Chromosomal landing pads and related uses
A chromosome and bacteriophage technology, applied in the direction of stably introducing foreign DNA into chromosomes, vectors, nucleic acid vectors, etc., can solve the problems of cumbersome, high-efficiency expression, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0095] Example 1 Identification of native chromosomal loci with strong transcriptional activity
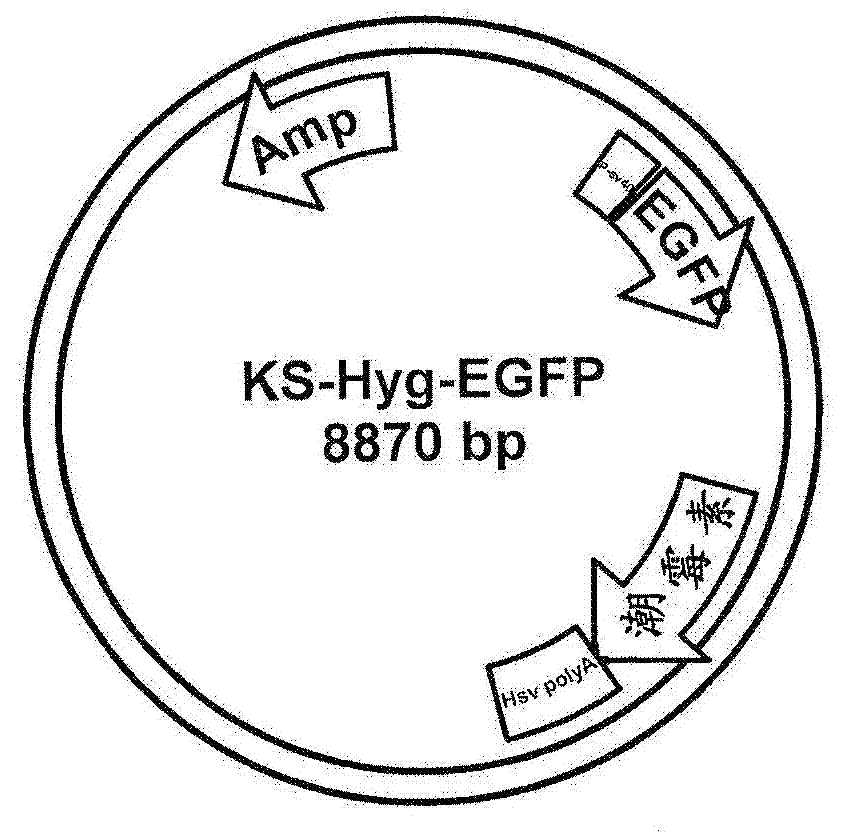
[0096] Plasmid for random integration into the CHO genome: modified based on the original attP-containing plasmid described by Thyagarajan et al. (2001, Mol Cell Biol. 21(12):3926-34). Specifically, the original plasmid was modified to replace the Zeocin marker with a neomycin marker. In addition, the firefly luciferase gene was replaced with the EGFP gene controlled by the SV40 promoter ( figure 1 ).
[0097] Stable transfection: The modified plasmid was purified using Qiagen Midiprep columns. 25 µg of DNA was digested overnight with the restriction enzyme BamHI to linearize the plasmid. The resulting linear DNA was then transfected into CHO cells to generate stable integration. Two days after transfection, the cells were split into new culture plates and hygromycin was added to the medium to select for stable integration events. Cell cultures were grown for 3 weeks and t...
Embodiment 2
[0105] Example 2 Insertion of landing pads at identified natural chromosomal integration sites
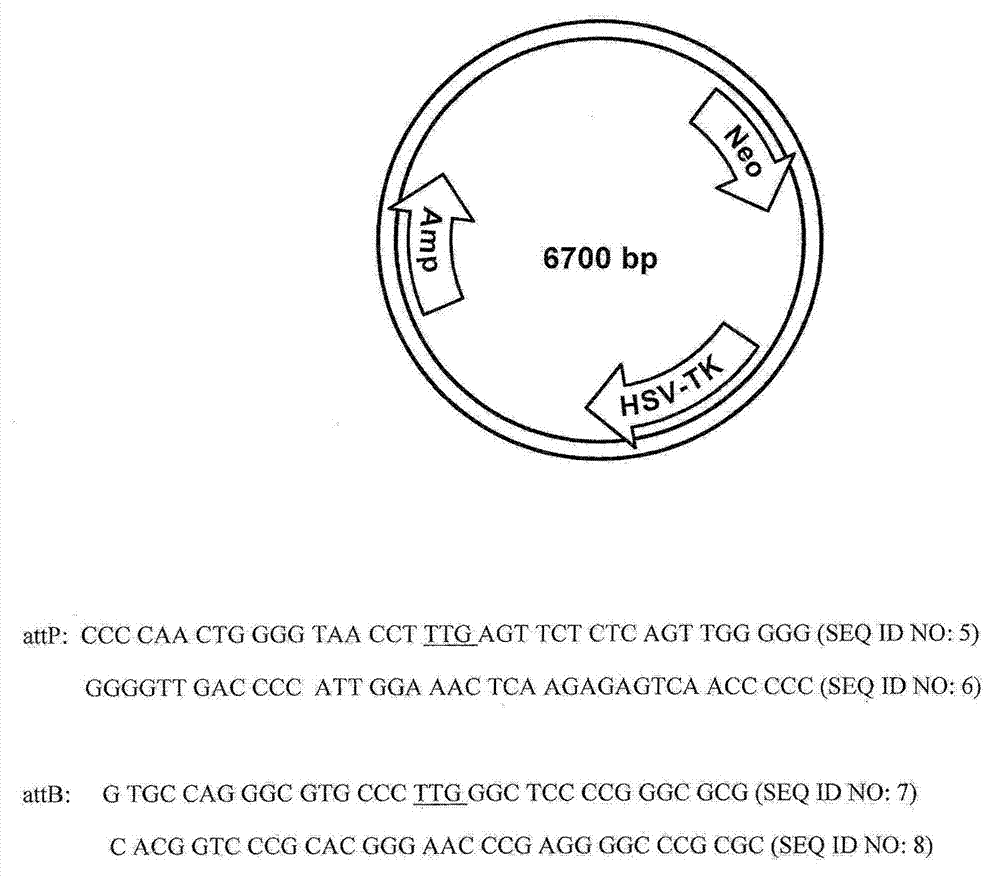
[0106] Homologous recombination: Genomic DNA flanking the integration site is identified and cloned into a plasmid containing positive and negative selection markers ( figure 2 ). For each site, a longer homology arm (3-4 kb in length) was cloned 5' of the neomycin resistance gene. A short homology arm (1.5-2 kb in length) was cloned 3' of the neomycin resistance gene. A single phage-binding site, attP, is located at the end of the long homology arm.
[0107] The homologous recombination plasmid was digested with NotI enzyme to linearize the plasmid; the long homology arm was at one end of the linear DNA. After transfection into CHO cells, neomycin was added to the medium to select for cells that integrated the linear DNA in the cell chromosome. A pool of resistant clones was obtained after 4-6 weeks. The cells are then negatively selected by adding ganciclovir to the mediu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com