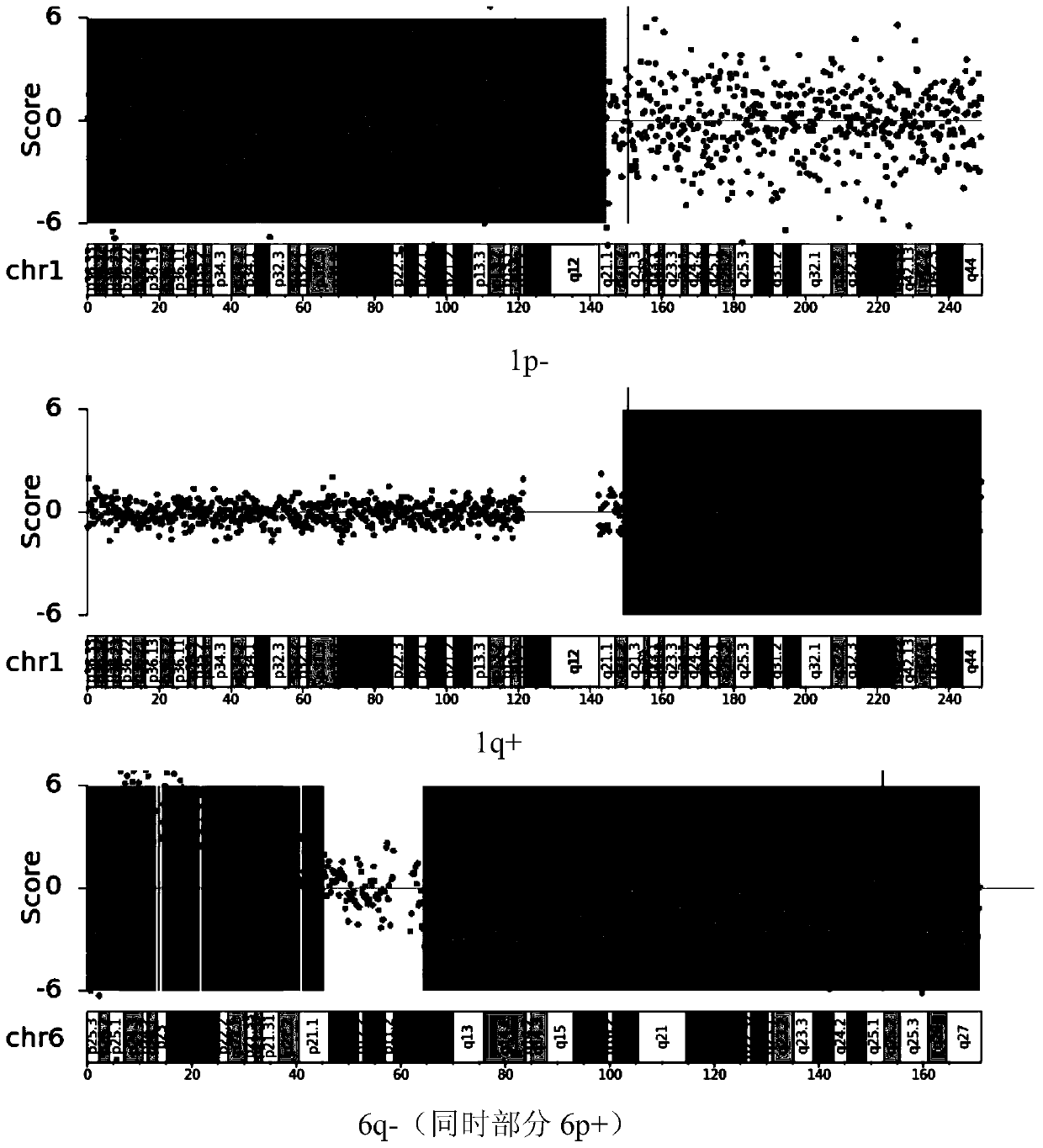
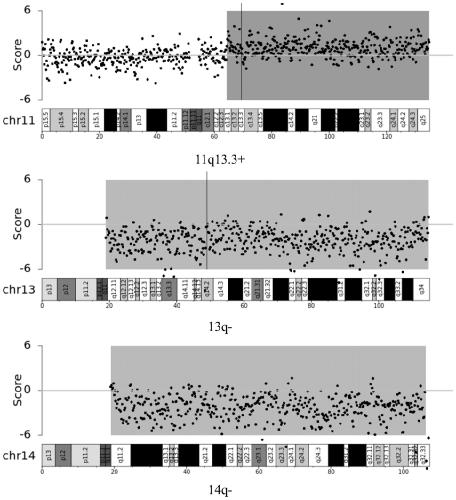
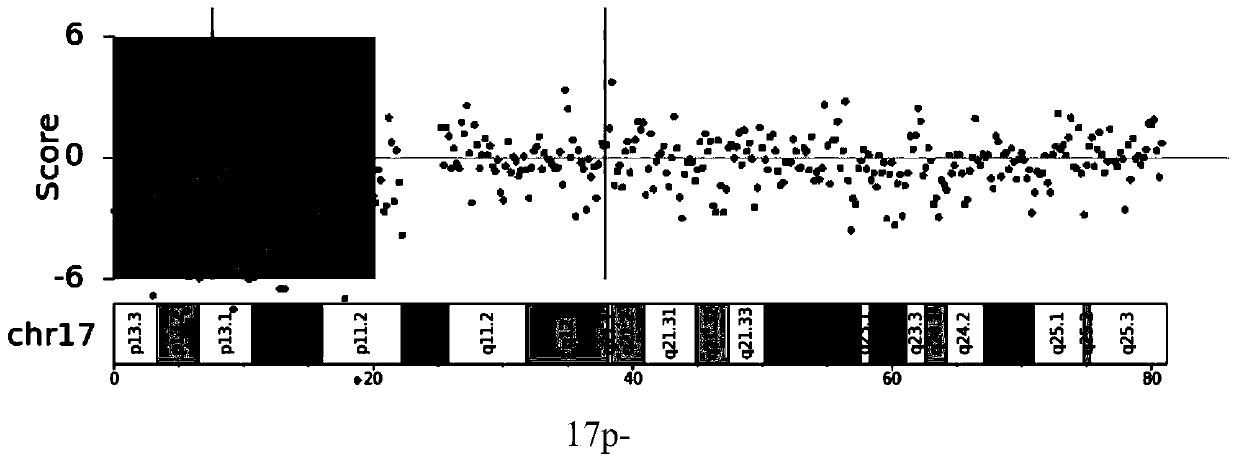
Application of chromosome instable mutation to preparing agent or kit for diagnosis, evaluation and prognosis of multiple myeloma
A multiple myeloma, chromosome technology, applied in the field of biotechnology and medical diagnosis, can solve the lack of multiple myeloma chromosome distribution characteristics and other problems, to achieve high sensitivity, simple operation, high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1: Preparation of library adapters
[0043] Linker sequences synthesized by third-party companies:
[0044] Linker-1 (SEQ ID NO.1):
[0045] 5'-CGTGATAGATCGGAAGAGCACACGTCTGAACTCCAGTC-3';
[0046] Linker-2 (SEQ ID NO.2):
[0047] 5'-ACACTCTTTCCCTACACGACGCTCTTCCGATCTATCACGT-3'.
[0048] The 5' end of Adapter-1 is modified by phosphorylation, and the 3' end of Adapter-2 is modified by phosphorothioation between G and T.
[0049] Use the annealing buffer (Annealing buffer) to dilute the dry powder of adapter-1 and adapter-2 to 20 μM respectively, mix the two dilutions in equal proportions, put the mixture in the PCR instrument, and run the program as shown in Table 1:
[0050] Table 1
[0051]
[0052]
[0053] The product was subjected to agarose electrophoresis, and the size range of the electrophoresis band was between 80-100bp.
Embodiment 2
[0055] Materials used:
[0056] human genomic DNA
[0057] DNA fragmentation reagent (Enzymatics)
[0058] DNA end repair, adding A, adding adapter enzyme reaction system (NEB)
[0059] PCR amplification reaction system (Kapa Biosystems)
[0060] Purification magnetic beads (Beckman)
[0061] Steps:
[0062] 1. Genome Fragmentation
[0063] Take 20ng of genomic DNA, use the enzyme digestion reaction system, configure the reaction system according to Table 2, and perform the reaction according to Table 3.
[0064] For cell-free DNA, such as cfDNA in blood, it does not need to undergo genome fragmentation and can directly enter the next step.
[0065] Table 2
[0066] DNA fragmentation reaction system Genomic DNA xμL interrupt enzyme 3μL Buffer 7μL nucleic acid free water 25-x μL total capacity 35μL
[0067] Shake to mix, centrifuge (avoid air bubbles) and run the following program on the PCR instrument:
[0068] table ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com