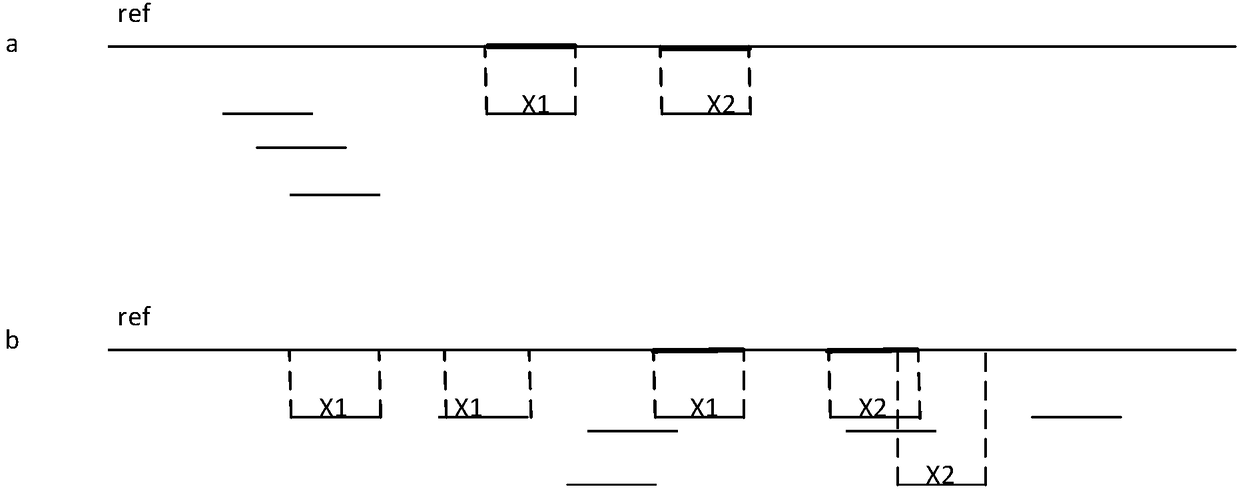
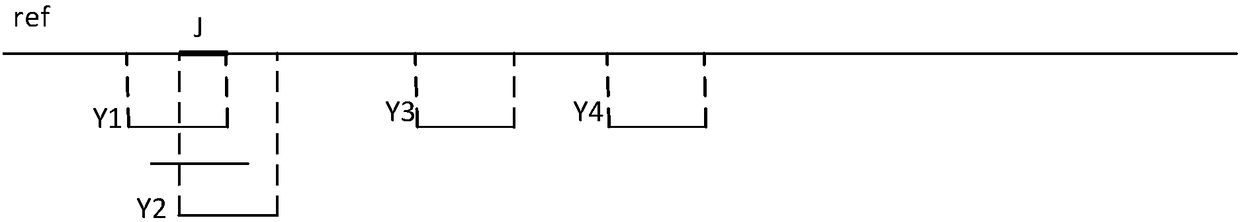
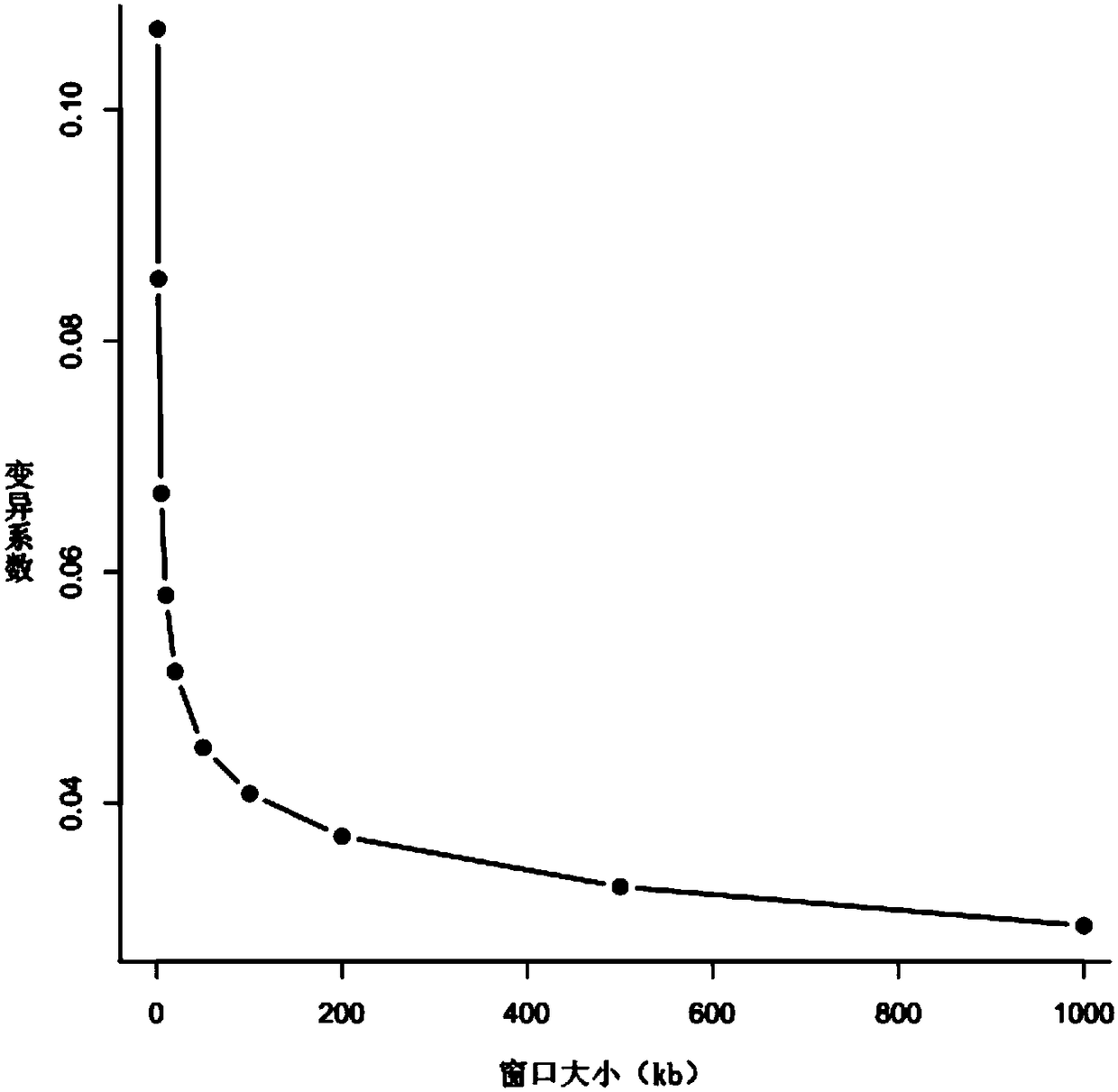
Chromosome aneuploidy detection method, device and system
An aneuploidy and chromosome technology, applied in the field of bioinformatics, can solve problems such as high morbidity and mortality, and achieve high sensitivity and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0141] The reference sequence used is a collection of regions of the human reference genome that do not contain the regions shown in Table 1 and satisfy the following: 1) the alignment ability is 1, 2) the sequencing depth is less than 6 times of the average sequencing depth, or the sequence has been sequenced The sequencing depth of regions with a depth percentile greater than 99 was assigned the 99th percentile sequencing depth value.
[0142] Both the control sample and the test sample were processed by:
[0143] 1. Sequencing, obtain the off-machine data, that is, obtain the set of reads; remove the reads less than 25bp;
[0144] 2. Obtain the alignment result (sam file), including obtaining unique reads (reads aligned to the unique position of the reference sequence) and the positions of these reads on the reference sequence / reference chromosome;
[0145] 3. Perform GC correction, including: cutting the reference sequence into 150Kbp windows / regions (Bin=150K); calculati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com