Fluorescent quantitative PCR probe primer group and kit for detecting high-frequency pathogenic variation of SLC22A5 gene
A SLC22A5-F1, fluorescence quantitative technology, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., to achieve clear pathogenicity, convenient and accurate interpretation of results, and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] A fluorescent quantitative PCR kit for detecting high-frequency pathogenic variants of the SLC22A5 gene, including a probe primer set designed for the detection site, and the probe primer set includes for amplifying the SLC22A5 gene NM_003060.3:exon8: c. The upstream and downstream primers of the 1400C>G variation site and the MGB probe for the variation site, the sequence of the probe primer set is shown in Table 1-2:
[0056] Table 1 Primer Sequence
[0057]
[0058] Table 2 MGB probe sequence
[0059]
[0060] The method for detecting the sample to be tested by using the above kit is as follows:
[0061] (1) Prepare reagents according to Table 3;
[0062] Table 3 Reagent preparation
[0063] components Volume ul Final concentration primer F (10μM) 1.8 0.9μM primer R (10μM) 1.8 0.9μM TaqMan Probe 1 (10μM) 0.4 0.2μM TaqMan Probe 2 (10μM) 0.4 0.2μM qPCR reaction buffer 10 Template (DNA solution) 2 ...
Embodiment 2
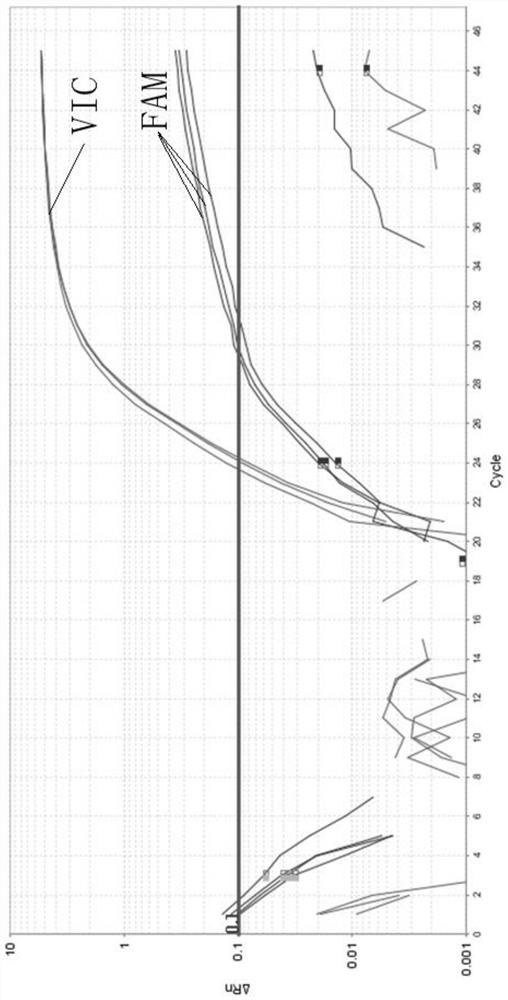
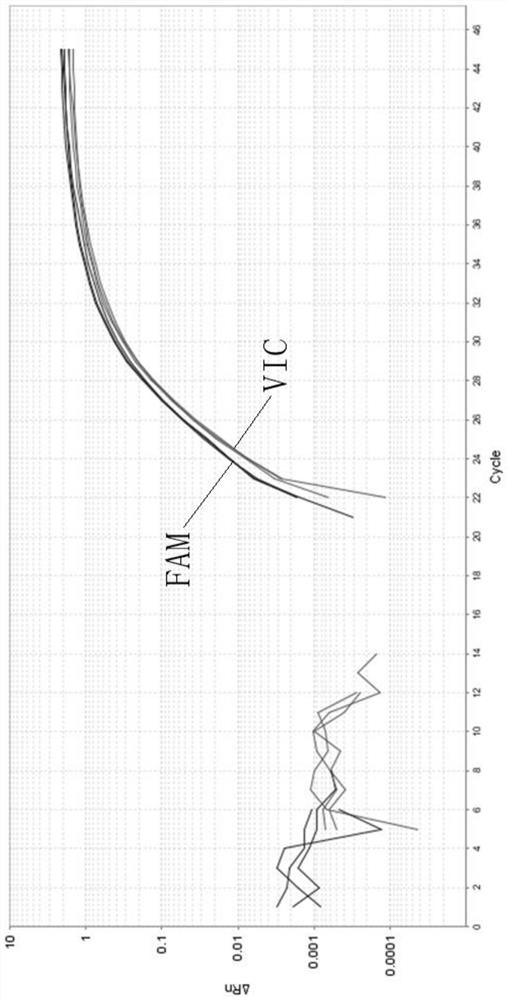
[0068] The kit described in Example 1 was used to detect high-frequency pathogenic variants of the SLC22A5 gene. When wild-type DNA was used as a template, the detection results were shown in Table 5:
[0069] Table 5 wild-type DNA detection results
[0070] Reaction well position sample name allele Reporting group Ct value A2 235-3 / 2 G FAM 31.54 A2 235-3 / 2 C VIC 24.31 B2 237-3 / 2 G FAM 29.56 B2 237-3 / 2 C VIC 23.72 C2 611-3 / 2 G FAM 29.93 C2 611-3 / 2 C VIC 24.18 D2 3 / 2-BLANK G FAM Undetermined D2 3 / 2-BLANK C VIC Undetermined
[0071] Among them, samples 235, 237 and 611 are wild-type human genomic DNA, and 3 / 2-BLANK is a blank control using purified water. The threshold line was set to 0.1. It can be seen from Table 5 that the average Ct value of the VIC signal is 24.07 with a standard deviation of 0.3106; while the average Ct value of the FAM signal is 30.34 with a standard deviati...
Embodiment 3
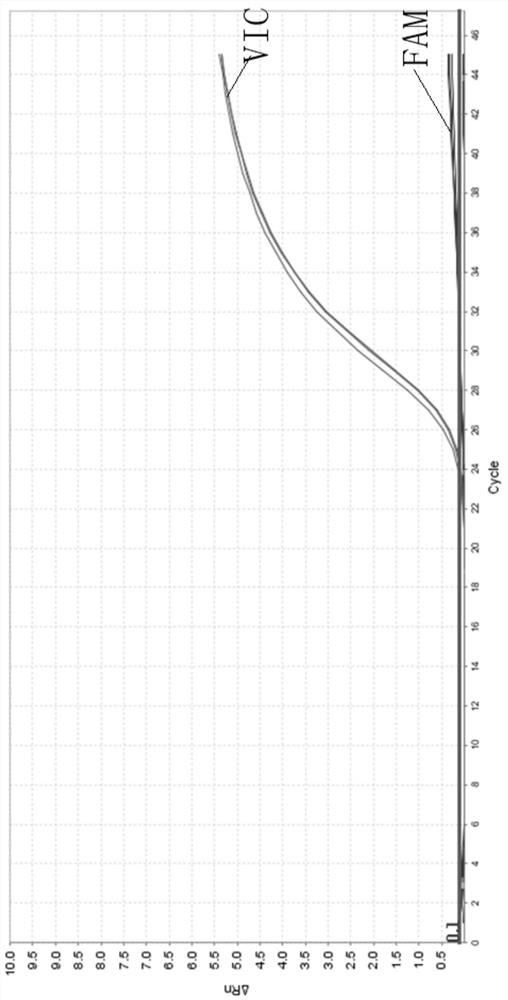
[0073] The kit described in Example 1 was used to detect high-frequency pathogenic variants of the SLC22A5 gene. When using heterozygous DNA as a template, the detection results are shown in Table 6:
[0074] Table 6 Heterozygous DNA detection results
[0075] Reaction well position sample name allele Reporting group Ct value A1 SLC22A5-1-1 G FAM 24.65 A1 SLC22A5-1-1 C VIC 25.12 B1 SLC22A5-2-2 G FAM 24.60 B1 SLC22A5-2-2 C VIC 25.20 C1 SLC22A5-3-2 G FAM 24.69 C1 SLC22A5-3-2 C VIC 25.25 D1 SLC22A5-BLANK FAM FAM Undetermined D1 SLC22A5-BLANK VIC VIC Undetermined
[0076] Among them, the samples SLC22A5-1-1, SLC22A5-2-2 and SLC22A5-3-2 are heterozygous human genomic DNA of the mutation site NM_003060.3:exon8:c.1400C>G, and SLC22A5-BLANK was purified using Water blank. The threshold line was set to 0.1. It can be seen from Table 6 that the average Ct value of the VIC signal i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com