Application of set of gene mutation sites in preparation of reagent or kit for diagnosing chronic pancreatitis (CP) and evaluating prognosis
A chronic pancreatitis and kit technology, applied in the fields of biotechnology and medical diagnosis, can solve the problems of not fully reflecting the real situation of pathogenic mutations, lack of them, etc., and achieve the effect of shortening the research period, high throughput, and deep coverage.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
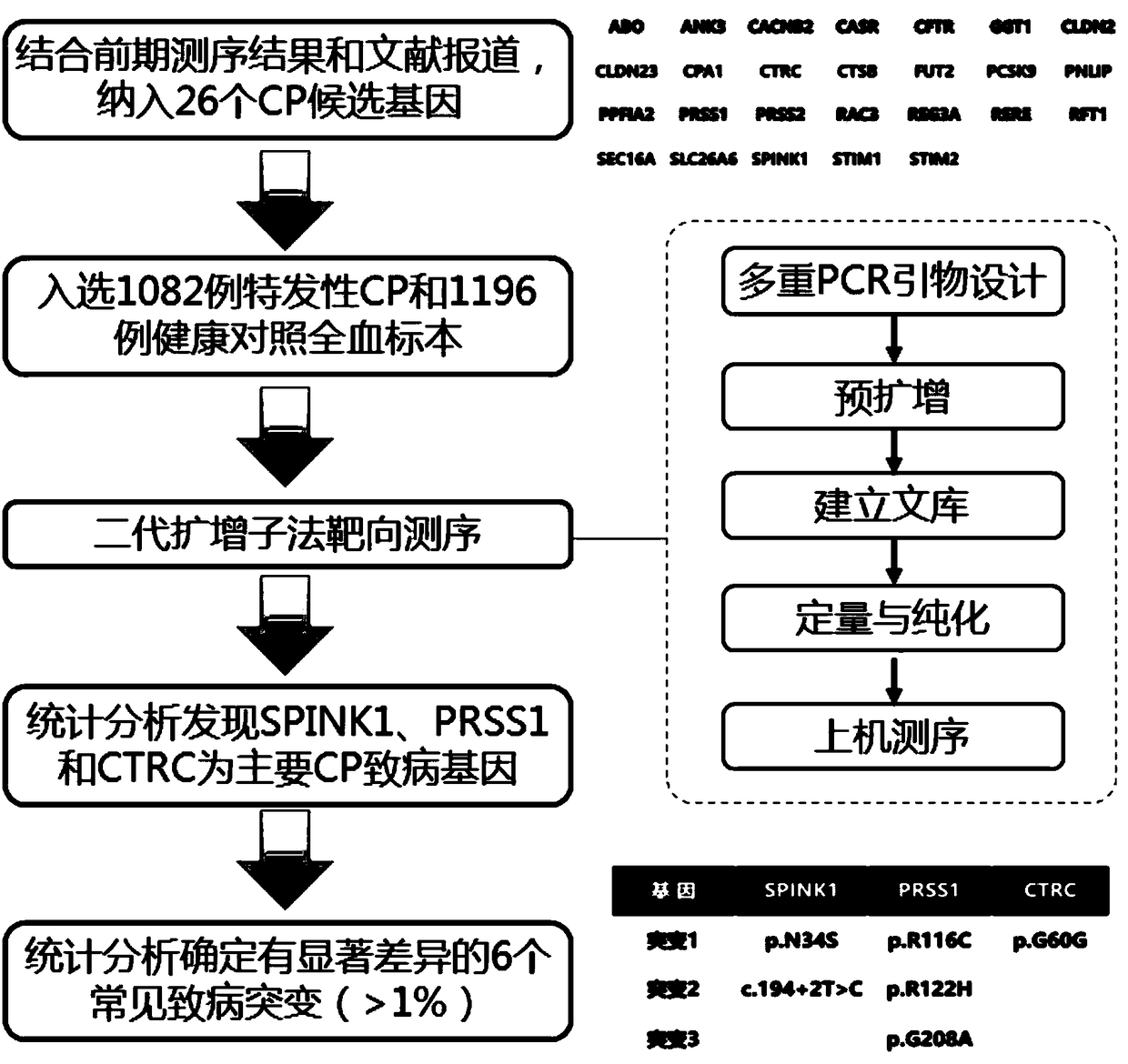
[0047] 1. Multiplex PCR primer design
[0048] Use Primer3 (http: / / simgene.com / Primer3) to design 26 gene coding sequences and exon / intron regions, and the 5' ends of the primers are synthesized using common linker sequences, that is, all 5' end primers contain CS1 (5 '-ACACTGACGACATGGTTCTACA-3'), all 3' primers contained CS2 (5'-TACGGTAGCAGAGACTTGGTCT-3'), all primers and some other primer pairs were divided into two multiplex primer pools (Table 2).
[0049] Table 2 List of primers for targeted sequencing of all exons of SPINK1, PRSS1 and CTRC
[0050]
[0051]
[0052]
[0053] 2. Pre-amplification of labeled gene amplicons
[0054] The labeled amplicons are first amplified in two multiplex PCR reactions as follows:
[0055]
[0056] The PCR reaction conditions are as follows:
[0057]
[0058] Treat the above 2.5μl PCR product as follows:
[0059]
[0060] First process at 37°C for 60min, then mix at 80°C for 20min. The processed samples were diluted...
Embodiment 2
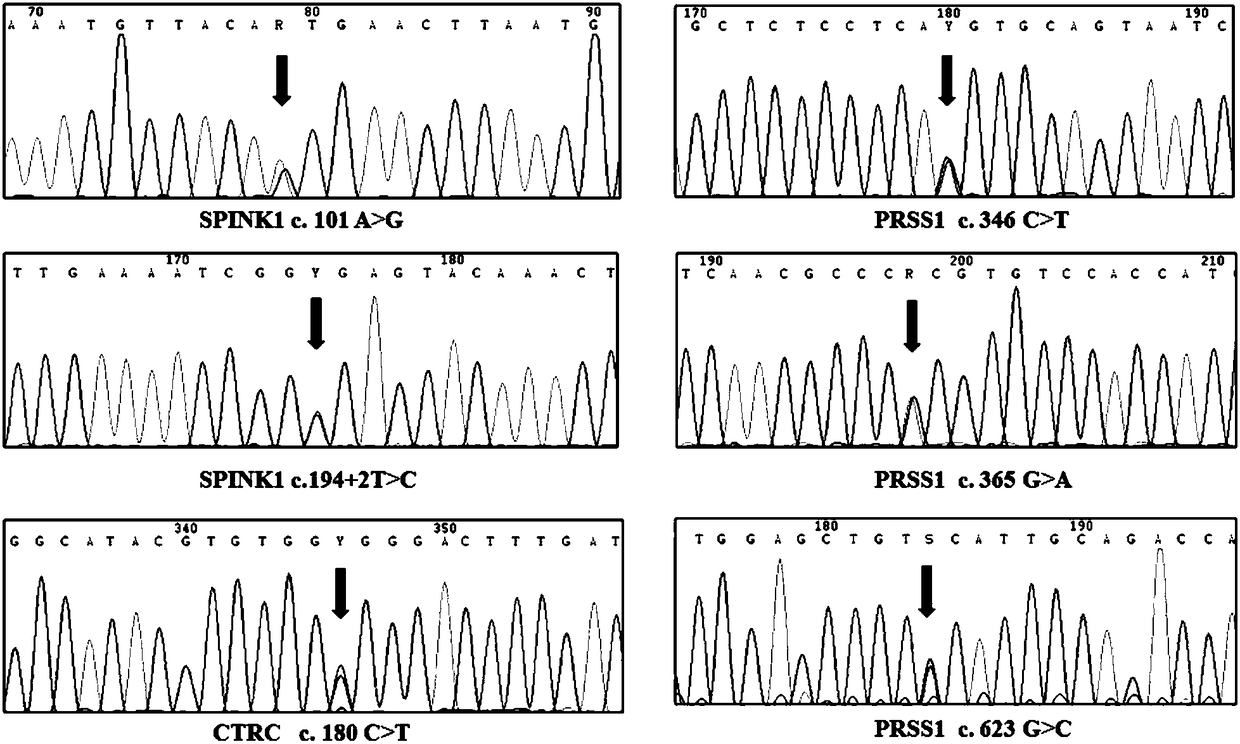
[0071] Statistical analysis was used to evaluate the performance of single mutation detection and combined detection of 6 mutations. As shown in Table 3, c.101A>G, c.194+2T>C, c.346C>T, c.365G>A, c.623G>C, c.180C>T mutations alone were used to detect chronic pancreas The sensitivities of inflammation were 2.2%, 37.6%, 1.3%, 2.2%, 8.6%, 1.8%, 46.1%, respectively, and the sensitivity of combined detection of chronic pancreatitis was 46.1%, which were significantly higher than those of the single mutation detection group (p <0.001).
[0072] Table 3 The results of individual detection of 6 mutations and joint detection in patients with chronic pancreatitis
[0073]
[0074] All mutations were verified by PCR-sequencing method, and the PCR reaction system was carried out using HotStarTaq enzyme (Qiagen, Germany), as follows:
[0075]
[0076] 1) For c.101A>G, c.194+2T>C on the SPINK1 gene:
[0077] The detection forward primer used is CCAATCACAGTTATTTCCCCAGAG (SEQ ID NO.1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com