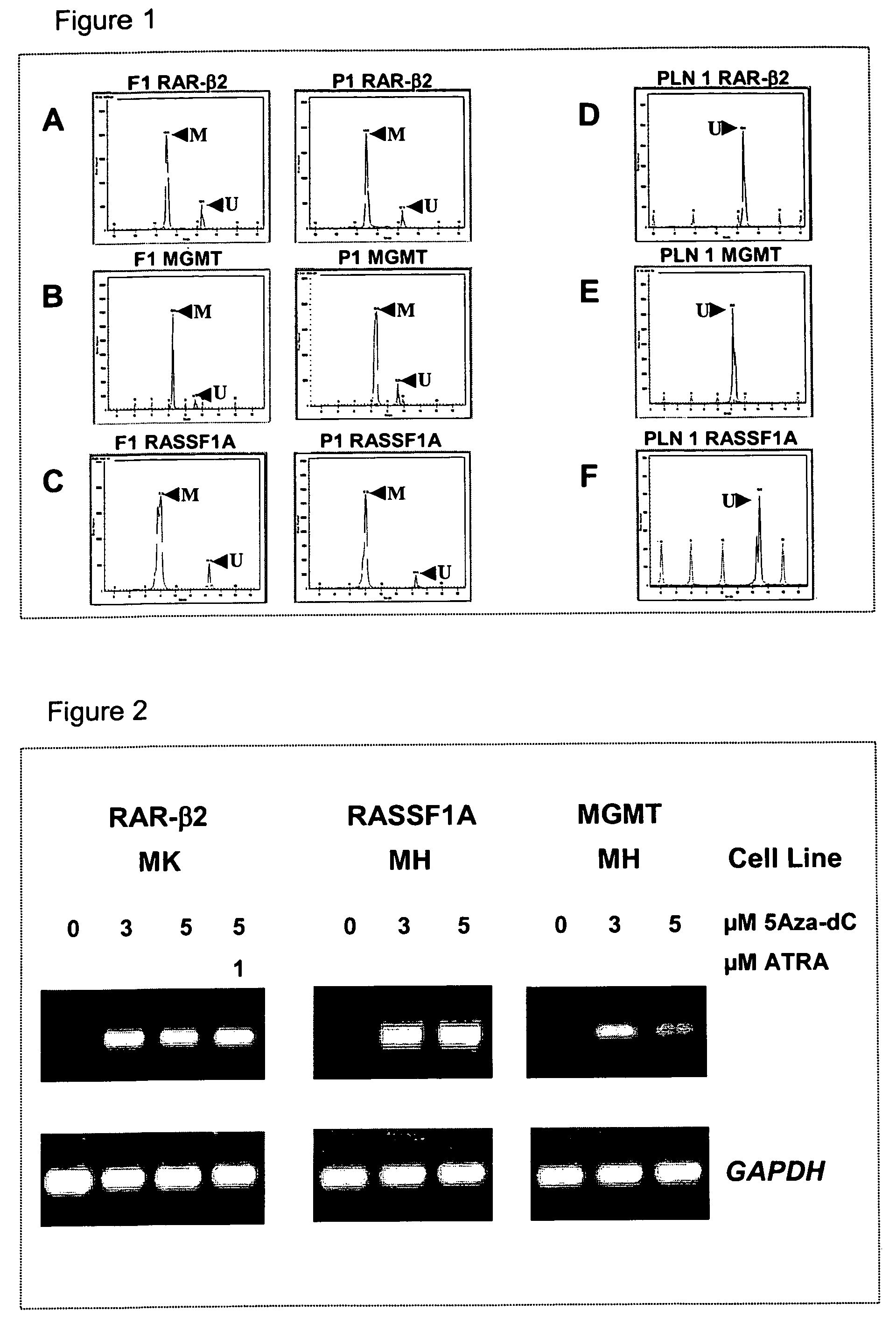
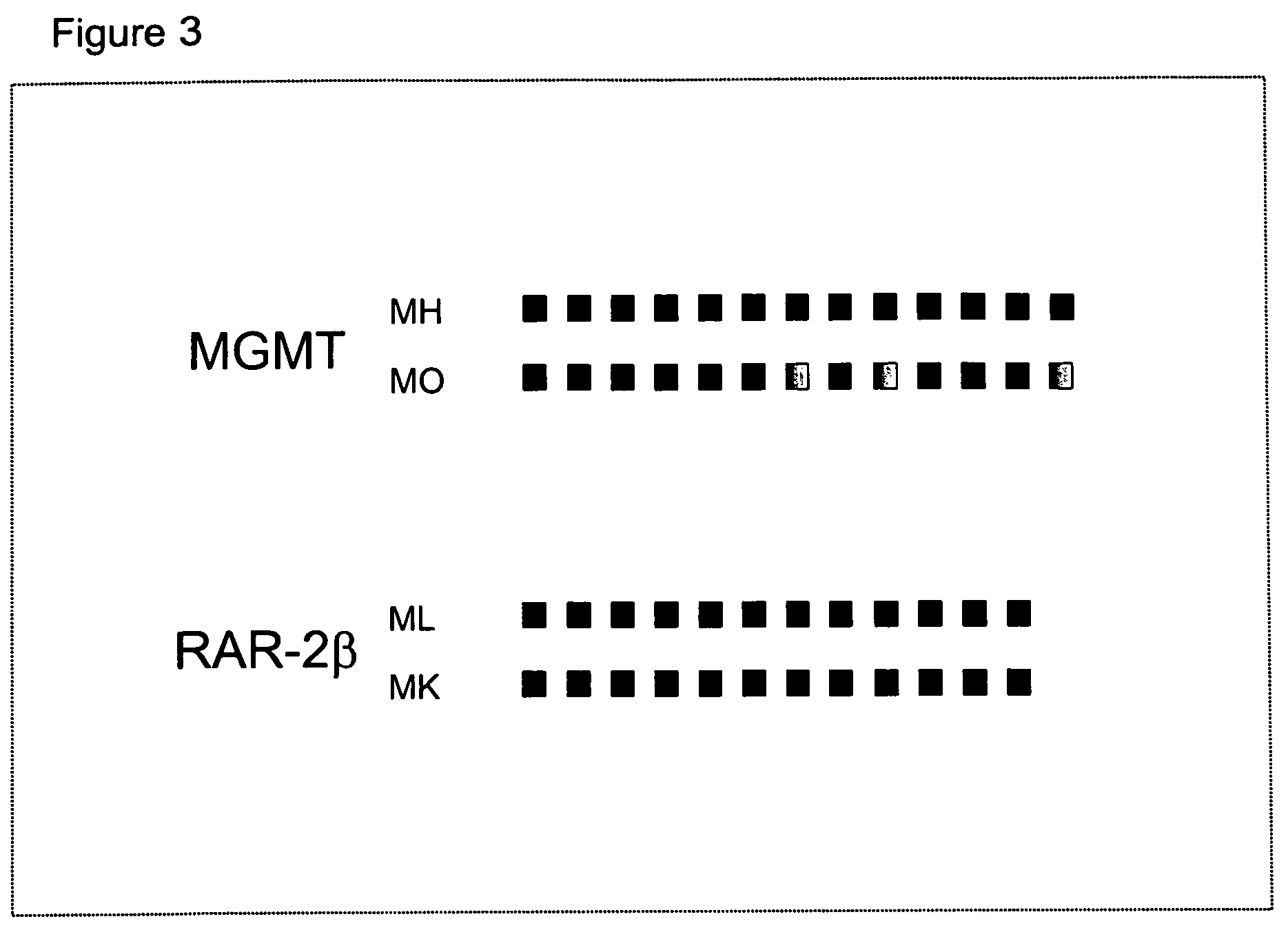
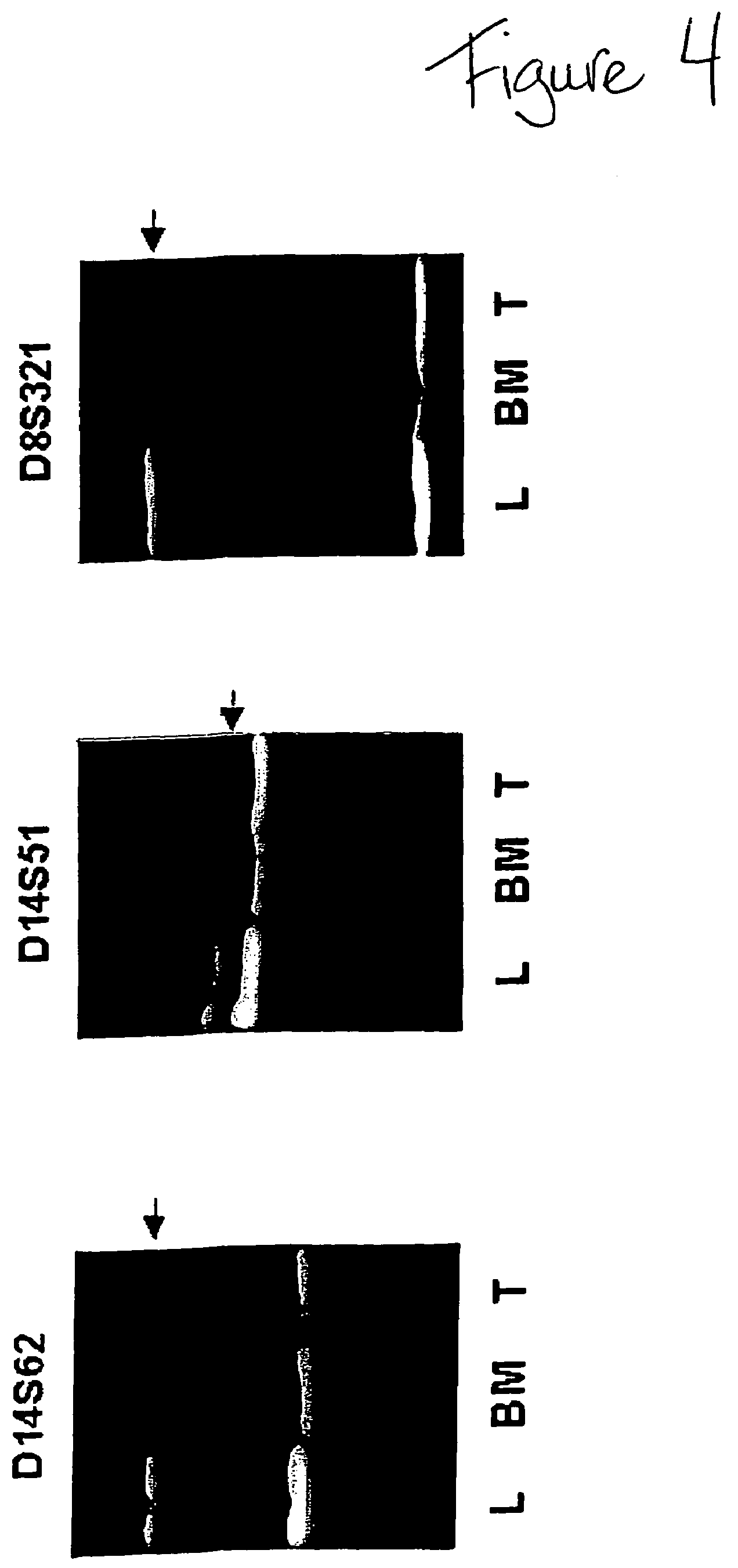
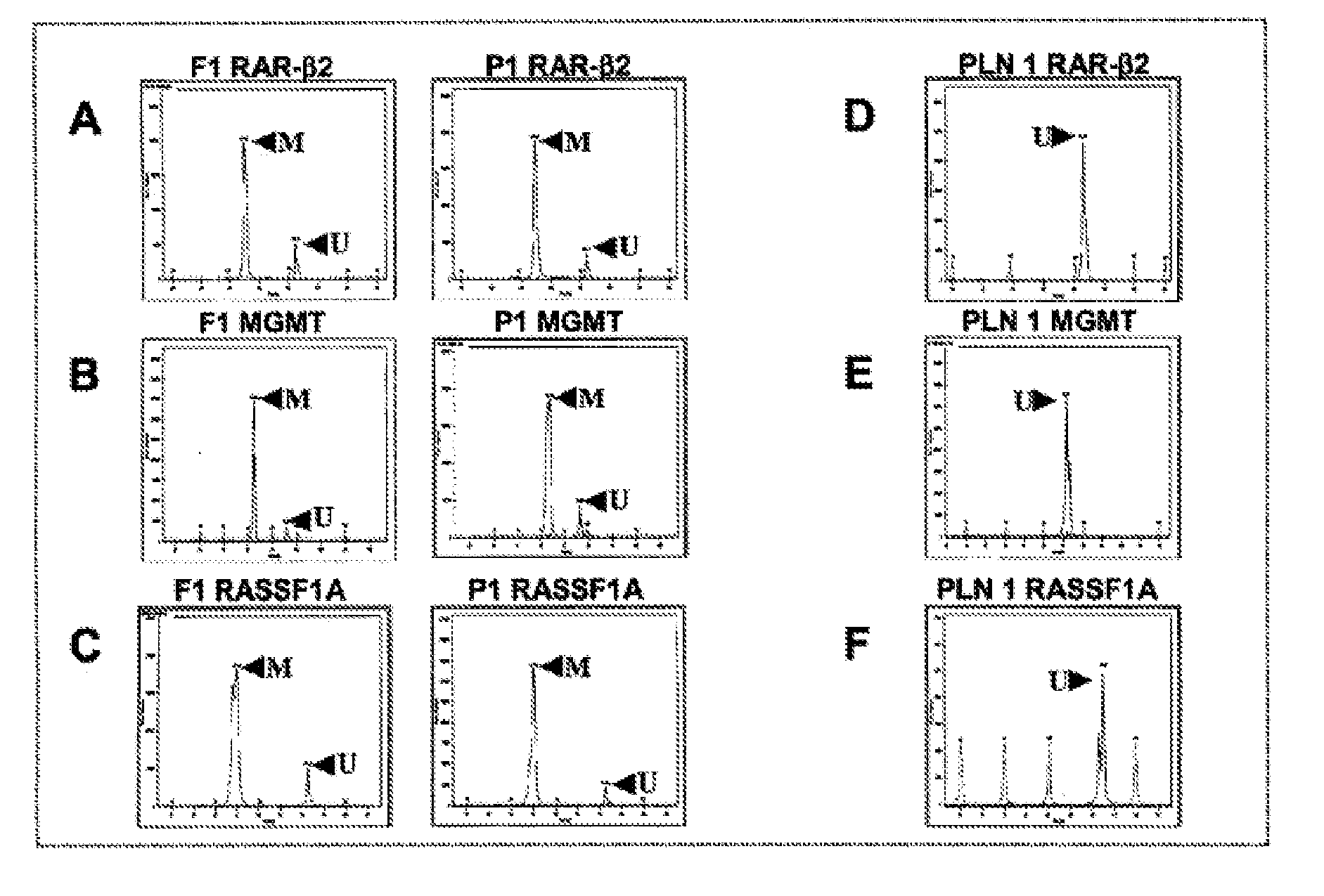
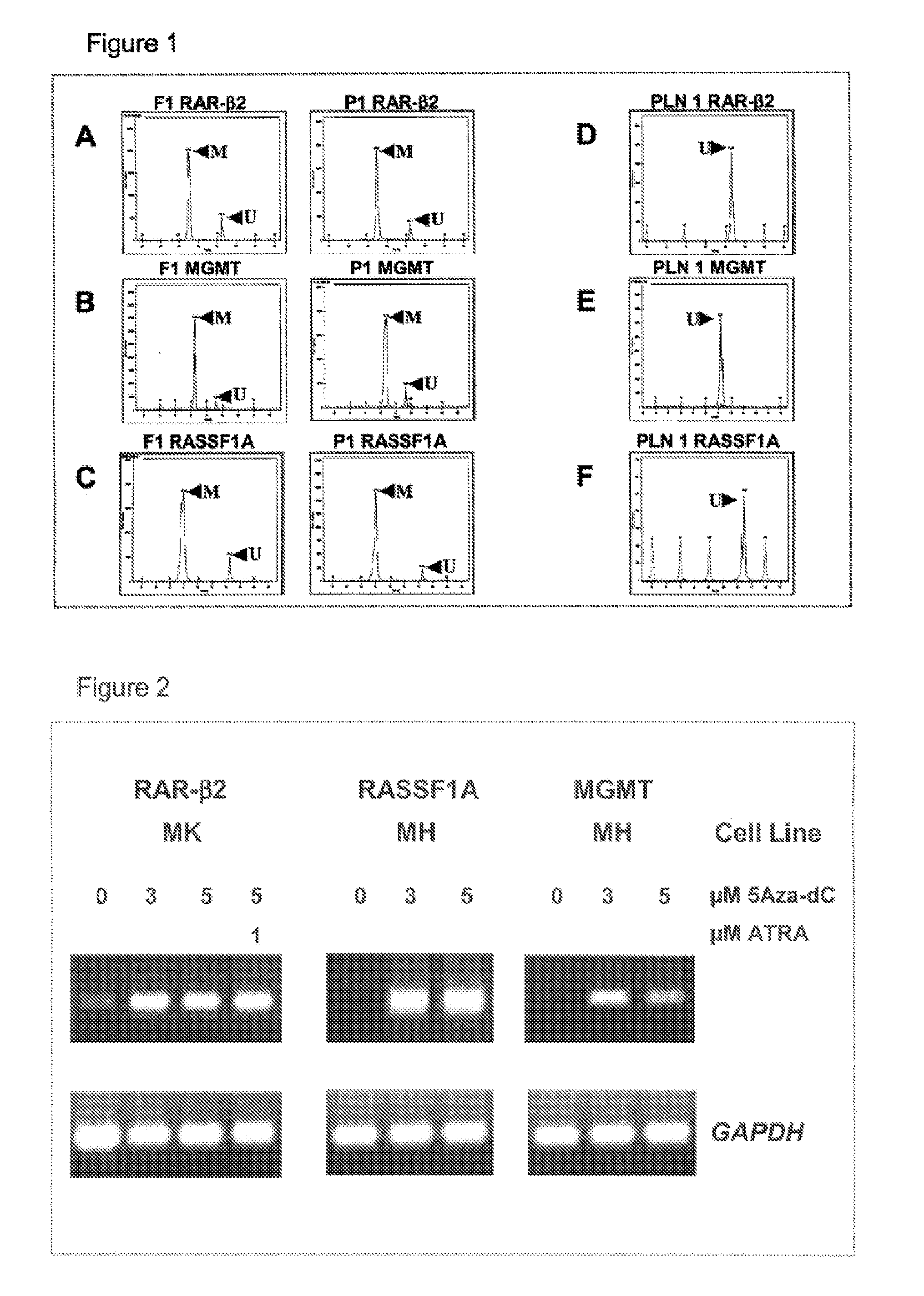
DNA markers for management of cancer
ActiveUS7718364B2Stage increaseMinimally invasiveSugar derivativesMicrobiological testing/measurementBlastomaSpecific chromosome
A method is provided for assessing allelic losses and hypermethylation of genes in CpG tumor promotor region on specific chromosomal regions in cancer patients, including melanoma, neuroblastoma breast, colorectal, and prostate cancer patients. The method relies on the evidence that free DNA and hypermethylation of genes in CpG tumor promotor region may be identified in the bone marrow, serum, plasma, and tumor tissue samples of cancer patients. Methods of melanoma, neuroblastoma, colorectal cancer, breast cancer and prostate cancer detection, staging, and prognosis are also provided.
Owner:JOHN WAYNE CANCER INST
Genome editing using targeting endonucleases and single-stranded nucleic acids
Owner:SIGMA ALDRICH CO LLC
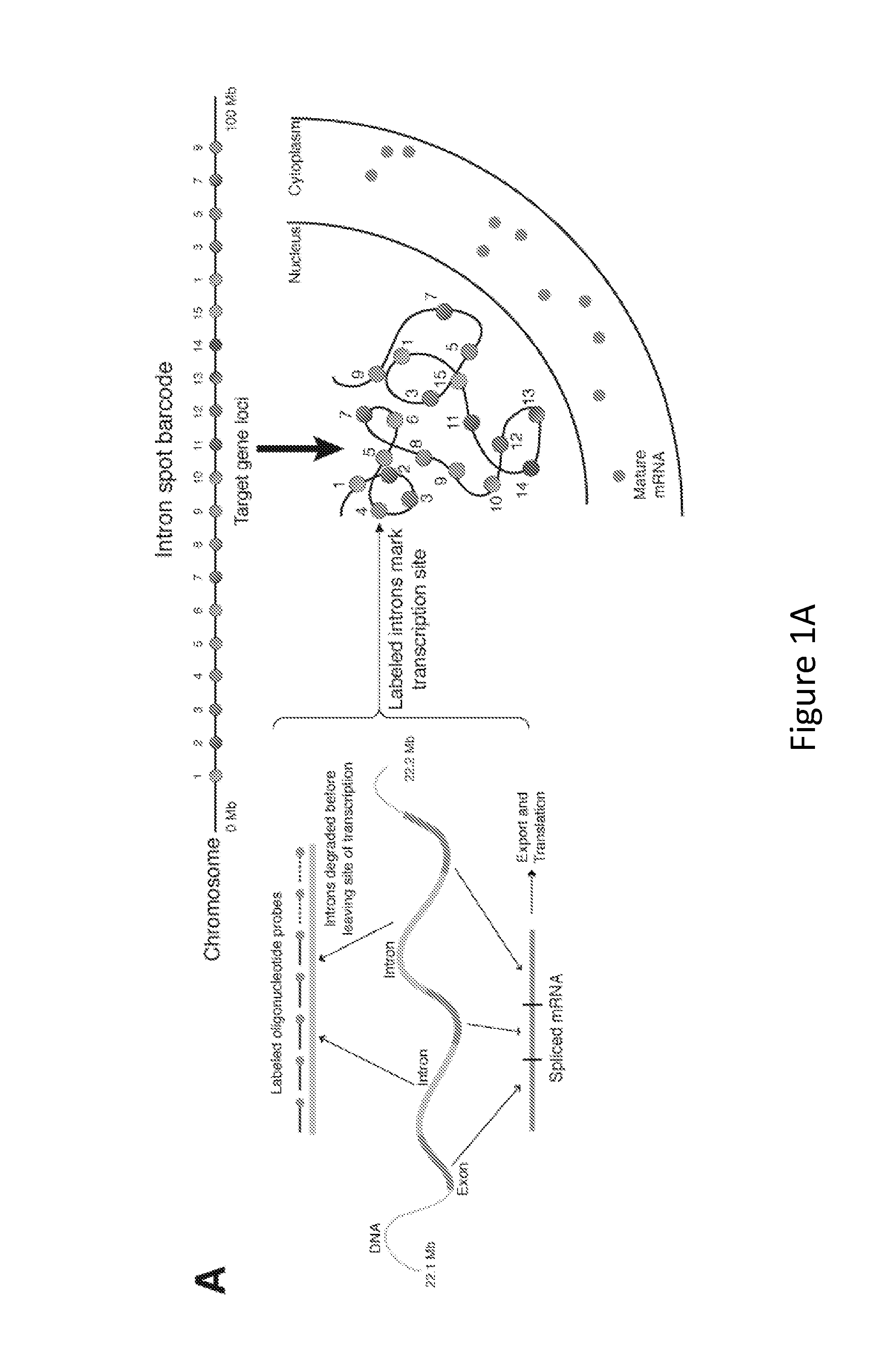
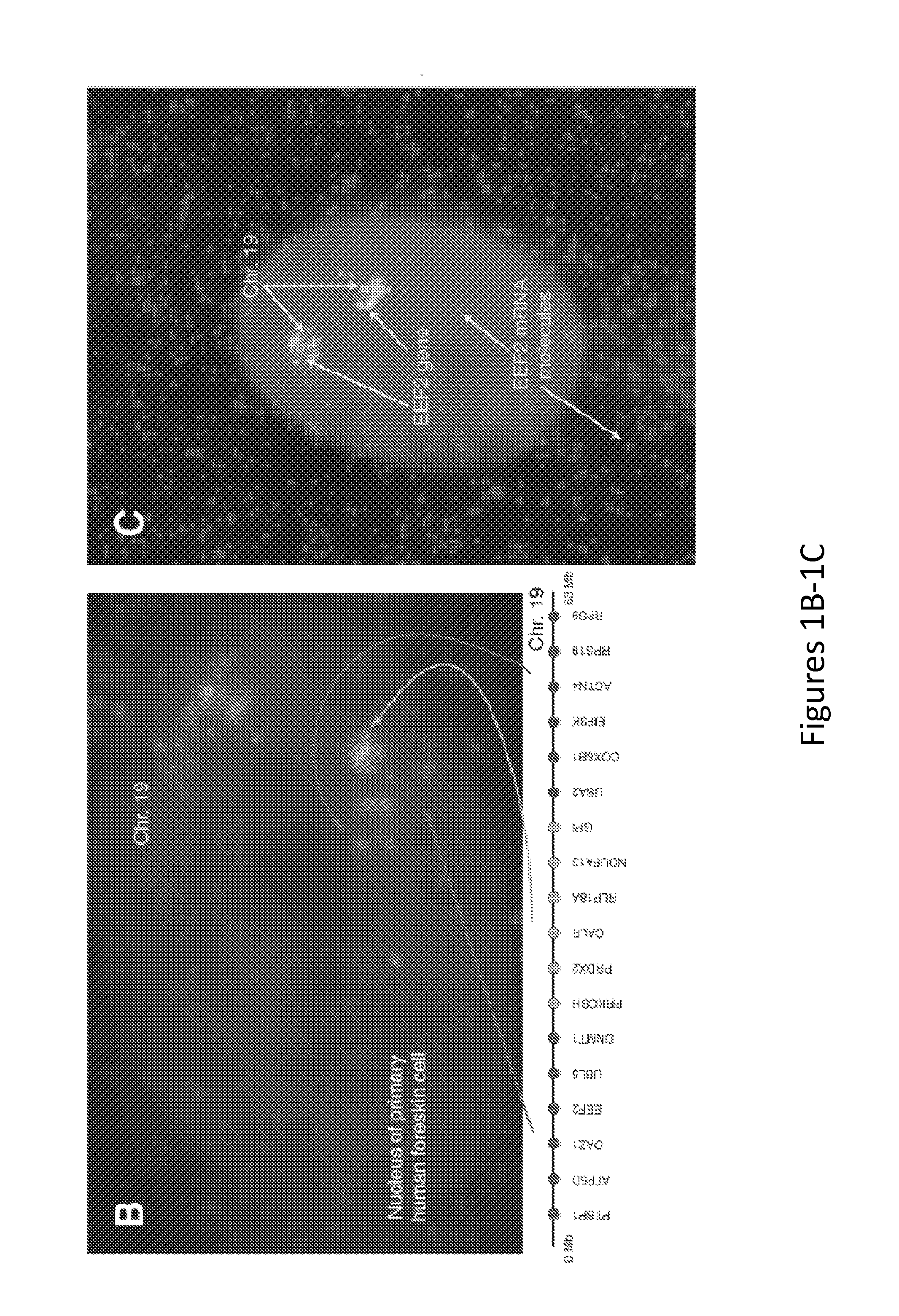
Method for Detecting Chromosome Structure and Gene Expression Simultaneously in Single Cells
The present invention relates to systems and methods for measuring chromosome structure and gene expression. In one embodiment, the present invention includes an assay for determining the spatial organization of gene expression in the nucleus. The present invention also includes a method based on fluorescence in situ hybridization (FISH) that simultaneously yields information on the physical position and expression of individual genes. By lighting up a large number of targets on a particular chromosome using a bar-coding scheme, the large scale structure of an entire chromosome can be determined.
Owner:THE TRUSTEES OF THE UNIV OF PENNSYLVANIA
Set membership testers for aligning nucleic acid samples
Disclosed are methods and tools for rapidly aligning reads to a reference sequence. These methods and tools employ Bloom filters or similar set membership testers to perform the alignment. The reads may be short sequences of nucleic acids or other biological molecules and the reference sequences may be sequences of genomes, chromosomes, etc. The Bloom filters include a collection of hash functions, a bit array, and associated logic for applying reads to the filter. Each filter, and there may be multiple of these used in a particular application, is used to determine whether an applied read is present in a reference sequence. Each Bloom filter is associated with a single reference sequence such as the sequence of a particular chromosome. In one example, chromosomal abundance is determined by aligning reads from a sequencer to multiple chromosomes, each having an associated Bloom filter or other set membership tester.
Owner:VERINATA HEALTH INC
Method for generating cloned animals using chromosome shuffling
InactiveUS20040139489A1Population uniformImprove quality controlGenetic material ingredientsGenetic engineeringSpecific chromosomeMammal
The present invention concerns the use of chromosomal replacement techniques in the context of producing cloned and transgenic animals, in order to correct chromosome abnormalities or alter autosomal genotypes, and provide for novel breeding pairs by replacing the sex chromosome in animals to be cloned. Replacement of a sex chromosome, or an X or Y chromosome, will result in animals that are autosomally isogenic and sexually non-isogenic (AISN), with "autosomally isogenic" meaning that the paired sets of autosomes (non-sex chromosomes) in each animal are isogenic or identical. Also included in the invention are animals that are both "autosomally" and "allelically" isogenic whereby each particular pair of chromosomes is internally isogenic or identical within a single animal as well as between animals. Such animals are particularly useful in generating a line of cloned mammals using sexual reproduction, without having to undergo nuclear transfer in order to propagate cloned animals.
Owner:ADVANCED CELL TECH INC
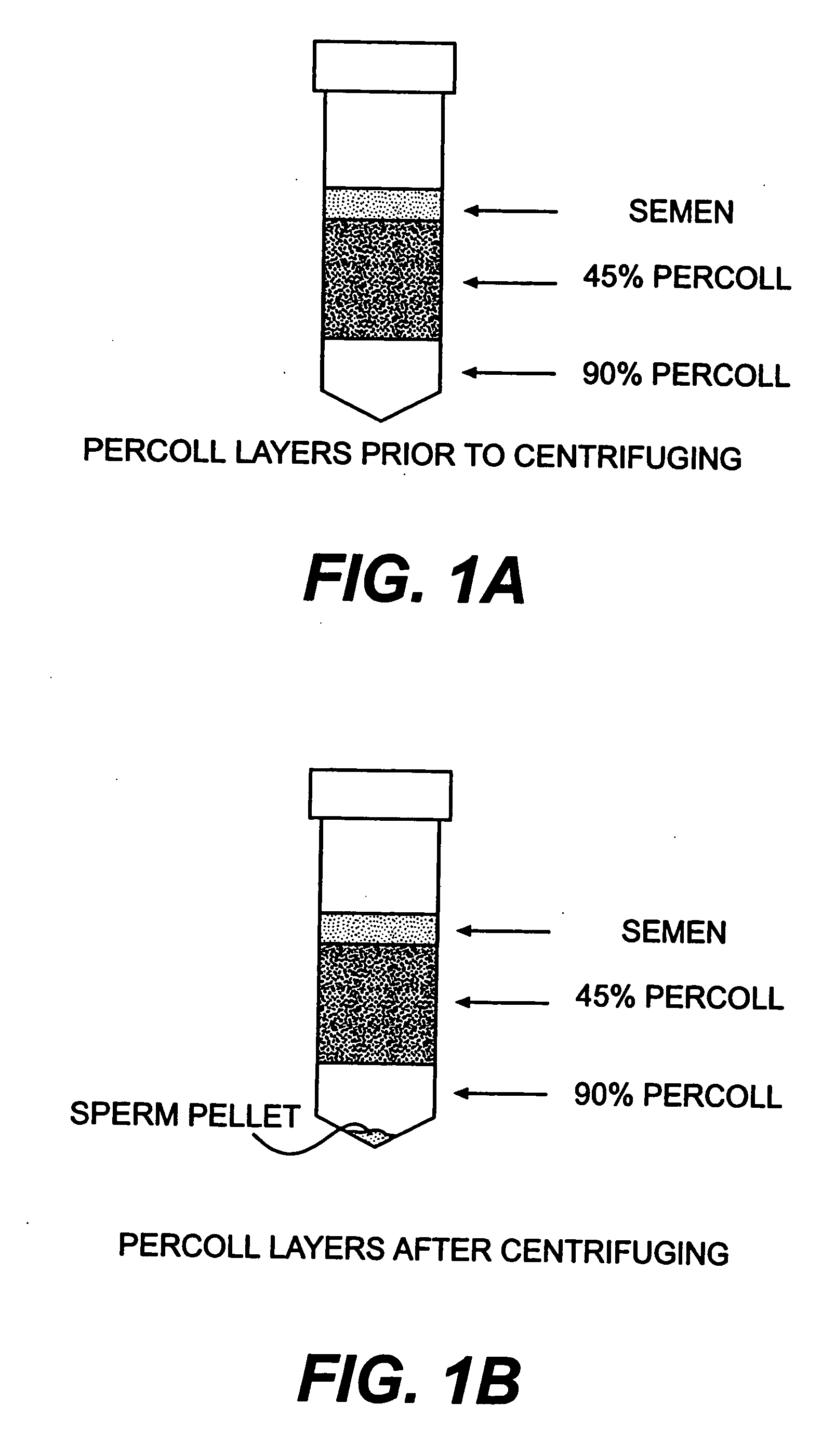
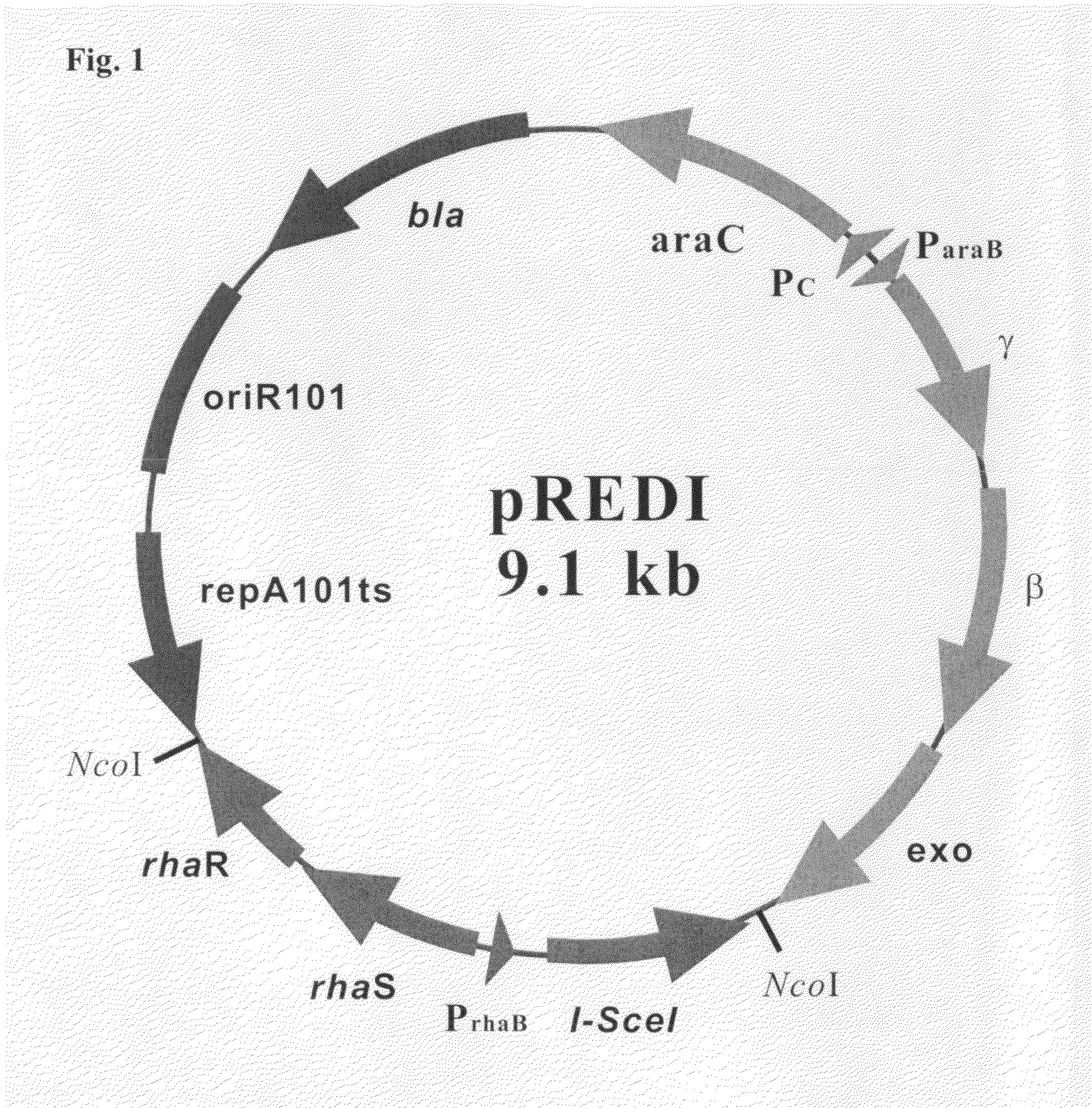
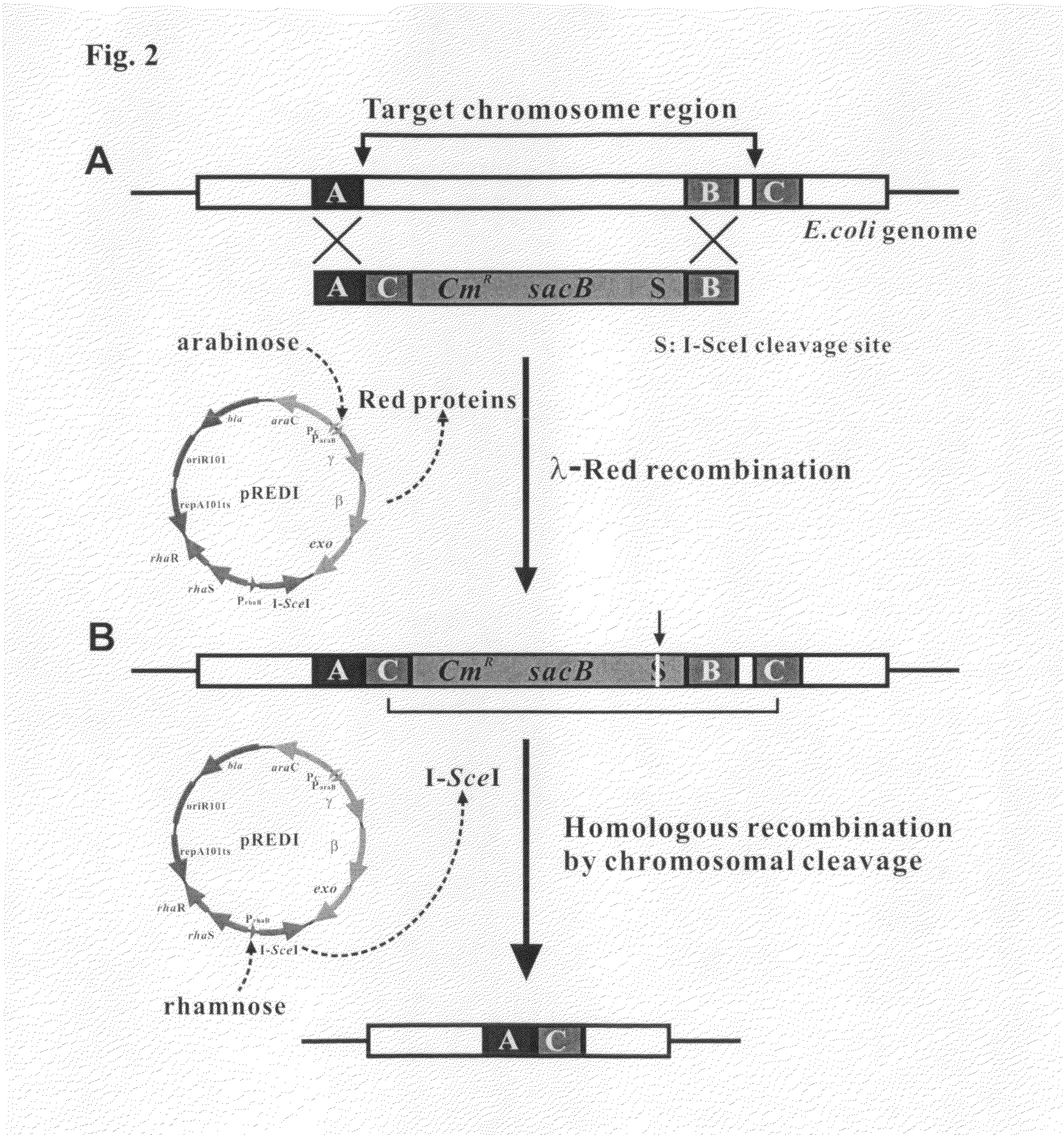
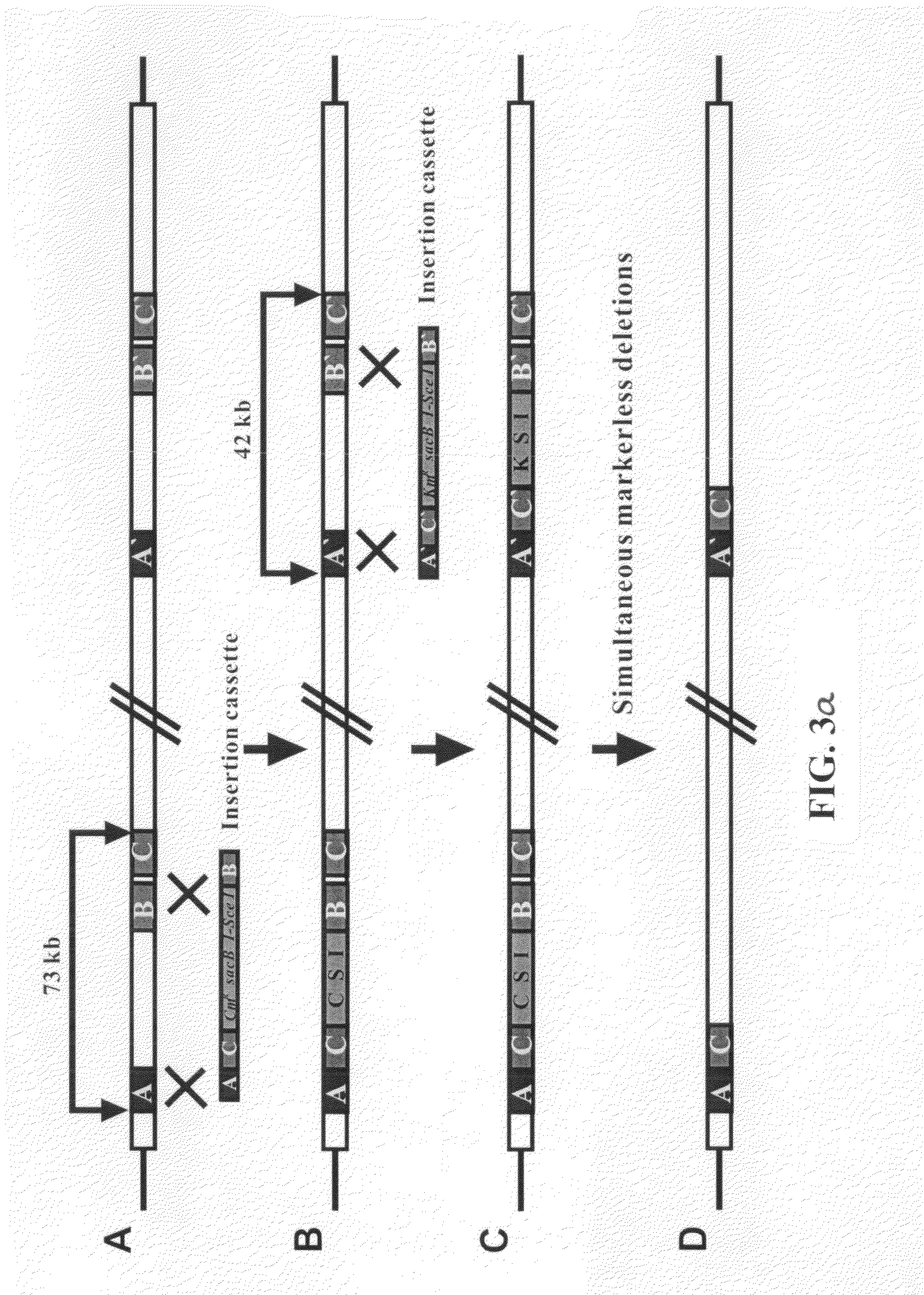
Recombinant vector for deleting specific regions of chromosome and method for deleting specific chromosomal regions of chromosome in the microorganism using the same
InactiveUS20090305421A1Rapid and efficient and successive deletionBacteriaNucleic acid vectorMicroorganismSpecific chromosome
Disclosed herein are a recombinant vector for deletion of specific chromosomal regions and a method for deletion of targeted microbial chromosomal regions using the same. Specifically, the recombinant vector comprises an arabinose-inducible promoter; a gene encoding a protein involved in lambda (λ)-red recombination; a rhamnose-inducible promoter; and a gene encoding the I-SceI endonuclease. The present invention enables a convenient, rapid and markerless successive deletion of specific genes of microbes, as compared to a conventional method.
Owner:KOREA ADVANCED INST OF SCI & TECH
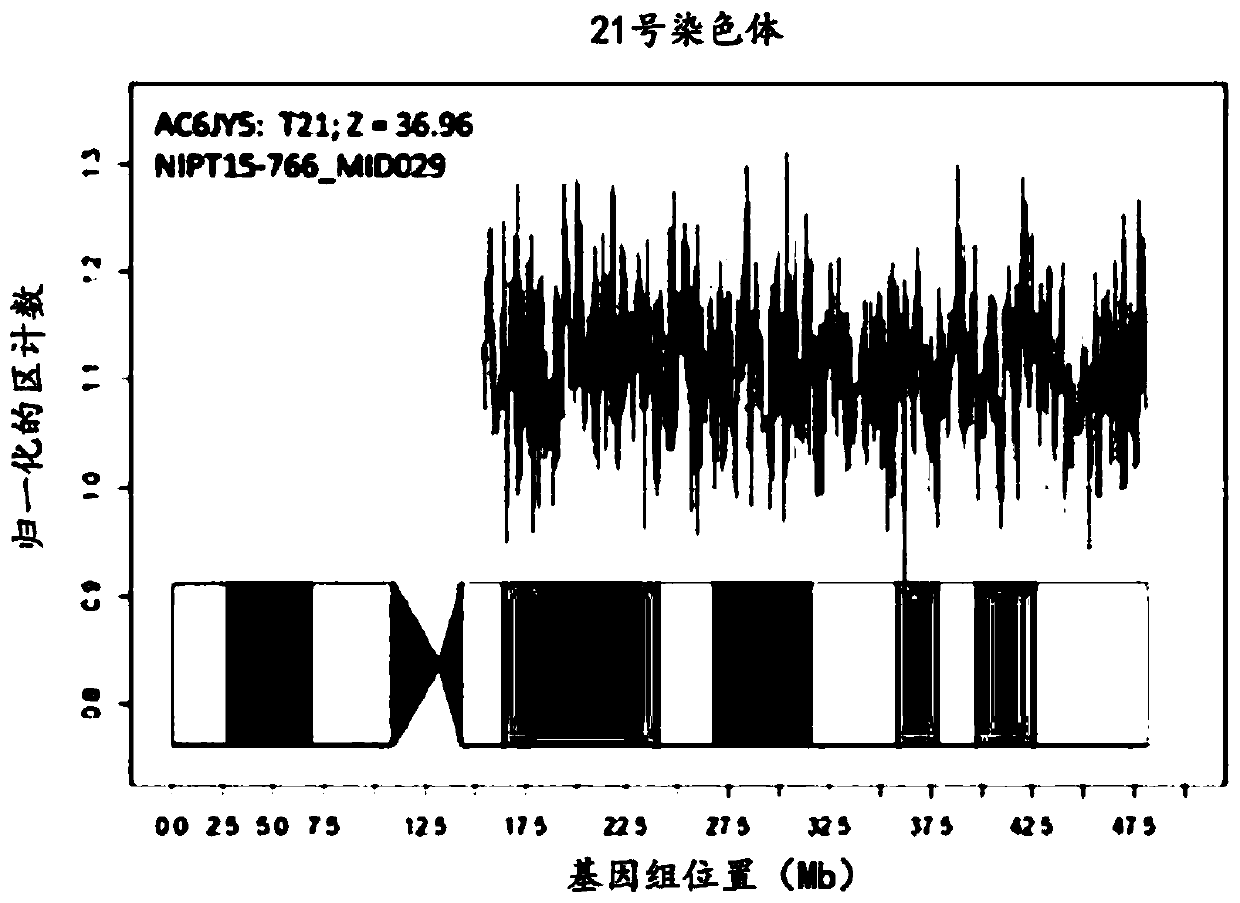
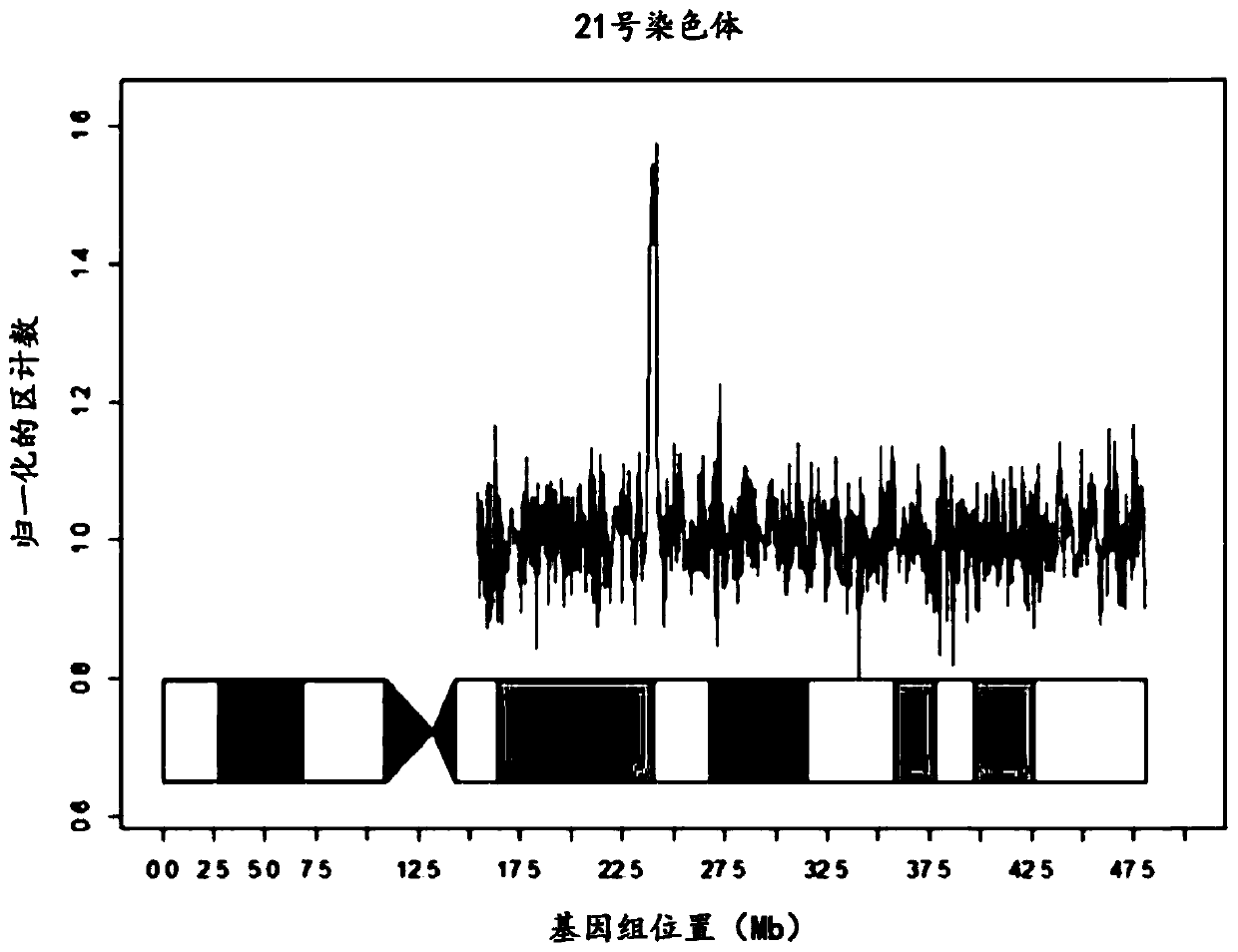
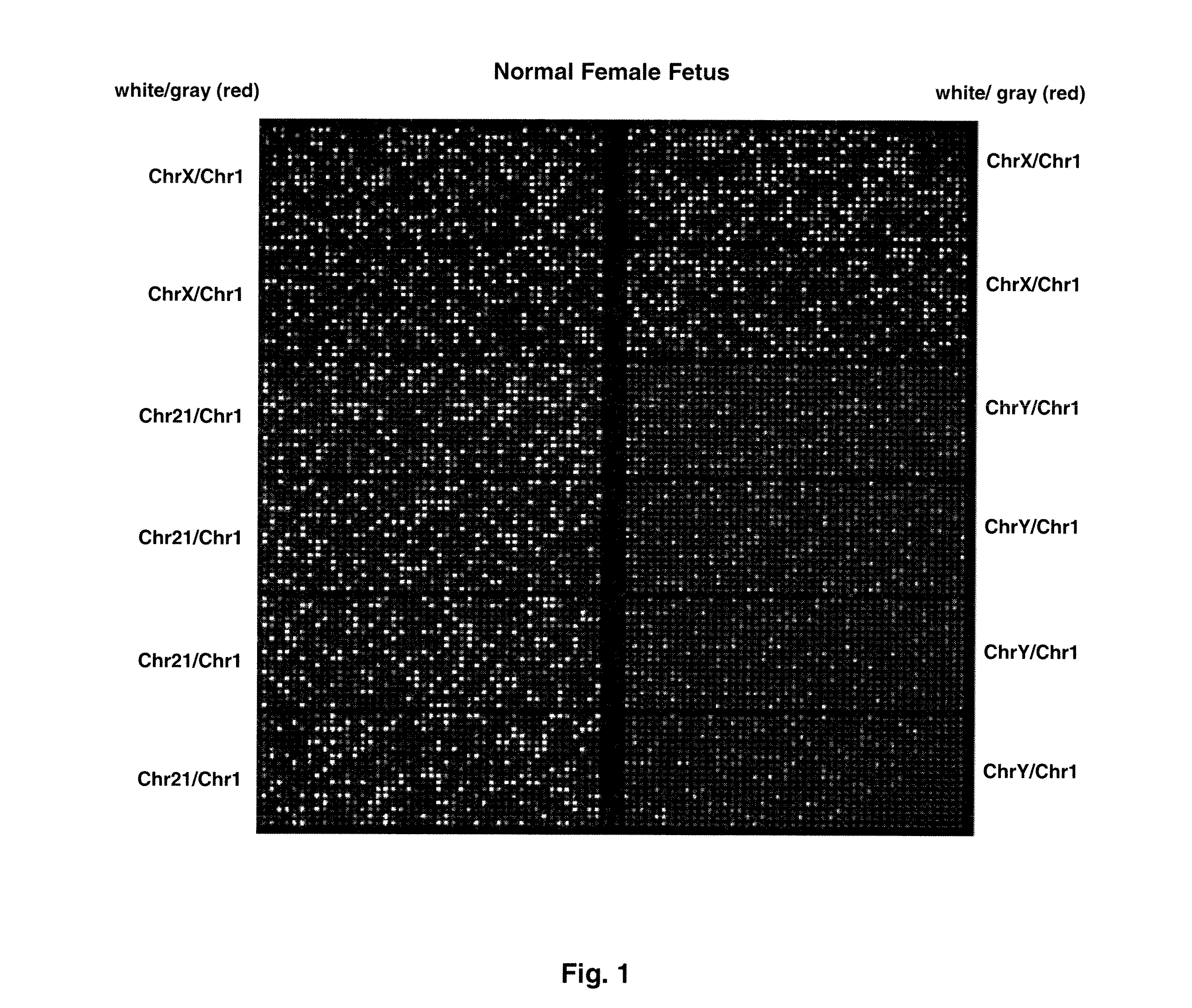
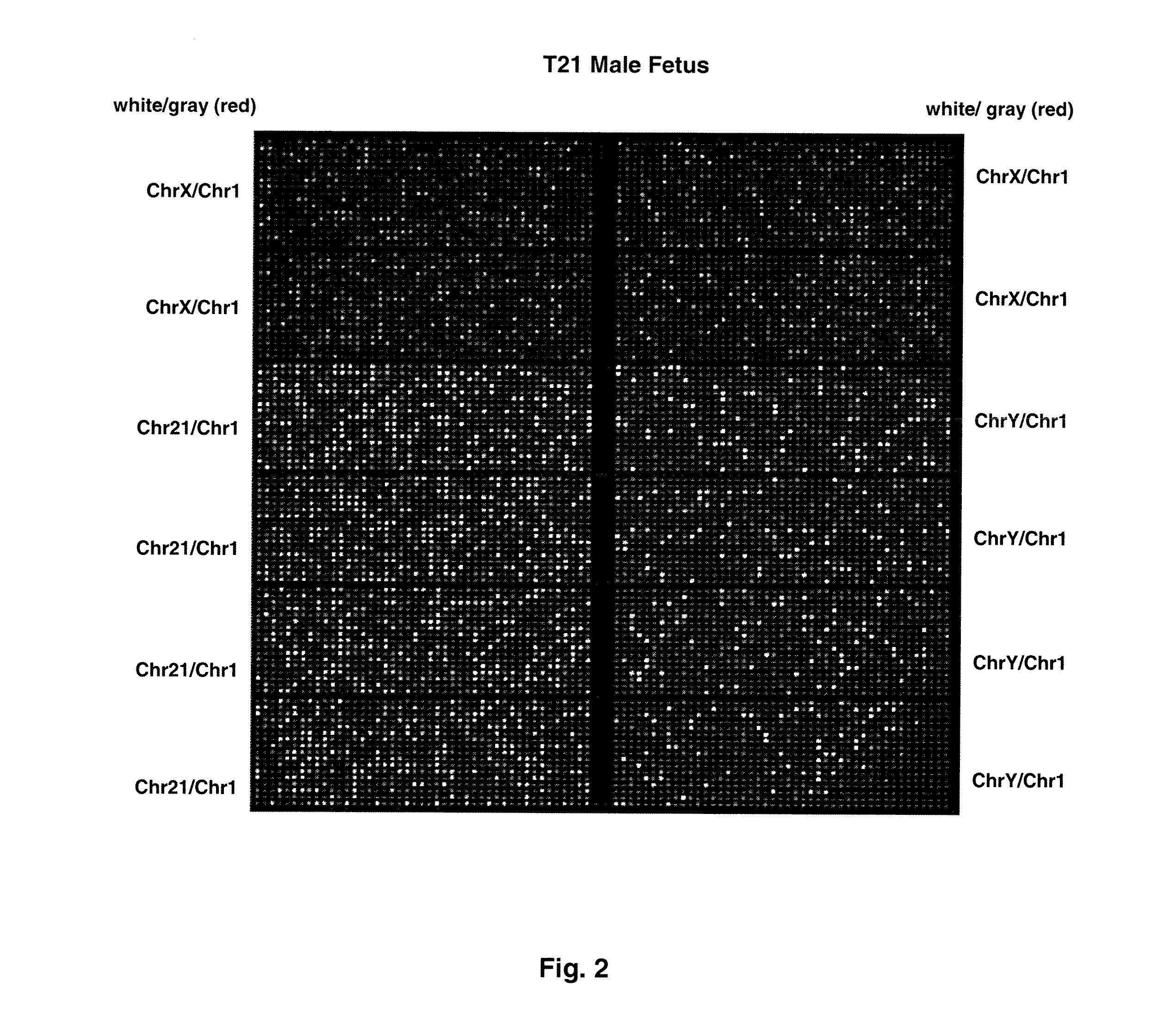
Chromosome aneuploidy detection method, device and system
PendingCN108595912AHigh sensitivityImprove accuracyBioreactor/fermenter combinationsBiological substance pretreatmentsSpecific chromosomeComputer science
The invention discloses a chromosome aneuploidy detection method, device and system. The method comprises the following steps of: sequencing at least one part of nucleic acids in a to-be-detected sample so as to obtain a sequencing result with reading segments; comparing the reading segments with a first reference sequence so as to obtain a comparison result, wherein the comparison result comprises information of positioning the reading segments in specific chromosomes; for first chromosomes, determining a number of reading segments positioned in the first chromosomes on the basis of the comparison result; and comparing the number of the reading segments positioned in the first chromosomes with a number of reading segments, positioned in corresponding first chromosomes, in a negative sample, so as to judge a number of the first chromosomes. By utilizing the method to carry out chromosome aneuploidy detection, the obtained detection results have relatively high sensitivity and correctness.
Owner:GENEMIND BIOSCIENCES CO LTD
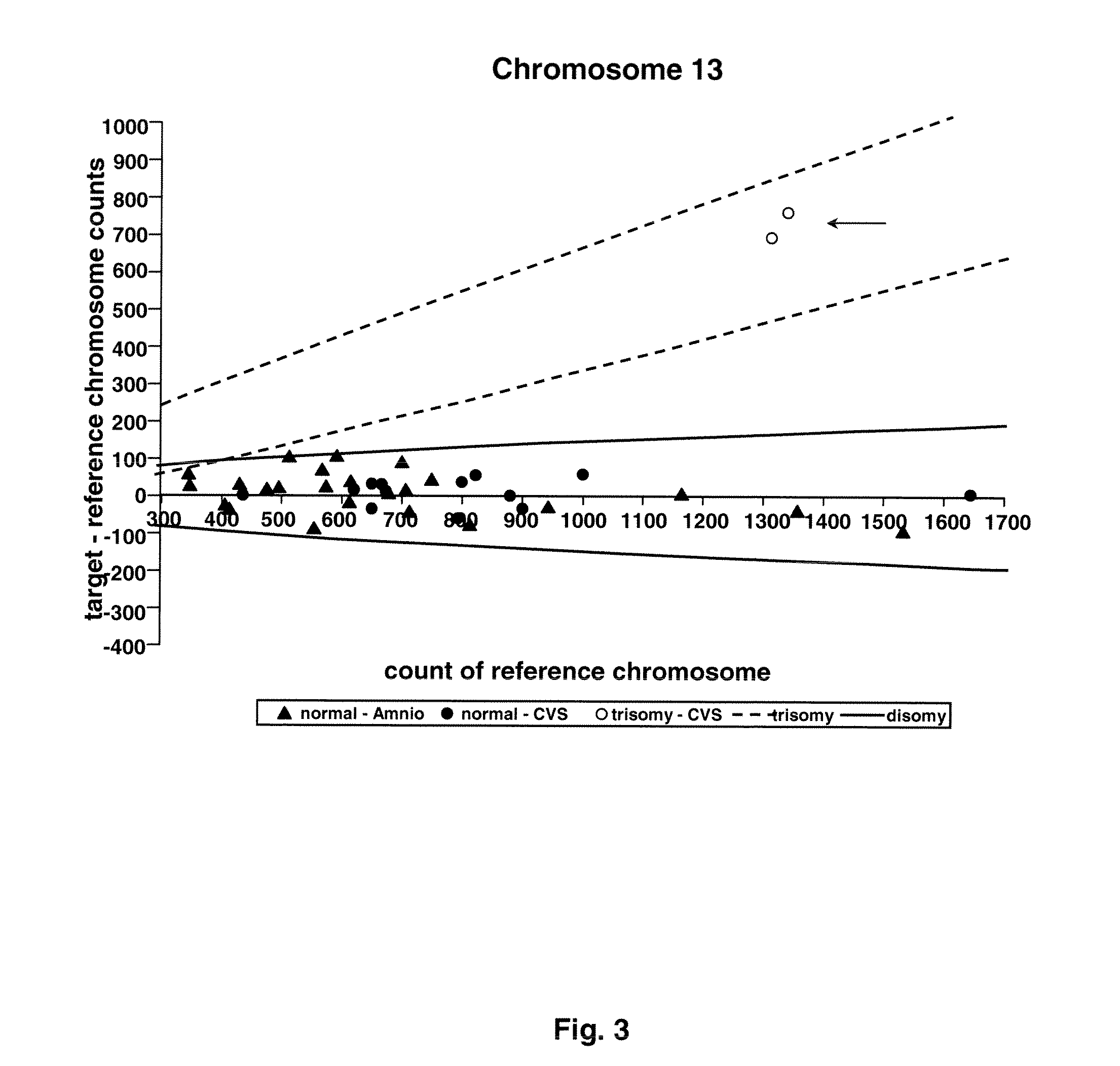
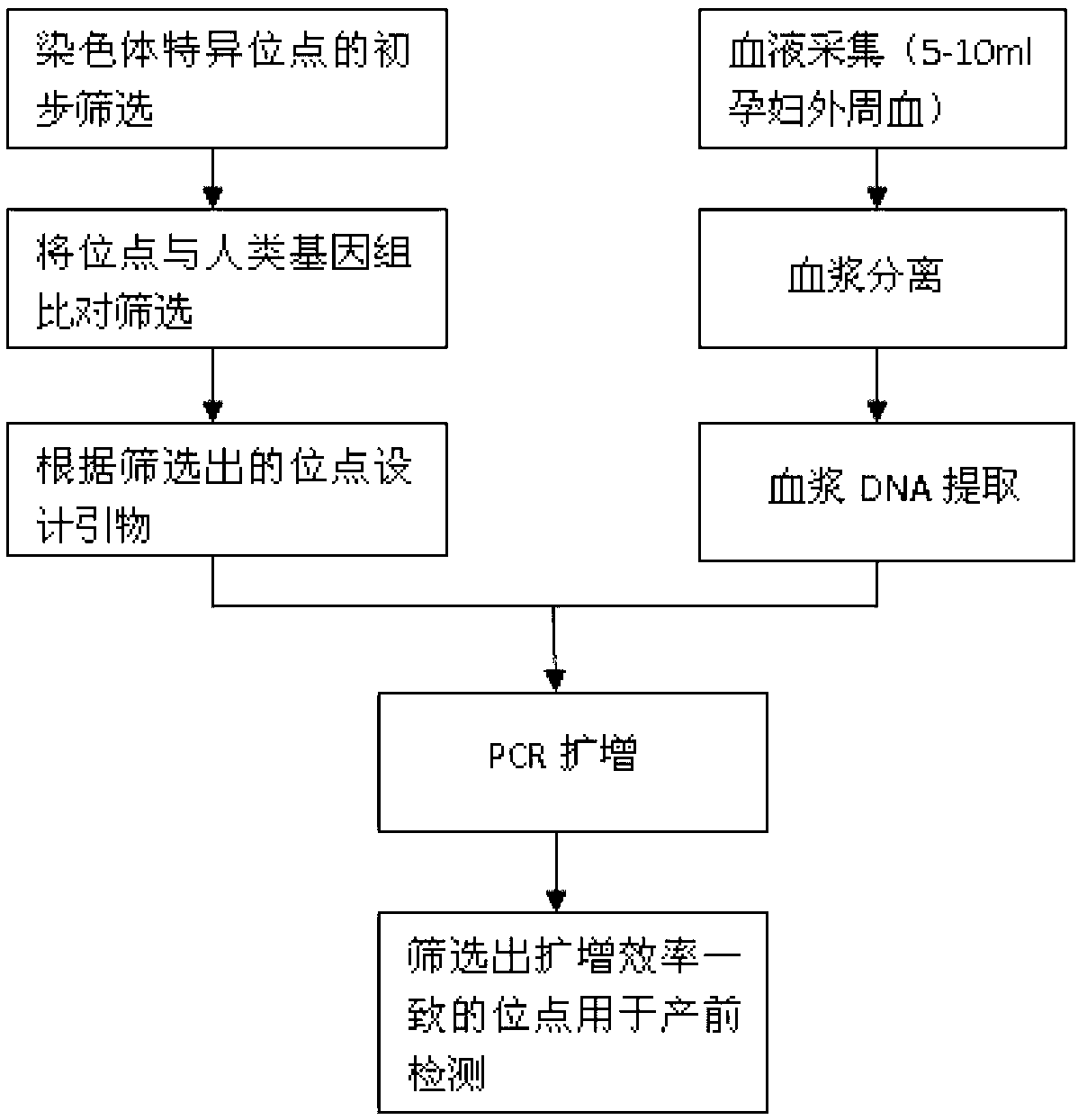
Method for non-invasive prenatal screening for aneuploidy
The present disclosure provides methods for non-invasive prenatal screening (NIPS) of fetal aneuploidies. The present methods are based on analyzing cell-free fetal DNA (cff DNA) found in a pregnant woman's circulation through the next generation sequencing (NGS) technology. Particularly, the present methods analyze the relative abundance of different fetal genomic fragments present in the maternal sample, where the fragments can be aligned to particular chromosomal locations of the fetal genome. The relative abundance information is indicative as to whether a particular chromosome is overrepresented or underrepresented in a fetal genome as compared to normal individuals, and thus can be used to detect fetal aneuploidy. Additionally, methods for increasing the positive predictive values (PPV) of NIPS by excluding false-positive detections are also provided.
Owner:QUEST DIAGNOSTICS INVESTMENTS INC
DNA markers for management of cancer
InactiveUS20100330567A1Stage increaseInvasive detectionSugar derivativesMicrobiological testing/measurementMelanomaNeurocytoma
A method is provided for assessing allelic losses and hypermethylation of genes in CpG tumor promotor region on specific chromosomal regions in cancer patients, including melanoma, neuroblastoma breast, colorectal, and prostate cancer patients. The method relies on the evidence that free DNA and hypermethylation of genes in CpG tumor promotor region may be identified in the bone marrow, serum, plasma, and tumor tissue samples of cancer patients. Methods of melanoma, neuroblastoma, colorectal cancer, breast cancer and prostate cancer detection, staging, and prognosis are also provided.
Owner:HOON DAVE S B +1
Application of chromosome instable mutation to preparing agent or kit for diagnosis, evaluation and prognosis of multiple myeloma
PendingCN110317876AHigh carry rateMicrobiological testing/measurementDNA/RNA fragmentationSpecific chromosomeIndividualized treatment
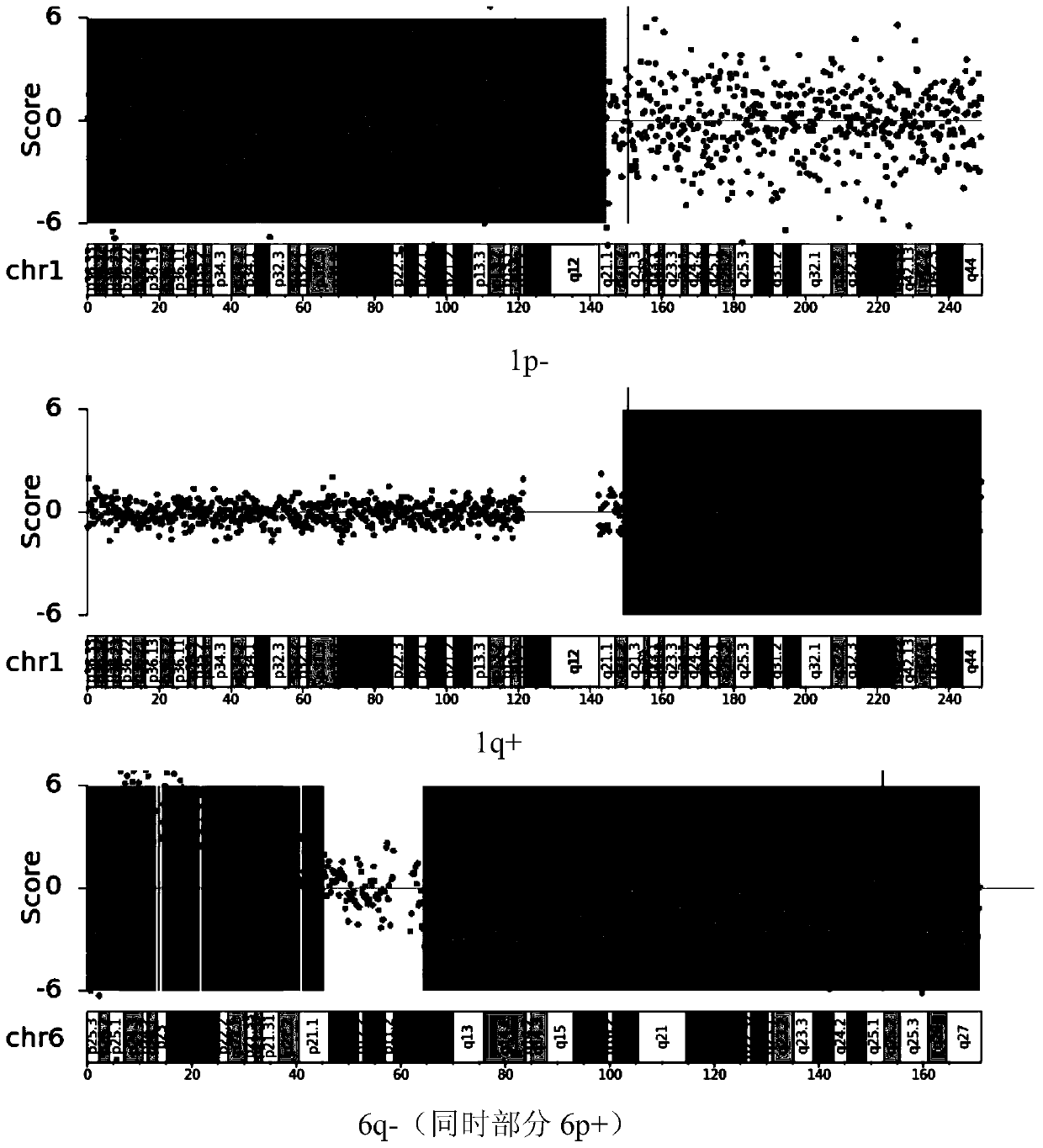
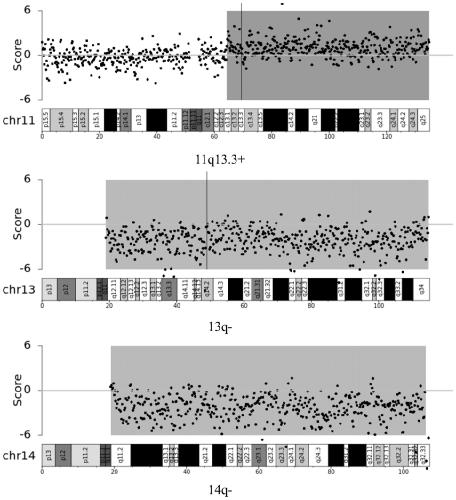
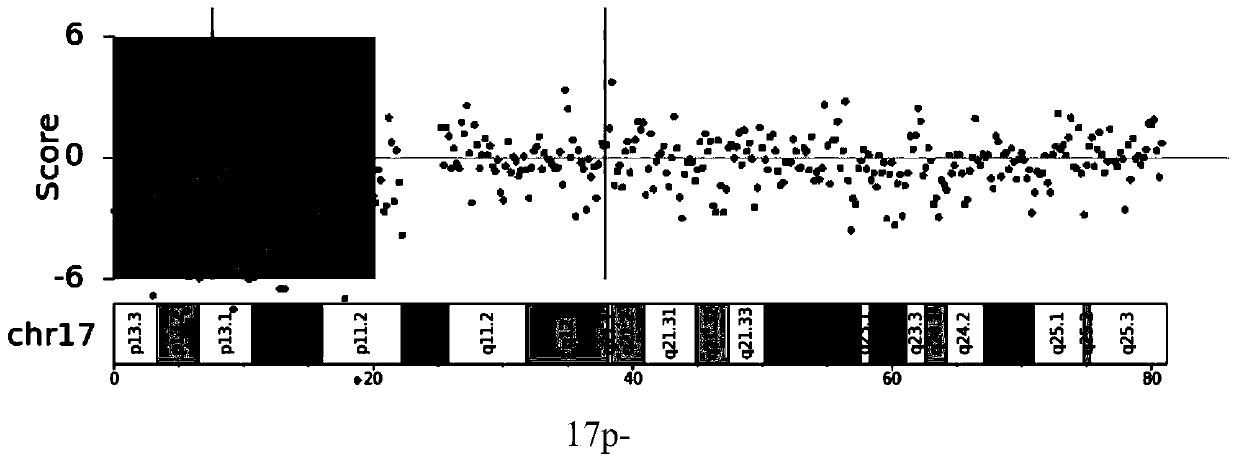
The invention relates to the field of biotechnology and medical diagnosis, in particular to application of chromosome instable regions to preparing an agent or kit for the diagnosis of multiple myeloma. There are seven chromosome instable regions 1p, 1q, 6q, 11q13.3, 13q, 14q and 17p. Samples of clinically confirmed multiple myeloma patients and healthy people are collected, the specific chromosome instability distribution characteristics of the multiple myeloma are expounded by comparing chromosome abnormity conditions of the tumor patients and the contrast through whole-genome sequencing, the seven common chromosome instable regions of the multiple myeloma patients are disclosed, and the total carrying rate is up to 89.3%, which has great significance in the clinical diagnosis, treatment, monitoring, prognosis and evaluation of the multiple myeloma and provides the scientific basis for conducting early diagnosis next and formulating an individualized treatment scheme.
Owner:SUZHOU HONGYUAN BIOTECH CO LTD
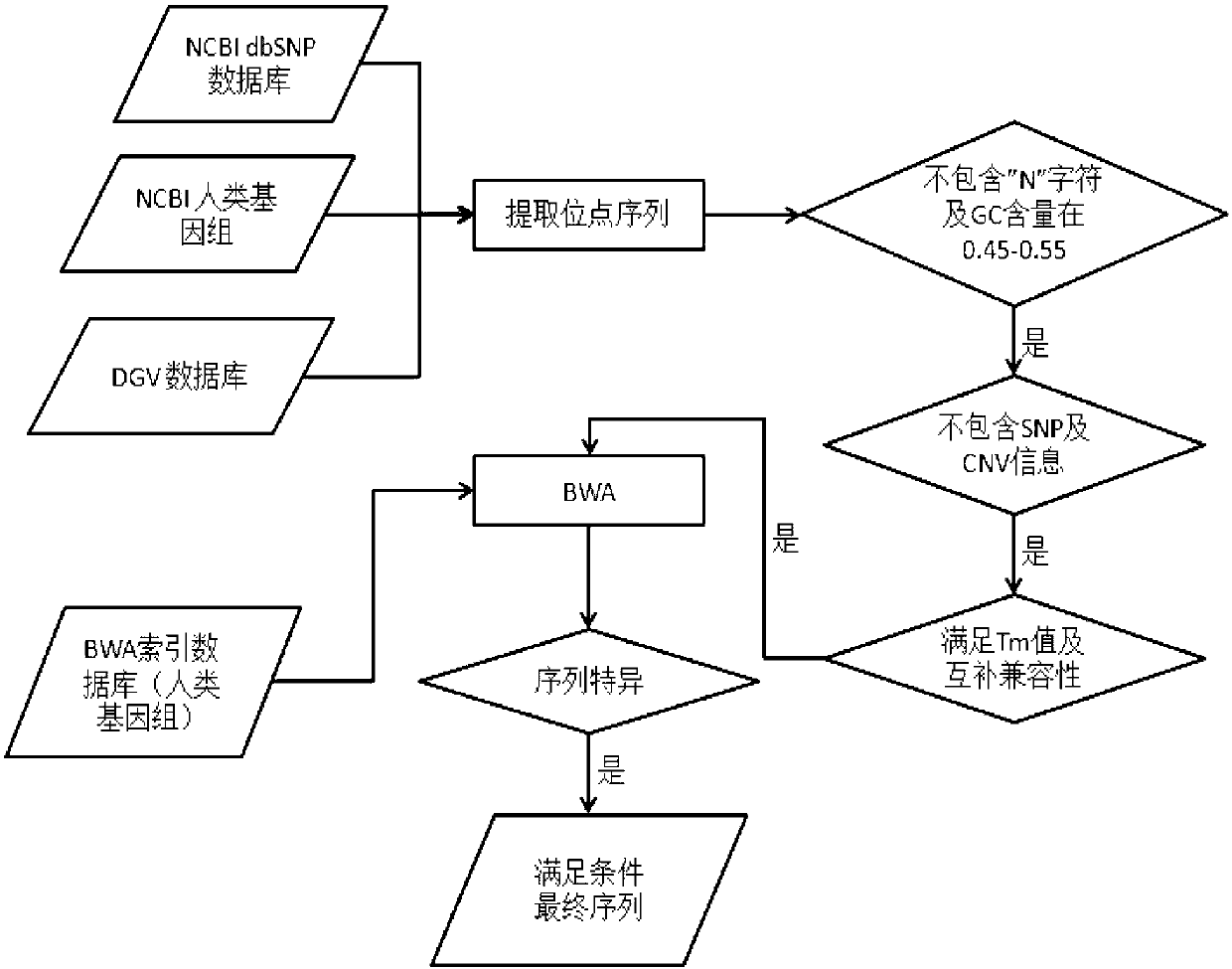
Chromosome specific loci screening method and application thereof
ActiveCN103215349AReduce in quantityStrong specificityMicrobiological testing/measurementSpecific chromosomeMedicine
The invention provides a chromosome specific loci screening method comprising the steps that: chromosome specific loci preliminary screening is carried out; chromosome specific loci comparison and removing are carried out; chromosome specific loci screening is carried out with a PCR primer design method; the loci screened by the PCR primers are finely screened with a real-time quantitative PCR reaction; and the like. The invention also provides the application of the screening method in prenatal testing. According to the chromosome specific loci screening method provided by the invention, specific locus sequence is screened on a specific chromosome by using common computer software. The operation is simple, cost is low, and the specific loci obtained by screening have small number and high specificity.
Owner:SUZHOU BASECARE MEDICAL DEVICE CO LTD
Direct molecular diagnosis of fetal aneuploidy
ActiveUS8574842B2Addressing slow performanceSugar derivativesMicrobiological testing/measurementSpecific chromosomeChromosomal region
Methods and materials for detection of aneuploidy and other chromosomal abnormalities using fetal tissue are disclosed. Results can be obtained rapidly, without cell culture. The method uses digital PCR for amplification and detection of single target sequences, allowing an accurate count of a specific chromosome or chromosomal region. Specific polynucleic acid primers and probes are disclosed for chromosomes 1, 13, 18, 21, X and Y. These polynucleic acid sequences are chosen to be essentially invariant between individuals, so the test is not dependent on sequence differences between fetus and mother.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
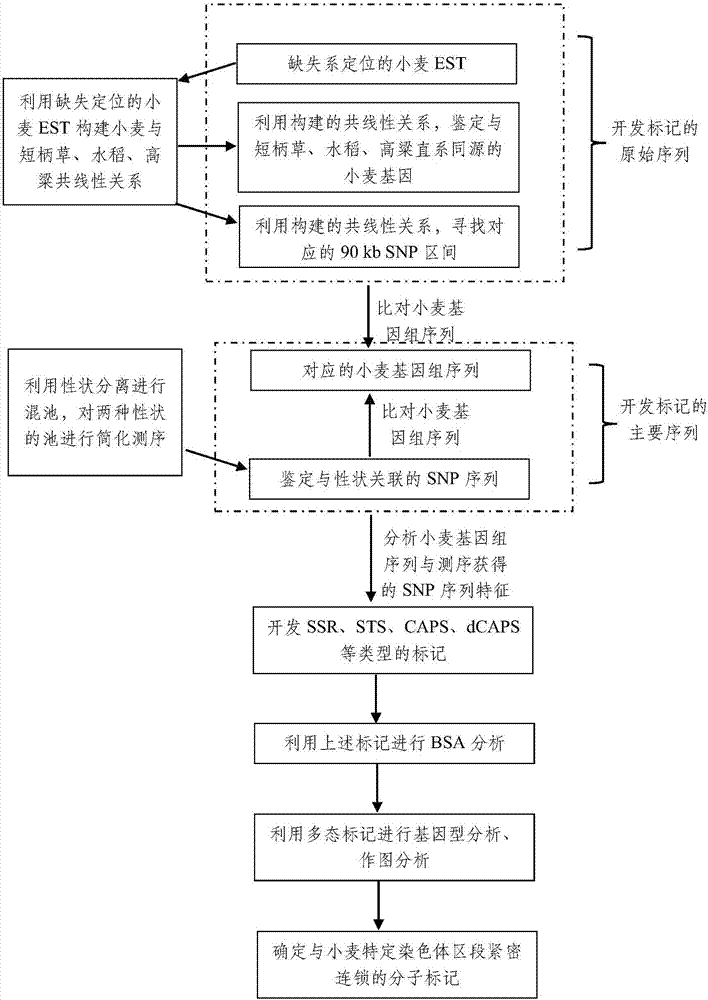
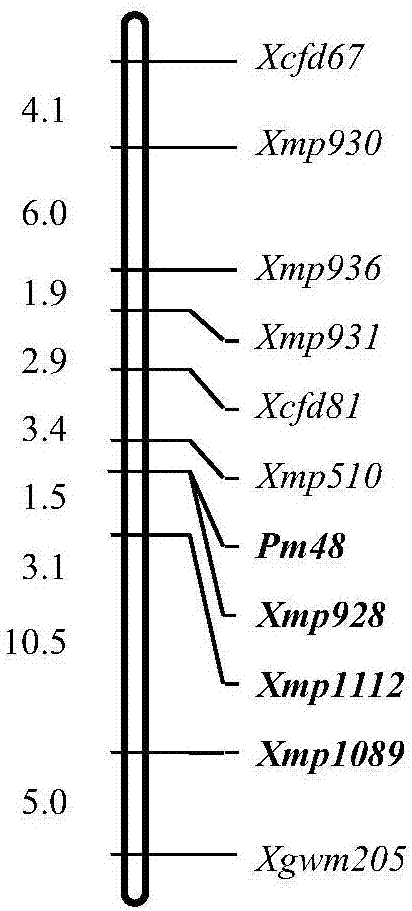
Method for developing molecular marker interlocked with specific chromosome region of wheat
PendingCN107354232AOvercoming density is not highMore difficult to avoidMicrobiological testing/measurementSpecific chromosomeAgricultural science
The invention discloses a method for developing a molecular marker interlocked with a specific chromosome region of wheat and relates to the technical field of agricultural living things. Orthologous genes determined by EST co-linear relation in wheat deletion positioning and corresponding SNP are utilized as original sequences for developing the markers, wheat genomic sequences obtained through sequence comparison and sequences obtained through sequencing serve as main sequences for developing the markers, sequence feature analysis is conducted on the obtained wheat genomic sequences and the sequences obtained through sequencing so as to design corresponding SSR, STS, CAPS and dCAPS markers; the developed markers sequentially undergo BSA analysis, genotype analysis and diagraph analysis, and the molecular marker interlocked with the specific chromosome region of wheat is obtained. By adopting the method, the shortcoming of the low density of the molecular marker of wheat is overcome, and the problem that the molecular marker of wheat in the specific chromosome region is difficult to mark is very well solved.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
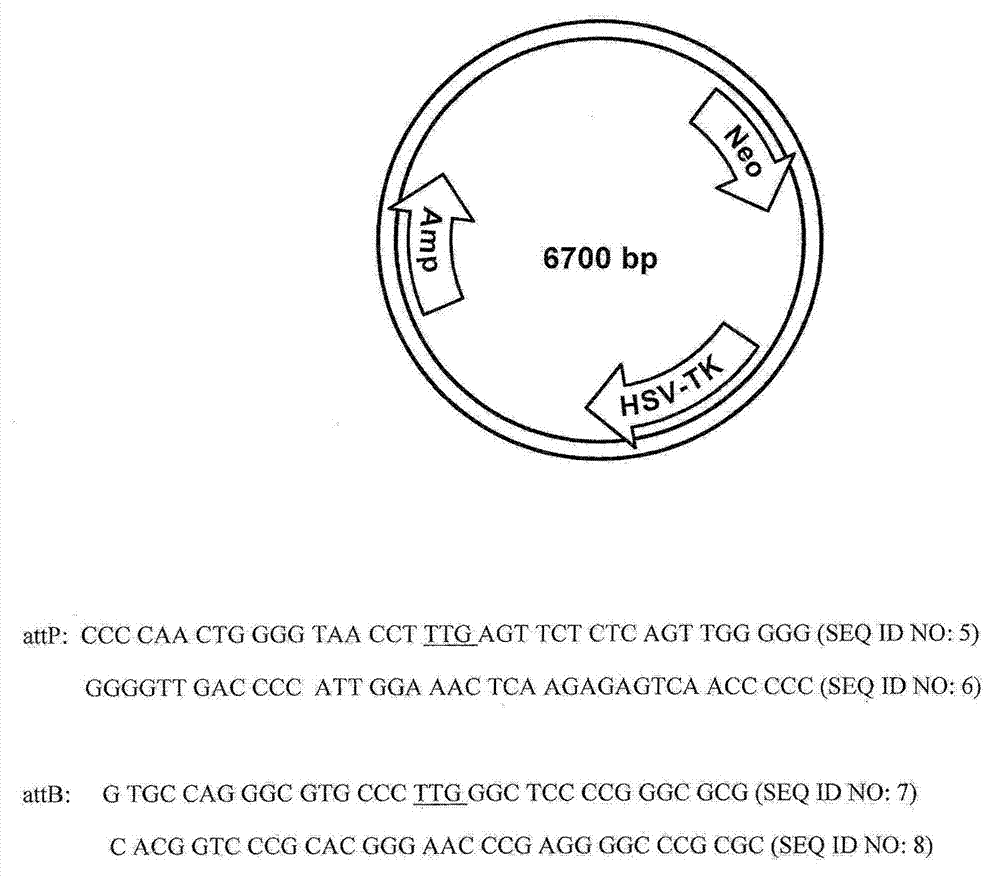
Chromosomal landing pads and related uses
Provided herein are methods for stable integration and / or expression of one or more recombinant polynucleotides in a host cell. The recombinant polynucleotides are typically integrated into the host genome at some native chromosomal integration sites. The integration can be mediated by homologous recombination or by using a hybrid recombinase targeting the specific chromosomal locations. The native chromosomal integration sites in the host cells, which support stable integration and strong transcription activities of foreign genes, are present within or adjacent to specific genes in the CHO genome, ankyrin 2 gene (Ank2), cleavage and polyadenylation specific factor 4 gene (Cpsf4), C-Mos gene, and Nephrocystin-1 / Mal gene. Also provided are methods and nucleic acid molecules for inserting site-specific recombination sequences (chromosomal landing pads) into these specific chromosomal locations, engineered host cells containing chromosomal landing pads, methods and compositions (e.g., kits) therefore.
Owner:THE SCRIPPS RES INST
Application of chromosome instable mutation to preparing agent or kit for diagnosis, evaluation and prognosis of urothelium carcinoma
PendingCN110317877AImprove accuracyHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationDiseaseSpecific chromosome
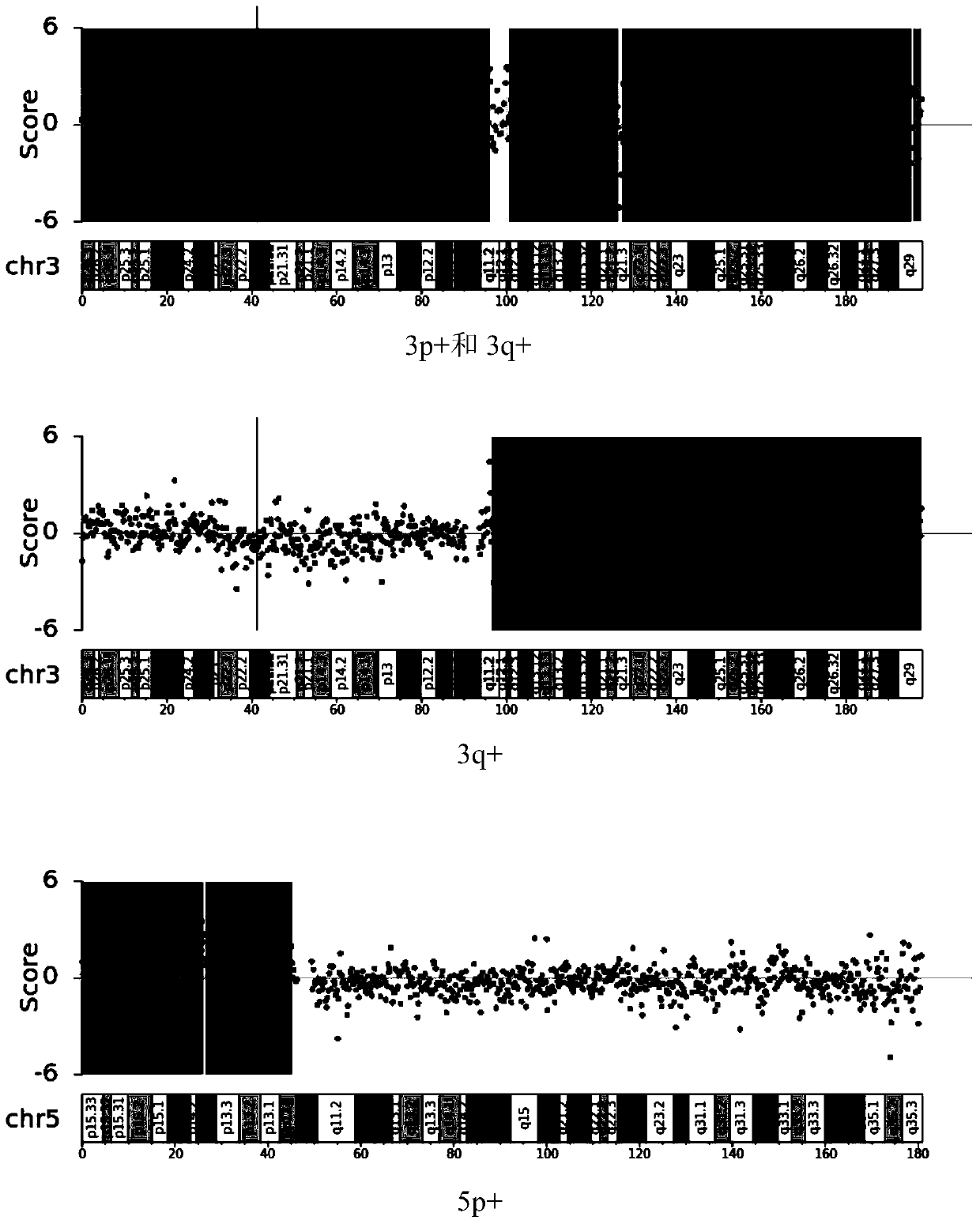
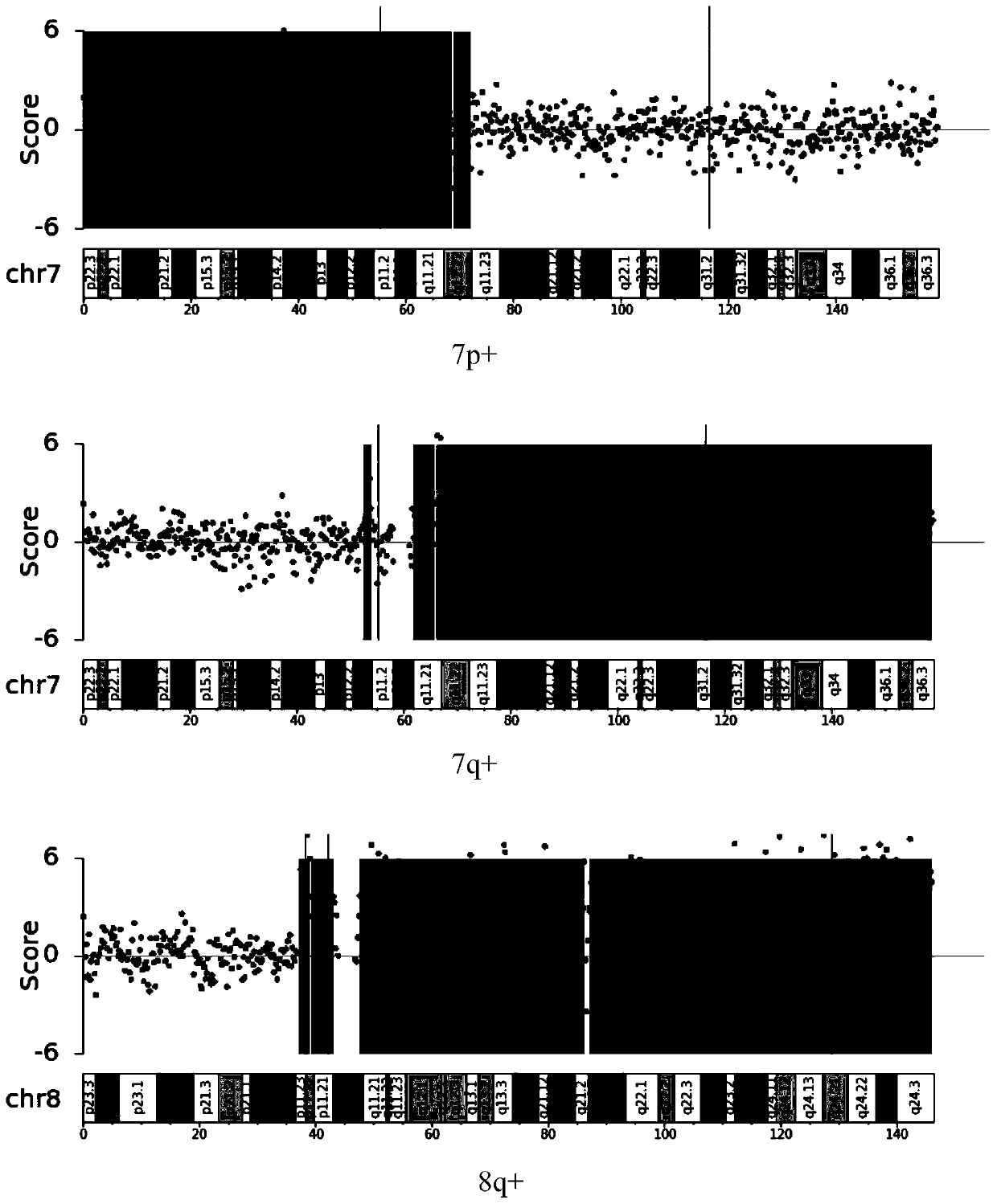
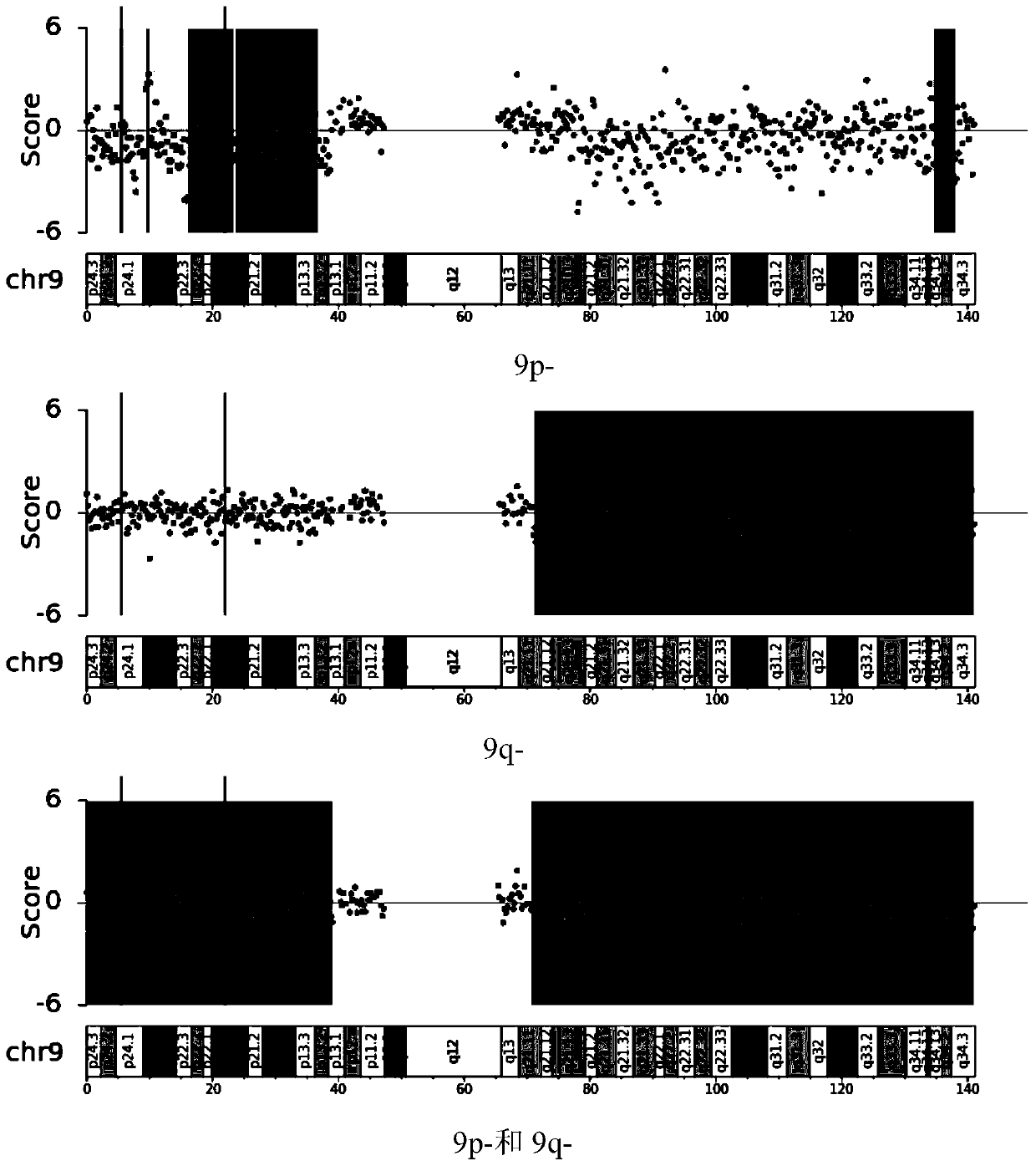
The invention relates to the field of biotechnology and medical diagnosis, in particular to application of chromosome instable regions to preparing an agent or kit for the diagnosis of urothelium carcinoma. There are ten chromosome instable regions 3p, 3q, 5p, 7p, 7q, 8q, 9p, 9q, 17p and 17q. Samples of clinically confirmed urothelium carcinoma patients, healthy people and non-urothelium-carcinomapatients are collected, the specific chromosome instability distribution characteristics of the urothelium carcinoma are expounded by comparing chromosome abnormity conditions of the tumor patients and the contrast through whole-genome sequencing, the ten common chromosome instable regions of the urothelium carcinoma patients are disclosed, and the total carrying rate is up to 93.4%, which has great significance in the clinical diagnosis, treatment, monitoring, prognosis and evaluation of the urothelium carcinoma and provides the scientific basis for conducting early diagnosis next and formulating an individualized treatment scheme.
Owner:SUZHOU HONGYUAN BIOTECH CO LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com