Preparation method and application of electrochemical sensor for non-coding RNA
An electrochemical and sensor technology, applied in the field of electrochemical biosensing, can solve the problems of cumbersome sample processing, high detection cost, and high maintenance cost, and achieve the effects of good chemical stability, low detection limit, and time saving.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Embodiment 1: The steps of the preparation method of the electrochemical sensor of the non-coding RNA are as follows:
[0050] 1. Ferric oxide (Fe 3 o 4 ) Preparation of nanospheres
[0051] Put 1.35 g FeCl 3 .6H 2 O was added to 40 mL of ethylene glycol to form a clear solution, and sodium acetate and polyethylene glycol were added, wherein the mass ratio of sodium acetate to ferric chloride was 3:1, and the mass ratio of polyethylene glycol to ferric chloride The ratio is 7:10; after stirring for 30 minutes, put it into a hydrothermal reaction kettle and heat it to 180°C, react for 7 hours, cool to room temperature to get a black precipitate, wash the precipitate with absolute ethanol, and treat it at 60°C for 10 hours to obtain Fe 3 o 4 nanospheres;
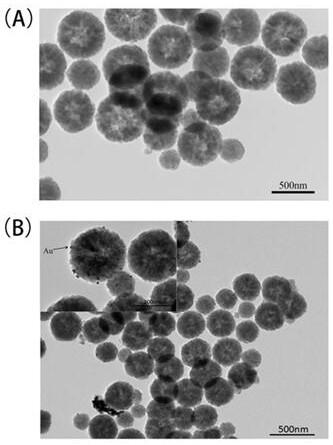
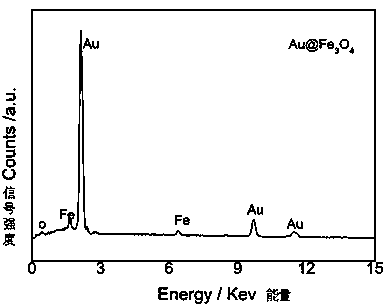
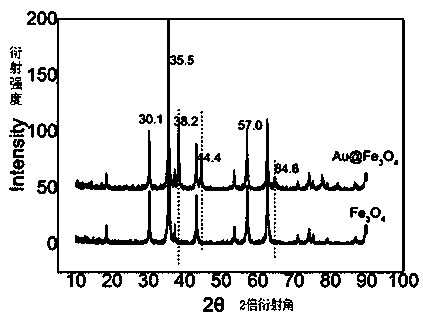
[0052] 2. Gold nanoparticle-loaded Fe3O4 nanocomposites (Au@Fe 3 o 4 ) preparation
[0053] Step (1) Fe 3 o 4 Disperse 10 mg of nanospheres in 10 mL of ultrapure water. After ultrasonic dispersion is unifor...
Embodiment 2
[0068] Example 2: Specific detection of electrochemical sensors
[0069] In this embodiment, the electrochemical sensor in Example 1 is used to detect four kinds of mismatched sequences, 1MT-S, 2-MT-S, 1MT-L, and 2MT-L; the concentration of mismatched sequences is diluted to 10 -10 M for electrochemical detection; refer to the method of step 4 (2) of the example to prepare the complex phosphate buffer dispersion, the difference is that the target RNA is replaced by the mismatched sequence in this example, and the prepared phosphate buffer dispersion Drop on and cover the surface of the screen printing electrode, measure DPV, DPV scanning range: 0~-0.5V;
[0070] The mismatched sequences are:
[0071] 1MT-S: CCG AAA GAG CGA GAC GCG TC CAT AAT CTG GTC TCT TCT TC;
[0072] 2MT-S: CCG AAA GAG CAA GAC GCG TC CAT AAT CTG GTC TCT TCT TC;
[0073] 1MT-L: ATG ATA TAG CCA GCT GCC TT TTA AGA GGT CTT ATC TGT TC;
[0074] 2MT-L: ATG ATA TAG CAA GCT GCC TT TTA AGA GGT CTT ATC TGT TC;
...
Embodiment 3
[0076] Example 3: Application of electrochemical sensor in the detection of CCND2-S and CCND2-L in human lung cancer cell H292 and human normal lung cell Beas-2B
[0077] (1) Human normal lung cells and human lung cancer cells were selected for RNA extraction, and the extraction method was as follows:
[0078] 1) Remove the medium in the cell culture flask, add 1mL PBS to wash the cells, and aspirate and discard;
[0079] 2) Add 0.5 mL Trizol, shake by hand for 5 min to lyse the cells, and collect them into a 1.5 mL centrifuge tube;
[0080] 3) Add 0.1 mL of chloroform, shake vigorously for 15 s, and place at room temperature for 2-3 min;
[0081] 4) Centrifuge at 12000 rpm at 4°C for 15 min; (start to prepare new tubes for centrifugation; prepare RNA collection column; prepare enzyme-RDD mixed system: 10 μL DNase I stock solution and 70 μL Buffer RDD);
[0082] 5) Transfer the supernatant to a new centrifuge tube, add an equal volume of 70% ethanol, turn it upside down to m...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com