Oligonucleotide probe and acquisition method thereof
An oligonucleotide probe and oligonucleotide technology, which is applied in the biological field, can solve the problems of long oligonucleotide probes, lack of chromosomal structure, and expensive synthesis routes, and achieve low cost, convenient design, and high efficiency Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
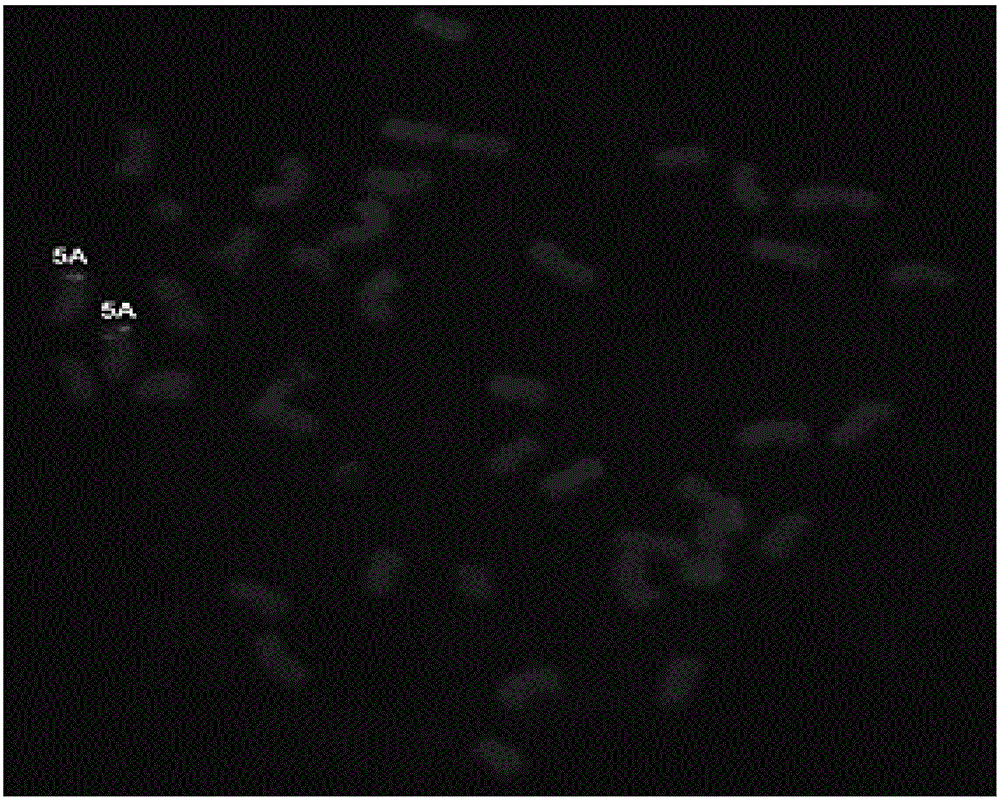
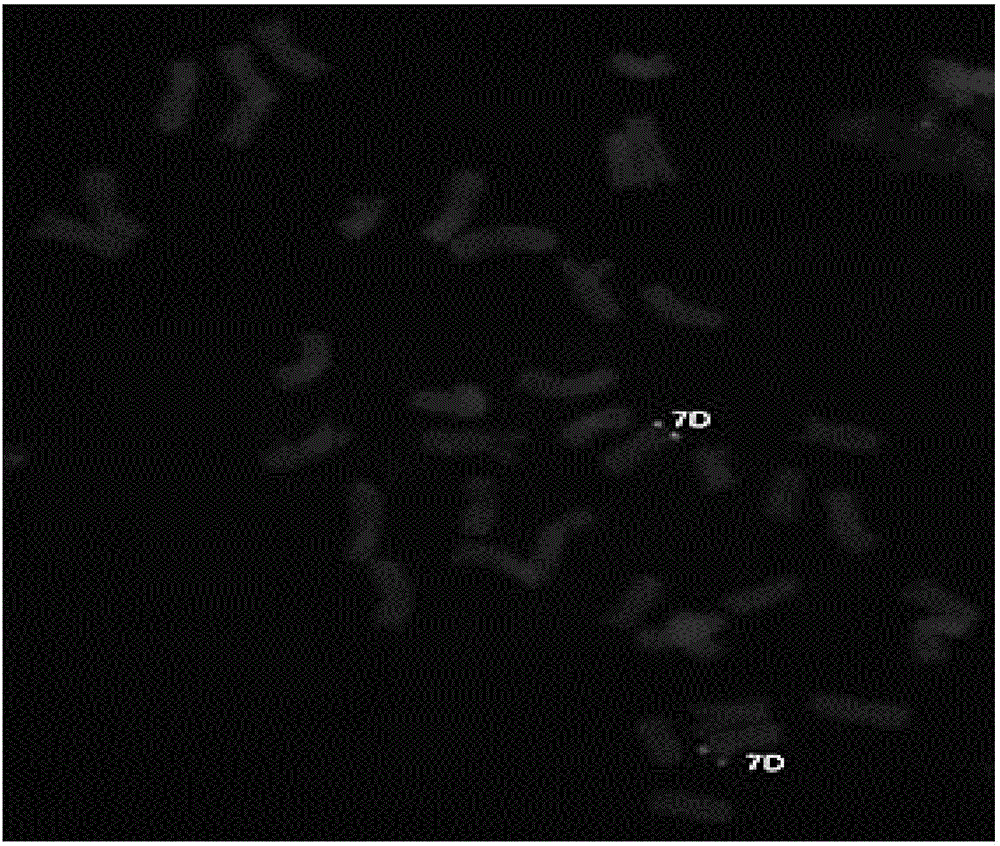
[0043] The method for obtaining oligonucleotide probes of the present invention is a method for obtaining oligonucleotide probes for non-denaturing fluorescence in situ hybridization (ND-FISH) identification of specific chromosomes of crops, and its specific steps are as follows:
[0044] (1) Download wheat and its close relatives from public databases such as NCBI (National Center for Biotechnology Information), IWGSC (International Wheat Genome Sequencing Consortium), MaizeGDB (Maize Genetics and Genomics Database), EnsemblPlants (Plant Genome Database), etc. For related species and the genome sequence of maize, use perl language programming to filter the original sequence, remove the sequence that is too short, and then use Refind and TRF software to find the tandem repeat sequence. In this step, tens of thousands of tandem repeat sequences were found.
[0045] (2) Use perl language programming to screen a large number of repetitive sequences initially found, and set four k...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com